Abstract
Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) is a potent cancer cell-specific apoptosis-inducing cytokine with little toxicity to most normal cells. Here, we report that gefitinib and TRAIL in combination produce a potent synergistic effect on TRAIL-sensitive human colon cancer HCT116 cells and an additive effect on TRAIL-resistant HT-29 cells. Interestingly, gefitinib increases the expression of cell surface receptors DR4 and DR5, possibly explaining the synergistic effect. Knockdown of DR4 and DR5 by siRNA significantly decreases gefitinib- and TRAIL-mediated cell apoptosis, supporting this idea. Because the inhibition of gefitinib-induced autophagy by 3-MA significantly decreases DR4 and DR5 upregulation, as well as reduces gefitinib- and TRAIL-induced apoptosis, we conclude that death receptor upregulation is autophagy mediated. Furthermore, our results indicate that death receptor expression may also be regulated by JNK activation, because pre-treatment of cells with JNK inhibitor SP600125 significantly decreases gefitinib-induced death receptor upregulation. Interestingly, SP600125 also inhibits the expression CHOP, yet CHOP has no impact on death receptor expressions. We also find here that phosphorylation of Akt and ERK might also be required for TRAIL sensitization. In summary, our results indicate that gefitinib effectively enhances TRAIL-induced apoptosis, likely via autophagy and JNK- mediated death receptor expression and phosphorylation of Akt and ERK.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL), a type II transmembrane protein from the TNF cytokine family, is a potent cancer cell- specific apoptosis-inducing agent that exhibits little or no effect on normal tissues [1–3]. TRAIL can interact with five distinct type I transmembrane receptors, two of which are death receptors, DR4 and DR5, and three of which are decoy receptors, DcR1, DcR2 and osteoprotegerin. Each of these death receptors contains a cytoplasmic functional death domain [4–6]. Upon binding death receptors, TRAIL triggers cell death via both extrinsic and intrinsic apoptosis pathways [7]. Because of this, monoclonal antibodies against death receptors have also demonstrated the potential as anti-cancer therapeutics [8–10]. However, the effectiveness of agonistic antibodies as monotherapies may be limited by the development of drug resistance, and agents that can enhance TRAIL-induced apoptosis or sensitize resistant cancer cells to TRAIL are urgently needed [11, 12].
Gefitinib (also known as ZD1839 or Iressa) is an orally administered, selective epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (EGFR-TKI) that blocks signal transduction pathways implicated in proliferation and survival of cancer cells [13]. Gefitinib has a broad anti-tumor effect against various types of solid tumors [14–16]. Moreover, EGFR regulates ERK and Akt that could limit apoptosis induced by cytotoxic agents [17–19]. Hence, gefitinib can be exploited in EGFR-dependent tumors in combination regimens [20, 21].
It has been reported that gefitinib sensitizes non-small cell lung cancer cells to TRAIL-induced apoptosis via upregulation of pro-apoptosis protein Bax and down-regulation of p-Akt [22]. Moreover, gefitinib can reverse TRAIL resistance in human bladder cell lines via inhibition of Akt-mediated X-linked inhibitor of apoptosis protein expression [23]. Gefitinib also enhances the anti-cancer effect of TRAIL in human esophageal squamous cell carcinoma by inhibiting p-Akt and inactivating Bcl-xL [24]. Furthermore, gefitinib causes upregulation of the long isoform of caspase-8 inhibitor FLICE-inhibitory protein (FLIP(L)) and makes non-transformed breast epithelial cells more resistant to TRAIL through extracellular signal-regulated kinase (ERK)1/2 pathway [25]. It is unknown, however, whether gefitinib can enhance TRAIL-induced apoptosis in human colon cancer cells. Although it was recently reported that gefitinib can enhance TRAIL anti-tumor activity in NSCLC cells by upregulