Abstract
Previous studies in several model organisms have revealed that members of the Forkhead (Fkh) transcription factor family have multiple functions. Drosophila Jumeau (Jumu), a member of this family, participates in cardiogenesis, hematopoiesis and immune system homeostasis. Here, we show that loss of jumu function positively regulates or triggers apoptosis via a JNK-dependent pathway in wing development. jumu mutants showed reduced wing size and increased apoptosis. Moreover, we observed a loss of the anterior cross vein (ACV) phenotype that was similar to that observed in wings in which JNK signaling has been ectopically activated. The JNK signaling markers puckered (puc) and p-JNK were also significantly increased in the wing discs of jumu mutants. In addition, apoptosis induced by the loss of jumu was rescued by knocking down JNK, indicating a role for JNK in reducing jumu-induced apoptosis. Jumu could also control wing margin development via the positive regulation of cut expression, and the observed wing margin defect did not result from a loss of jumu-induced apoptosis. Further, jumu deficiency in the pupal wing could induce multiple wing hairs via a Rho1-mediated planar cell polarity pathway, but abnormal Rho1 expression was not why jumu loss induced apoptosis via a JNK-dependent pathway in wing discs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Drosophila is a useful model system for studying the underlying molecular mechanisms of morphogenesis and patterning at both the cellular and tissue levels. In particular, much has been learned about the genetic basis of the development of the wing (the largest organ in Drosophila).
c-Jun NH2-terminal kinase (JNK) belongs to the mitogen-activated protein kinases (MAPKs) superfamily and is activated primarily by cytokines and exposure to environmental stress [1, 2]. Previous studies have shown that the ectopic activation of JNK leads to small, rough eyes and abnormal wing phenotypes. The overexpression of eiger (egr) or hemipterous (hep) activates the JNK pathway in the wing discs, resulting in cell death, a loss-of-anterior cross vein (ACV) phenotype and small wings [3, 4]. Moreover, reducing the level of puckered (puc), a negative regulator of JNK activity, results in robust cell death in third-instar larval wing discs and a loss-of-ACV phenotype in adult wings [3, 5, 6]. Apoptosis is a major form of programmed cell death in which cells activate a self-destruct process. During development and under stress, organisms must remove excess or damaged cells to maintain tissue homeostasis [7]. The JNK pathway plays a critical role in regulating cell death via the core apoptotic pathway [2, 8]. Recent work has shown that apoptotic cells can induce neighbouring surviving cells to increase their proliferation to compensate for cell loss, a phenomenon termed apoptosis-induced proliferation (AiP) [9, 10]. In Drosophila, AiP is a JNK-dependent process that leads to the production of mitogens, including wingless (Wg), decapentaplegic (Dpp), and spitz (Spi), for tissue repair and regeneration [11, 12]. AiP can also cause tissue overgrowth, a process that has important implications for cancer biology. One study showed that impaired Hippo signaling induces JNK activation through Rho1 and causes tissue overgrowth, which indicates an essential role for the JNK pathway in Hippo-signaling-related tumourigenesis [13].
jumeau (jumu) encodes a 720-amino-acid (aa) nuclear protein and is a member of the winged-helix/forkhead (WH/FKH) family of transcription factor genes in Drosophila. jumu is well conserved among insect species and exhibits a broad spectrum of functions. In addition, Jumu shows similarity to the mammalian winged-helix nude (whn) protein near its C-terminus. whn is required for the regulation of tissue-specific transcription and cell fate in thymus and hair development [14]. Jumu also plays a role in cell fate decisions during differentiation in neurogenesis as well as in compound eye, wing and bristle development. During neuronal differentiation, Jumu is required for the generation of asymmetric sibling localization and the segregation of Pon/Numb [15]. The phenotypes of jumu homozygous mutants include bristle disorders, variegated eyes and defective posterior wing margins. Moreover, they exhibit severely diminished vitality and fertility [16]. A recent study showed that Jumu and its checkpoint suppressor homologue (CHES-1-like) control the division of cardiac progenitors via a Polo-dependent pathway during Drosophila cardiogenesis [17]. Moreover, in mutants lacking jumu, the expression levels of the gene frizzled (fz), which encodes a receptor of the Wnt signaling protein wingless (Wg), were significantly reduced in the mesoderm of embryos [18]. Our previous work demonstrated that Jumu is involved in Drosophila hematopoiesis, mediating the proliferation and differentiation of blood cells [19]. In addition, jumu overexpression induces the deposition of hemocytes and the formation of melanotic nodules by activating the Toll pathway [20]. In particular, jumu plays an important role in the control of hematopoietic progenitors in the Drosophila lymph gland [21]. In this study, we observed small wings, the loss of the ACV, defective wing margins and multiple wing hairs in double heterozygous jumu mutants and jumu knockdown flies. Furthermore, we detected cell death in late third-instar larvae after reducing the expression of jumu. Simultaneously, we observed increased levels of puc and p-JNK, which are factors in the JNK pathway. Moreover, the loss of jumu affected Wg-producing cells and led to decreased Cut levels. During the pupal stages, jumu positively regulated Rho1 to refine multiple bundles into a single growing hair. Therefore, we have identified previously unknown functions for Jumu in modulating JNK-dependent cell death and other processes in wing development.
Materials and methods
Fly stocks
The following transgenic lines were used in our study: jumu RNAi (v12610, jumu-i) was obtained from the Vienna Drosophila RNAi Stock Center (VDRC). jumuP (GE27806) was purchased from GenExel (Daejeon, South Korea). jumuDf3.4 and UAS-jumu were gifts from Michelson [17, 22]. dTAK1 RNAi (National Institute of Genetics, 5115R2), hep RNAi (VDRC, v47507), dTRAF1 RNAi [23], JNK RNAi (VDRC, v34138), bskDN [23] and UAS-puc [24] were gifts from Xu. MS1096-Gal4 [25, 26] was a gift from Liu [27]. dTRAF1 RNAi dTRAF2 RNAi and DRONCDN were gifts from Xue [3, 23, 28]. en-Gal4 was obtained from the Tsinghua Fly Center. Other strains used in this study included Rho1CA [29], UAS-p35 (BL5072) [30], UAS-cut (BL36496), dpp-lacZ [31], puc-lacZ [24] and w1118. All genotypes were bred into the w1118 background.
Immunohistochemistry
Wing imaginal discs obtained from third-instar larvae were fixed in 4% paraformaldehyde for 30 min at room temperature. The wing discs were then placed in blocking buffer (PBS plus 0.1% Tween 20 and 5% normal goat serum) for 1 h at room temperature. For the preparation of pupal wings, late third-instar larvae were selected and allowed to develop for 24–36 h APF at 29 °C. Whole pupae were removed from their pupal cases and fixed in 3.7% formaldehyde for 2 h at room temperature. The pupal wings were dissected in 0.3% PBST (0.3% Triton X-100 in PBS) and incubated in blocking buffer (PBS containing 0.3% Triton X-100, 2% BSA, and 2% normal goat serum) for 1 h. The wings were incubated in primary antibodies overnight at 4 °C and then incubated with secondary antibodies according to standard methods. Finally, the wings were mounted in Vectashield fluorescent mounting medium (Vector Laboratories) or Prolong Diamond Antifade Mountant (Molecular Probes). The tissues were analyzed using an LSM 510 META confocal microscope (Zeiss) or an Axioskop 2 plus microscope (Zeiss). The following primary antibodies were used: mouse anti-β-gal (Promega), mouse anti-Wg, mouse anti-Cut, mouse anti-DE-cad, mouse anti-βPS (Developmental Studies Hybridoma Bank), rabbit anti-dMyc (Santa Cruz), rabbit anti-p-JNK, rat anti-Jumu [21], rabbit phospho-H3 (1:800, Upstate) and 7-AAD (Life Technologies). Secondary antibodies were conjugated with Alexa Fluor 488 and Alexa Fluor 568 (Molecular Probes) and used at 1:200 dilution. All of the experiments were independently repeated at least three times.
TUNEL assay for wing imaginal discs
TUNEL assays were performed using an In Situ Cell Death Detection Kit (Roche Applied Science) according to the manufacturer’s instructions. Third-instar larval wing discs were dissected in ice-cold PBS and fixed in 4% paraformaldehyde in PBS for 30 min at room temperature. The samples were then washed 4 times in 0.4% PBST (0.4% TritonX-100 in PBS) and permeabilized by incubation in a PBS containing 100 mM sodium citrate and 0.1% Triton X-100 for 3 min on ice. After extensive washing with PBS, the samples were submerged in a terminal deoxynucleotidyl-transferase-mediated dUTP nick-end labeling (TUNEL) reaction solution and incubated in a 37 °C chamber for 2 h. After being washed three times with PBS and mounted in mounting medium, the wing discs were observed using a confocal laser microscope (Carl Zeiss, Germany).
Statistical analysis
Images were acquired using a Zeiss fluorescence microscope. All numerical data, including wing size, cell number and intensity values, were analyzed using ImageJ software. The statistical analyses were performed using a two-tailed unpaired Student t test with Prism software (GraphPad 6.0). The results were considered statistically significant when P < 0.05. ****, ***, ** and * indicate P < 0.0001, P < 0.001, P < 0.01 and P < 0.05, respectively. “ns” indicates no significant difference. The error bars in the graphs indicate the SEMs.
Results
Depletion of jumu induces abnormal wing phenotypes in adult flies
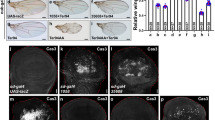
Because all jumu homozygous null mutants died during embryogenesis, we examined the wings of jumu heterozygous mutant adults to further analyse the functions of Jumu in wing development. The wings of jumu heterozygous mutant adults showed obvious reductions in size (Fig. S1g–j). The wings of adult jumu heterozygotes showed a nearly 10% reduction in size, whereas jumu double heterozygous mutants showed wing size reductions of more than 14% (Fig. 1a, b, g, i). Accordingly, jumu double mutants displayed greatly reduced adult eye size (Fig. 1e, f, j). In addition, jumuDf3.4/jumuP double heterozygous mutant adults exhibited normal wing vein locations (Fig. 1a, b), but unexpectedly, the wings of adults displayed a loss of wing margin structures (Fig. 1a′, b′). Notably, multiple hairs formed on the wings (Fig. 1a″, b″). These results indicate that changes in jumu expression substantially disturb the growth and development of the wings.
Adult wing phenotypes of jumu mutants. a, c Control wings of a w1118 or MS1096/w1118 (MS1096>+, control) fly. Normal wing; note the prominent veins (structural struts) and intact wing edge or margin. b, d, a′–d″ Loss of jumu resulted in abnormal wing phenotypes. b Wing from jumu double heterozygous mutant (jumuDf3.4/jumuP); note the nick at the wing margin. d RNAi-mediated downregulation of jumu in the wing blade area via MS1096-Gal4 produced a loss-of-ACV (over 55%) phenotype in adult wings, indicated by the arrow. a′–d″ Comparison with the control; reduced jumu expression resulted in a loss of the wing margin and in multiple hairs, as indicated with asterisks. e, f Micrographs showing adult Drosophila eyes. f The small, rough eye phenotype of the jumu mutant. g, h An analysis of the overlap of wings from adult females showed that jumu mutants had smaller wings than the controls. i Quantification of wing size in females based on the data in g and h. j Quantification of eye size in females based on the data in e and f. Scale bars: 200 µm (a–d), 50 µm (a′–d′) and 20 µm (a″–d″)
To determine whether the abnormal wing phenotypes were caused by autonomous loss of the Jumu protein, we used MS1096-Gal4 to ubiquitously knock down jumu in the wing blade. The resulting flies displayed a 23% smaller wing size (Fig. 1h, i) and more cells with multiple hairs than the control flies (Fig. 1c″, d″). Accordingly, we observed a loss of wing margin tissue in MS1096>jumu RNAi mutants (Fig. 1c′, d′). Unexpectedly, MS1096>jumu RNAi flies exhibited a loss-of-ACV phenotype (Fig. 1d, over 55%). To provide further confirmation that Jumu is involved in wing development, we used transgenic UAS-jumu [17, 21, 22] flies in which the entire jumu coding region was ectopically expressed. The wing morphological defects detected in MS1096>jumu RNAi flies were clearly rescued by jumu overexpression (Fig. S1a, b, d, f, g). jumuP [21] contains a P-element insertion in the 5′ UTR region of jumu that encodes a Gal4-responsive enhancer. Similar to previous results, overexpressing jumuP using the MS1096-Gal4 driver as the background for jumu RNAi rescued the abnormal wing phenotypes (Fig. S1a–c, e, g). These observations suggest that Jumu performs an important role in development, particularly in wing development. Furthermore, the loss-of-ACV phenotype in the jumu RNAi wings was similar to that observed in JNK signaling gain-of-function wings [3,4,5,6]. We therefore propose that Jumu may be involved in JNK signaling during Drosophila wing development.
Regenerative apoptosis-induced proliferation is triggered by loss of Jumu
To further investigate the biological function of Jumu during wing development, we first identified the expression patterns of the Jumu protein in larval and pupal wings by staining with anti-Jumu antibodies. In wild-type larvae, Jumu was expressed in the wing blade and showed high expression at the dorsal/ventral (D/V) wing margin (Fig. S2a, c). In addition, Jumu was expressed in intervein cells and localized to the nucleus during the pupal stages (Fig. S2f, f′). However, the signal was nearly abolished in jumu double heterozygotes and MS1096>jumu RNAi mutants (Fig. S2b, d). In en>jumu RNAi flies, Jumu was specifically depleted in the posterior compartment (Fig. S2e, e′). Adult wing size was reduced in the jumu mutant flies, which led us to speculate that the deletion of jumu resulted in cell death during the larval stage. We detected many 7-AAD-positive cells in the late third-instar larval wing discs of jumu mutants (Fig. 2a–d, m). This result suggested that the wing blade cells were dying or dead and had lost their normal functions. We also used TUNEL assays to detect apoptotic cells and found that many cells in the wing blade were apoptotic (Fig. 2e–h, n). To determine whether Jumu is involved in apoptosis in a cell-autonomous manner, in addition to its role in the regulation of tissue repair processes, we used the en-Gal4 driver to knock down jumu in the posterior compartments of the wing discs. Only the posterior part of the wing disc, rather than the whole wing blade region, exhibited dead cells (Fig. S3d, e). Simultaneously, we analysed wing blade cell proliferation using anti-phospho-histone H3 antibodies (PH3), which stain dividing cells in M phase. There were significantly more PH3-positive cells in jumuDf3.4/jumuP double heterozygous and MS1096>jumu RNAi mutant wing discs than in the controls (Fig. 2i–l, o). We further examined the late third-instar larval wing blade using anti-Wg antibodies and found that Wg expression levels were significantly increased (Fig. 2i′–l′, p). The depletion of jumu in the posterior half of the wing disc under en-Gal4 control increased the number of PH+ cells and Wg intensity in the posterior compartment (Fig. S3a–c).
Loss of jumu promotes cell death by affecting cell cycle progression. a–d Wing discs of the indicated genotypes analysed via 7-AAD staining. Red spots indicate dead cells with disrupted membrane integrity. e–h Wing discs of the indicated genotypes analysed via TUNEL staining. Red spots indicate apoptotic cells. i–l Analysis of mitosis and wing blade development in the indicated genotypes. Cell proliferation was monitored using an anti-PH3 antibody (green); wing blade morphology was detected via Wingless staining (Wg, red). The number of PH3+ cells in the area containing the wing blade and the levels of Wg were significantly increased in the loss-of-jumu flies. i′–l′ High magnification revealed a widespread Wg signal along the strip of the D/V boundary in jumu mutant wing discs compared with those of the controls. m–p Quantification of dead cells, apoptotic cells, PH3+ cells and Wg labeling intensity in wing discs from females (fold difference from control). Scale bar: 50 µm
JNK signaling has been proposed to stimulate surviving cells neighbouring apoptotic cells, thereby inducing an increase in cell proliferation to compensate for cell loss [9, 10]. Reductions in jumu expression in larval wing discs caused ectopic cell proliferation and increased Wg levels. These results suggest that the mechanism through which Jumu controls tissue regeneration is mainly mediated by the regulation of cell proliferation by the Wg mitogen in the imaginal discs. Taken together, these findings suggest that Jumu may be involved in JNK signaling during Drosophila wing development.
Loss of jumu induces JNK pathway activation
To examine whether JNK signaling plays a role in the loss of jumu-induced cell death, we used a lacZ insert in the puc gene, a transcriptional target of the JNK pathway [24]. We found that knocking down jumu in the wing blade of the wing disc resulted in greater expression of puc-lacZ (Fig. 3b, l) than of the MS1096-Gal4 control (Fig. 3a, l), suggesting that the loss of jumu promotes JNK pathway activation. The activity of Drosophila JNK was simultaneously assessed by immunostaining the discs with a phospho-specific antibody that recognizes the active form of JNK. The suppression of the jumu gene in the wing blade resulted in a significant increase in p-JNK levels (Fig. 3c, d, m). Moreover, the depletion of jumu in the posterior half of the wing disc under en-Gal4 control increased the activity of the JNK pathway in the posterior compartment (Fig. S3j–m). These results collectively indicated that jumu is an inhibitor of JNK.
Loss of jumu induces JNK pathway activation and reduces Cut levels during wing development. a, b The expression of the JNK target puckered (puc, labeled red) was determined using the lacZ reporter gene. c, d The activity of Drosophila JNK was assessed by immunostaining the discs with a phospho-specific antibody that recognizes the active form of JNK (p-JNK). e–h The levels of Cut (red) were examined via anti-Cut staining. e′–h′ High magnification revealed a loss of the Cut signal along the D/V boundary in jumu mutant wing discs compared with those of the controls, as indicated with arrows. i, g Low jumu expression resulted in the wing margin defects, k which were significantly suppressed by the overexpression of cut. l–m Quantification of puc total intensity, p-JNK-positive cells and cut total intensity in the wing blade. Scale bars: 50 µm (a–h, i–k) and 10 µm (e′–h′)
JNK is indispensable for apoptosis induced by the loss of Jumu
The remarkable contribution of Jumu to apoptotic cell death prompted us to conduct a genetic modifier screen that was designed to search for additional JNK pathway components mediating MS1096>jumu RNAi-induced apoptosis. To fully inhibit JNK activity, we utilized UAS-puc, bskDN, JNK RNAi, hep RNAi, dTRAF1 RNAi, dTRAF2 RNAi and dTAK1 RNAi combined with MS1096>jumu RNAi (Fig. 4a–h, the percentage indicates the loss-of-ACV phenotype). As expected, in the reduced-JNK-signaling background, the MS1096-Gal4-driven decrease in jumu levels rescued the wing size and loss-of-ACV phenotypes. More importantly, in the dTRAF2 RNAi and dTAK1 RNAi mutant backgrounds, the jumu-knockdown-induced abnormal wing phenotypes were completely suppressed (Fig. 4g, h, m). This finding clearly demonstrated that the JNK pathway is indispensable for apoptosis induced by the loss of jumu. The wings of UAS-puc, bskDN, JNK RNAi, hep RNAi, dTRAF1 RNAi, dTRAF2 RNAi and dTAK1 RNAi flies, driven by MS1096-Gal4, exhibited normal adult wing phenotypes (Fig. S4). We detected almost no dead cells in the late third-instar larval wing discs of MS1096>dTAK1 RNAi mutants with a jumu knocked-down background (Fig. S3h, i). Moreover, inhibiting apoptosis by suppressing DRONC or ectopically activating p35 also rescued the abnormal wing phenotypes of MS1096>jumu RNAi flies (Fig. 4i–m). Considering these results, we concluded that jumu loss activates JNK and that this activation is essential for loss-of-Jumu-induced apoptosis in Drosophila.
jumu antagonizes the JNK signaling pathway, and its loss mediates caspase-dependent cell death. Light micrographs of adult Drosophila wings are shown (the percentage indicates the loss-of-ACV phenotype). a Low jumu expression resulted in the clear loss of the ACV (by 55.2%) and a reduction in wing size, b–h, j, l which were significantly suppressed by the loss of JNK signaling or the suppression of cell death. g, h The ACV-defective wing phenotype induced by MS1096>jumu RNAi was strongly suppressed by dTRAF2-i or dTAK1-i. j, l The suppression of apoptotic signals via DRONCDN or p35 overexpression rescued wing size and the defective ACV. i, k The overexpression of DRONCDN or p35 via MS1096-Gal4 resulted in normal wing phenotypes. Quantification of observed wing sizes from female adults for all combinations is shown in m (fold difference compared with MS1096>+, red dotted line). Scale bar: 200 µm
Reducing jumu in Drosophila wings results in a loss of Wnt signaling responses
Compared with the control, the jumuDf3.4/jumuP double heterozygous and MS1096 > jumu RNAi mutant adults exhibited wing margin defects (Fig. 1a–d′). Originally, we speculated that these defects might be explained by apoptosis in the jumu mutant wing discs. However, the MS1096>jumu RNAi-induced wing margin defects were not rescued by knocking down JNK signaling or inhibiting cell death (Fig. 4). Additionally, there were no dead cells along the pupal wing margin (data not shown). Since such phenotypes can also arise from defective Wg signaling, we analysed the expression of a Wg target in the wing imaginal discs of late third-instar larvae. We observed the downregulation of the short-range Wg target gene cut in the jumu mutant wing discs (Fig. 3e–h, n). High magnification revealed a loss of the Cut signal along the strip of the D/V boundary (Fig. 3e′–h′). The depletion of jumu in the posterior half of the wing disc under en-Gal4 control inhibited the level of cut in the posterior compartment (Fig. S3f, g). To further confirm the role of cut in the regulation of wing margin development in the MS1096>jumu RNAi mutants, we performed a rescue experiment with a UAS-cut transgenic line. As expected, the overexpression of cut effectively reduced the wing margin defects in the MS1096>jumu RNAi mutants (Fig. 3i–k). In wild-type discs, the majority of anti-Wg staining was localized close to the stripe of Wg-producing cells, and the concentration of Wg rapidly declined with the distance from the source of production (Fig. 2i′, k′). Unexpectedly, jumu loss resulted in the erosion of the Wg gradient, and high magnification of the anti-Wg staining revealed that Wg was widespread (Fig. 2j′, l′). Thus, the loss of jumu changes the way Wg-producing cells release Wg. This change apparently involves increased secretion and the production of a more-mobile form of Wg, affecting gradient formation. In addition, a previous study reported that Jumu can control the Wnt signaling pathway by regulating Fz levels during cardiac progenitor development [18]. Taken together, Jumu may positively regulate cut levels to affect Drosophila wing margin development through a Wnt-dependent pathway.
Jumu regulates Drosophila wing planar polarity through Rho1
Recent research has indicated that the Rho-family GTPases, including RhoA, Rac, and Cdc42, play a central role in JNK signaling during both morphogenesis and apoptosis [32]. In addition, Rho1, the Drosophila RhoA homologue, promotes apoptosis by activating the JNK pathway [33]. Rho1 also plays a role in wing planar polarity, and aberrant Rho1 activity leads to multiple wing hairs [34]. The results described above showed that the loss of jumu in the adult wing caused the formation of multiple hairs (Fig. 1b″, d″). This phenotype is related to wing planar polarity, as mutations in tissue polarity genes lead to hairs forming at alternative cellular locations and the formation of multiple hairs. Published images of developing wing hairs usually show fixed, phalloidin-stained (phalloidin specifically stains F-actin) wings in an intermediate stage of hair growth, while at later stages, only a single large region (bundle) of F-actin is observed, which appears to fill the hair (Fig. 5a). To further examine whether Jumu can regulate wing hair formation, we stained growing hairs for F-actin 32 h APF and consistently observed that the loss of jumu in MS1096>jumu RNAi pupal wings led to substantially more actin bundles than in the controls (Fig. 5b). Moreover, en>jumu RNAi mutant wings presented a stronger phenotype than that observed in the wings of MS1096>jumu RNAi flies (Fig. 5c, d, f, h). One interesting finding was that the DE-cad-stained cell membrane showed a dramatic downregulation of DE-cadherin (Fig. 5a′, b′). Cadherins are central to the formation of adherens junctions (AJs) and are prominent markers of AJs [35]. In some cells, no cadherin staining remained, while others showed prominent gaps in DE-cadherin staining (Fig. 5b′, arrows). We also immunostained the wings of en>jumu RNAi flies that exhibited a specific, posterior compartment knockdown phenotype. The most strongly affected cells also showed gaps in DE-cadherin staining (Fig. 5c′, g, i) that appeared similar to what we observed following the knockdown of jumu expression driven by MS1096-Gal4. This result suggested that Jumu might also be involved in the maintenance of epithelial cell structure and AJs.
Effects of jumu loss on pupal wings. a, a′, d, e Immunochemical staining for F-actin (green) and DE-cadherin (red) showing wing hairs and wing cell shape in a 32 h APF control pupal wing. b, c, f, h Arrows indicate cells with multiple hairs/actin bundles in MS1096>jumu RNAi or en>jumu RNAi flies. b′, c′, g, i Knocking down jumu resulted in large cells in pupal wings, where multiple hairs/actin bundles are marked by DE-cadherin and the levels of DE-cadherin were significantly decreased, as indicated with arrows. a″–c″ Compared with the control, reduced jumu expression resulted in a greater reduction in Rho1 (red). a–b″ and d–i, Scale bars: 20 µm. c–c″, scale bars: 50 µm
Based on the results described above, we identified a role for Jumu in wing planar polarity and found that Jumu had multiple functions. As expected, these phenotypes were similar to those observed in association with abnormal Rho1 activity in pupal wings [34]. Hence, we hypothesized that jumu affects wing planar polarity via Rho1. When we stained jumu knockdown pupal wings with anti-Rho1 antibodies, we found that jumu knockdown resulted in a dramatic downregulation of Rho1 (Fig. 5a″–c″). These experiments show that Jumu functions upstream of Rho1, a Drosophila actin cytoskeleton regulator, to affect wing planar polarity.
jumu is epistatic to Rho1
To further confirm the involvement of Rho1 in the regulation of wing planar polarity in the MS1096>jumu RNAi and en>jumu RNAi mutants, we performed a rescue experiment with a Rho1CA mutant. The overexpression of Rho1 effectively reduced the increase in multiple hairs in the MS1096>jumu RNAi and en>jumu RNAi mutants (Fig. 6a–i, a′–h′); however, the elevation of Rho1 expression did not rescue the small wing size or loss-of-ACV phenotypes of the jumu knockdown flies (Fig. 6d, h), and we did not detect any differences between the jumu knockdown compartment and the control in terms of Rho1 levels (Fig. S5n, n′) during the larval stages. Taken together, these results suggest that a loss of jumu in wing development can decrease Rho1 expression in the pupal wing blade cells and that insufficient amounts of Rho1 prevent the polymerization of multiple actin bundles, consequently inducing multiple wing hairs on adult wings. Rho1 plays an important role in regulating the formation of multiple hairs in jumu mutant flies, independent of JNK signaling, and Jumu is important for refining multiple bundles into a single growing hair.
The epistatic relationship between jumu and Rho1 is related to multiple wing hairs. a–a′, e–e′ Control wings of MS1096>+ or en>+flies. b–b′, f–f′MS1096>jumu RNAi or en>jumu RNAi resulted in multiple wing hairs, as indicated by asterisks. c–c′, g–g′ Wings of Rho1-overexpressing, MS1096>Rho1CA or en>Rho1CA flies. d–d′, h–h′MS1096>jumu RNAi; Rho1CA, en>jumu RNAi; Rho1CA suppressed the multiple wing hair phenotype in adult wings. i Quantification of wing cells with multiple hair cells. a–h Scale bars: 200 µm. a′–h′ Scale bars: 20 µm
Discussion
In the present study, we used double heterozygous jumu mutants and jumu RNAi hairpins combined with the GAL4/UAS system to deplete jumu expression in the Drosophila wing. We present evidence that the impact of Jumu depletion on the size of the wing and the formation of the ACV, wing margin and hairs is moderate but consistent. Jumu depletion reduced the size of the adult wing. jumu mutants displayed autonomous apoptosis in the wing blade in an area similar to the region of Jumu activity. In addition, we observed many PH3-positive cells and increased Wg levels in jumu mutant flies. Moreover, the adult wing showed a loss-of-ACV phenotype, and target genes of JNK signaling were significantly upregulated. These results argue against a direct relationship between loss-of-Jumu-induced cell death and the JNK pathway within the developing wing blade. However, a decrease in Jumu levels in the wing discs induced a wing margin defect resulting from the loss of Wnt signaling responses. Furthermore, during pupal development, Jumu was found in intervein cell regions, where we suggest that it functions in wing hair polarity to promote actin polymerization and refine multiple bundles into a single growing hair via a Rho1-dependent pathway.
The loss of jumu throughout the wing disc caused a decrease in wing size in adults. Numerous factors participate in organ growth within developing tissues. Wing size depends on the conditions of wing disc growth during the larval stages. For example, cell proliferation and cell death mediate wing disc growth in different ways. The wing disc has a distinctive shape and size, indicating that its growth is regulated before the onset of morphogenesis [36]; hence, Dpp and Wg signaling can induce the formation of an additional wing blade [37]. The morphogen Dpp is expressed along the anterior/posterior (A/P) compartment boundary, where it forms a gradient during wing disc development [38,39,40]. Dpp has long been thought to exclusively promote growth and proliferation. This view is based on observations that the overexpression or enhancement of dpp in wing discs leads to overgrowth or enlarged wings in adult flies [25, 41,42,43]. Knocking down Dpp signaling leads to the development of small discs, reducing adult wing size [44, 45]. The Wg morphogen is secreted across two to three cell widths straddling the D/V boundary of the wing disc and forms a gradient that helps to regulate adult wing development [46,47,48]. Reduced Wg signaling leads to the development of small wings and defective wing margins [10]. We observed a marked increase in the number of PH3-positive cells and the level of Wg in jumu mutant flies, which indicates that Jumu opposes Dpp or Wg to inhibit cell proliferation in wing discs. However, the levels of Dpp were not significantly altered and were similar to the levels observed in the controls (Fig. S5o, p). The involvement of Wg signaling in compensatory proliferation has been explored by examining its expression in apoptotic cells, and in these cells, ectopic Wg signaling induces compensatory proliferation during mitosis [10, 49]. Based on this information, we examined jumu mutant wing discs and found many dead cells via 7-AAD staining (Fig. 2a–d). As expected, TUNEL assays revealed that the number of apoptotic cells significantly increased when the Wg intensity increased in jumuDf3.4/jumuP double heterozygous and jumu RNAi-induced wing discs. Moreover, we also found that the cell size in jumu-knockdown pupal wings was clearly larger than that in the controls (Fig. 5e, g, i). These results demonstrate that the observed small wing size is related to apoptotic signaling (Fig. 2e–h). The JNK pathway is closely linked to cell death regulation in the core apoptotic pathway. When cell death is induced by JNK, apoptotic cells secrete Wg, Dpp or Spi, which are involved in regenerative responses to maintain tissue homeostasis [11, 12]. Therefore, we analysed the downstream transcriptional targets of the JNK pathway in jumu knockdown flies and found that the levels of puc and p-JNK were clearly increased (Fig. 3a–d). These data further confirm that jumu likely induces apoptosis and proliferation via a JNK-dependent pathway.
Several factors are involved in regulating the size and shape of an organ. Previous studies have suggested that Myc plays pivotal roles in promoting cell growth, proliferation and apoptosis in organ development [50, 51]. The loss of dMyc leads to the development of smaller wings and delayed patterning [52]. Moreover, our recent work demonstrated that Jumu could control the proliferation of lymph gland cells by regulating dMyc expression [21]. In this study, we analysed dMyc levels in wing discs via immunohistochemical staining. However, we did not observe any differences between the jumu knockdown and control wing discs (Fig. S5a–c′), indicating that the small wing size of the jumu RNAi flies was not mediated by dMyc.
Members of the Rho GTPase family control the polymerization of actin and the assembly of focal complexes at the plasma membrane in response to extracellular signals [53, 54]. Several studies have indicated that Rho1 uniquely and specifically regulates apoptosis-induced compensatory proliferation in Drosophila epithelia through a JNK-dependent pathway. Previous observations showed that reducing the expression of Rho1 is sufficient to activate the JNK pathway and that overexpressing Rho1 induces apoptosis in imaginal wing disc epithelia [33, 55, 56]. The specific disruption of the Cdc42/Par6/aPKC polarity complex leads to Rho1-JNK-dependent growth [57]. Rho1-induced apoptosis is generally coupled to effects on cell adhesion. Studies have shown that abnormal levels of Rho1 affect the organization of F-actin [58]. In Rho1 mutants, DE-cadherin is downregulated or shows a disturbed pattern of localization [34, 59]. We analysed Rho1 levels in wing discs via immunohistochemical staining. However, we did not observe any differences between jumu knockdown and control wing discs (Fig. S5n, n′), indicating that the apoptosis observed in jumu RNAi flies is not caused by alterations in Rho1 levels. We also detected F-actin, βPS and DE-cadherin in the wing discs of jumu knockdown flies, all of which were found to retain a normal pattern (Fig. S5d–m′). Thus, the results demonstrated that Rho1 downregulation due to reduced levels of jumu only occurred during the pupal stages.
In summary, jumu has multiple functions in Drosophila wing development. Loss-of-Jumu-induced apoptosis causes compensatory cell proliferation in Drosophila epithelia through a JNK-dependent pathway. In addition, a decrease in Jumu levels in the wing discs induces a wing margin defect due to a loss of Wnt signaling responses. jumu appears to function as a positive regulator of hair morphogenesis. These multiple functions of Jumu lead to the wide range of phenotypes seen in jumu mutants.
References
Davis RJ (2000) Signal transduction by the JNK group of MAP kinases. Cell 103:239–252
Weston CR, Davis RJ (2007) The JNK signal transduction pathway. Curr Opin Cell Biol 19:142–149
Wu C, Chen C, Dai J, Zhang F, Chen Y, Li W, Pastor-Pareja JC, Xue L (2015) Toll pathway modulates TNF-induced JNK-dependent cell death in Drosophila. Open Biol 5:140171. https://doi.org/10.1098/rsob.140171
Sun G, Irvine KD (2013) Ajuba family proteins link JNK to hippo signaling. Sci Signal 6:ra81. https://doi.org/10.1126/scisignal.2004324
Ma X, Xu W, Zhang D, Yang Y, Li W, Xue L (2015) Wallenda regulates JNK-mediated cell death in Drosophila. Cell Death Dis 6:e1737. https://doi.org/10.1038/cddis.2015.111
Hwang S, Song S, Hong YK, Choi G, Suh YS, Han SY, Lee M, Park SH, Lee JH, Lee S, Bang SM, Jeong Y, Chung WJ, Lee IS, Jeong G, Chung J, Cho KS (2013) Drosophila DJ-1 decreases neural sensitivity to stress by negatively regulating Daxx-like protein through dFOXO. PLoS Genet 9:e1003412. https://doi.org/10.1371/journal.pgen.1003412
Fuchs Y, Steller H (2011) Programmed cell death in animal development and disease. Cell 147:742–758. https://doi.org/10.1016/j.cell.2011.10.033
Umemori M, Habara O, Iwata T, Maeda K, Nishinoue K, Okabe A, Takemura M, Takahashi K, Saigo K, Ueda R, Adachi-Yamada T (2009) RNAi-mediated knockdown showing impaired cell survival in Drosophila wing imaginal disc. Gene Regul Syst Bio 3:11–20
Dichtel-Danjoy ML, Ma D, Dourlen P, Chatelain G, Napoletano F, Robin M, Corbet M, Levet C, Hafsi H, Hainaut P, Ryoo HD, Bourdon JC, Mollereau B (2013) Drosophila p53 isoforms differentially regulate apoptosis and apoptosis-induced proliferation. Cell Death Differ 20:108–116. https://doi.org/10.1038/cdd.2012.100
Herrera SC, Martín R, Morata G (2013) Tissue homeostasis in the wing disc of Drosophila melanogaster: immediate response to massive damage during development. PLoS Genet 9:e1003446. https://doi.org/10.1371/journal.pgen.1003446
Fan Y, Wang S, Hernandez J, Yenigun VB, Hertlein G, Fogarty CE, Lindblad JL, Bergmann A (2014) Genetic models of apoptosis-induced proliferation decipher activation of JNK and identify a requirement of EGFR signaling for tissue regenerative responses in Drosophila. PLoS Genet 10:e1004131. https://doi.org/10.1371/journal.pgen.1004131
Pérez-Garijo A, Shlevkov E, Morata G (2009) The role of Dpp and Wg in compensatory proliferation and in the formation of hyperplastic overgrowths caused by apoptotic cells in the Drosophila wing disc. Development 136:1169–1177. https://doi.org/10.1242/dev.034017
Ma X, Chen Y, Xu W, Wu N, Li M, Cao Y, Wu S, Li Q, Xue L (2015) Impaired Hippo signaling promotes Rho1-JNK-dependent growth. Proc Natl Acad Sci USA 112:1065–1070. https://doi.org/10.1073/pnas.1415020112
Nehls N, Pfeifer D, Schorpp M, Hedrich H, Boehm T (1994) New member of the winged-helix protein family disrupted in mouse and rat nude mutations. Nature 372:103–107
Cheah PY, Chia W, Yang X (2000) Jumeaux, a novel Drosophila winged-helix family protein, is required for generating asymmetric sibling neuronal cell fates. Development 127:3325–3335
Strödicke M, Karberg S, Korge G (2000) Domina (Dom), a new Drosophila member of the FKH/WH gene family, affects morphogenesis and is a suppressor of position-effect variegation. Mech Dev 96:67–78
Ahmad SM, Tansey TR, Busser BW, Nolte MT, Jeffries N, Gisselbrecht SS, Rusan NM, Michelson AM (2012) Two forkhead transcription factors regulate the division of cardiac progenitor cells by a polo-dependent pathway. Dev Cell 23:97–111. https://doi.org/10.1016/j.devcel.2012.05.011
Ahmad SM, Bhattacharyya P, Jeffries N, Gisselbrecht SS, Michelson AM (2016) Two Forkhead transcription factors regulate cardiac progenitor specification by controlling the expression of receptors of the fibroblast growth factor and Wnt signaling pathways. Development 143:306–317. https://doi.org/10.1242/dev
Jin LH, Shim J, Yoon JS, Kim B, Kim J, Kim-Ha J, Kim YJ (2008) Identification and functional analysis of antifungal immune response genes in Drosophila. PLoS Pathog 4:e1000168. https://doi.org/10.1371/journal.ppat.1000168
Zhang G, Hao Y, Jin LH (2016) Overexpression of jumu induces melanotic nodules by activating Toll signaling in Drosophila. Insect Biochem Mol Biol 77:31–38. https://doi.org/10.1016/j.ibmb.2016.08.002
Hao Y, Jin LH (2017) Dual role for Jumu in the control of hematopoietic progenitors in the Drosophila lymph gland. Elife 6. pii: e25094. https://doi.org/10.7554/eLife.25094
Zhu X, Ahmad SM, Aboukhalil A, Busser BW, Kim Y, Tansey TR, Haimovich A, Jeffries N, Bulyk ML, Michelson AM (2012) Differential regulation of mesodermal gene expression by Drosophila cell type-specific forkhead transcription factors. Development 139:1457–1466. https://doi.org/10.1242/dev.069005
Willsey HR, Zheng X, Carlos Pastor-Pareja J, Willsey AJ, Beachy PA, Xu T (2016) Localized JNK signaling regulates organ size during development. Elife 5:e11491. https://doi.org/10.7554/eLife.11491
Martín-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, Martinez-Arias A (1998) puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev 12:557–570
Capdevila J, Guerrero I (1994) Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J 13:4459–4468
Guillén I, Mullor JL, Capdevila J, Sánchez-Herrero E, Morata G, Guerrero I (1995) The function of engrailed and the specification of Drosophila wing pattern. Development 121:3447–3456
Liu Z, Matsuoka S, Enoki A, Yamamoto T, Furukawa K, Yamasaki Y, Nishida Y, Sugiyama S (2011) Negative modulation of bone morphogenetic protein signaling by Dullard during wing vein formation in Drosophila. Dev Growth Differ 53:822–841. https://doi.org/10.1111/j.1440-169X.2011.01289.x
Hay BA (2000) Understanding IAP function and regulation: a view from Drosophila. Cell Death Differ 7:1045–1056
Rothenfluh A, Threlkeld RJ, Bainton RJ, Tsai LT, Lasek AW, Heberlein U (2006) Distinct behavioral responses to ethanol are regulated by alternate RhoGAP18B isoforms. Cell 127:199–211
Hay BA, Wolff T, Rubin GM (1994) Expression of baculovirus P35 prevents cell death in Drosophila. Development 120:2121–2129
St Johnston RD, Hoffmann FM, Blackman RK, Segal D, Grimaila R, Padgett RW, Irick HA, Gelbart WM (1990) Molecular organization of the decapentaplegic gene in Drosophila melanogaster. Genes Dev 4:1114–1127
Mathew SJ, Haubert D, Krönke M, Leptin M (2009) Looking beyond death: a morphogenetic role for the TNF signalling pathway. J Cell Sci 122:1939–1946. https://doi.org/10.1242/jcs.044487
Neisch AL, Speck O, Stronach B, Fehon RG (2010) Rho1 regulates apoptosis via activation of the JNK signaling pathway at the plasma membrane. J Cell Biol 189:311–323. https://doi.org/10.1083/jcb.200912010
Yan J, Lu Q, Fang X, Adler PN (2009) Rho1 has multiple functions in Drosophila wing planar polarity. Dev Biol 333:186–199. https://doi.org/10.1016/j.ydbio.2009.06.027
Gumbiner BM (2005) Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol 6:622–634
Serrano N, O’Farrell PH (1997) Limb morphogenesis: connections between patterning and growth. Curr Biol 7:R186–R195
Campbell G, Weaver T, Tomlinson A (1993) Axis specification in the developing Drosophila appendage: the role of wingless, decapentaplegic, and the homeobox gene aristaless. Cell 74:1113–1123
Akiyama T, Kamimura K, Firkus C, Takeo S, Shimmi O, Nakato H (2008) Dally regulates Dpp morphogen gradient formation by stabilizing Dpp on the cell surface. Dev Biol 313:408–419
Entchev EV, Schwabedissen A, González-Gaitán M (2000) Gradient formation of the TGF-beta homolog Dpp. Cell 103:981–991
Teleman AA, Cohen SM (2000) Dpp gradient formation in the Drosophila wing imaginal disc. Cell 103:971–980
Affolter M, Basler K (2007) The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat Rev Genet 8:663–674
Martín-Castellanos C, Edgar BA (2002) A characterization of the effects of Dpp signaling on cell growth and proliferation in the Drosophila wing. Development 129:1003–1013
Nellen D, Burke R, Struhl G, Basler K (1996) Direct and long-range action of a DPP morphogen gradient. Cell 85:357–368
Künnapuu J, Björkgren I, Shimmi O (2009) The Drosophila DPP signal is produced by cleavage of its proprotein at evolutionary diversified furin-recognition sites. Proc Natl Acad Sci USA 106:8501–8506. https://doi.org/10.1073/pnas.0809885106
Spencer FA, Hoffmann FM, Gelbart WM (1982) Decapentaplegic: a gene complex affecting morphogenesis in Drosophila melanogaster. Cell 28:451–461
Neumann CJ, Cohen SM (1997) Long-range action of Wingless organizes the dorsal-ventral axis of the Drosophila wing. Development 124:871–880
Strigini M, Cohen SM (2000) Wingless gradient formation in the Drosophila wing. Curr Biol 10:293–300
Zecca M, Basler K, Struhl G (1996) Direct and long-range action of a wingless morphogen gradient. Cell 87:833–844
Martín FA, Peréz-Garijo A, Morata G (2009) Apoptosis in Drosophila: compensatory proliferation and undead cells. Int J Dev Biol 53:1341–1347. https://doi.org/10.1387/ijdb.072447fm
Huang J, Feng Y, Chen X, Li W, Xue L (2017) Myc inhibits JNK-mediated cell death in vivo. Apoptosis 22:479–490. https://doi.org/10.1007/s10495-016-1340-4
de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA (2004) Drosophila myc regulates organ size by inducing cell competition. Cell 117:107–116
Wu DC, Johnston LA (2010) Control of wing size and proportions by Drosophila myc. Genetics 184:199–211. https://doi.org/10.1534/genetics.109.110379
Hall A (1994) Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol 10:31–54
Chant J, Stowers L (1995) GTPase cascades choreographing cellular behavior: movement, morphogenesis, and more. Cell 81:1–4
Bloor JW, Kiehart DP (2002) Drosophila RhoA regulates the cytoskeleton and cell-cell adhesion in the developing epidermis. Development 129:3173–3183
Vidal M, Larson DE, Cagan RL (2006) Csk-deficient boundary cells are eliminated from normal Drosophila epithelia by exclusion, migration, and apoptosis. Dev Cell 10:33–44
Warner SJ, Yashiro H, Longmore GD (2010) The Cdc42/Par6/aPKC polarity complex regulates apoptosis-induced compensatory proliferation in epithelia. Curr Biol 20:677–686. https://doi.org/10.1016/j.cub.2010.03.025
Chountala M, Vakaloglou KM, Zervas CG (2012) Parvin overexpression uncovers tissue-specific genetic pathways and disrupts F-actin to induce apoptosis in the developing epithelia in Drosophila. PLoS ONE 7:e47355. https://doi.org/10.1371/journal.pone.0047355
Magie CR, Pinto-Santini D, Parkhurst SM (2002) Rho1 interacts with p120ctn and alpha-catenin, and regulates cadherin-based adherens junction components in Drosophila. Development 129:3771–3782
Acknowledgements
We thank Alan M Michelson for supplying us with the fly strains used in this study. We gratefully acknowledge Vienna Drosophila RNAi Stock Center, Tsinghua Drosophila model animal center, GenExel Stock Center and Developmental Studies Hybridoma Bank for providing fly lines and antibodies.
Funding
This work was supported by the National Natural Science Foundation of China (31772521) and Fundamental Research Funds for the Central Universities (2572018CG05, 2572015AA10).
Author information
Authors and Affiliations
Contributions
XCW, investigation, visualization, writing—original draft; LZ, review; LHJ, supervision, funding acquisition, project administration—review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing or financial interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, X.C., Liu, Z. & Jin, L.H. Drosophila jumu modulates apoptosis via a JNK-dependent pathway and is required for other processes in wing development. Apoptosis 24, 465–477 (2019). https://doi.org/10.1007/s10495-019-01527-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-019-01527-x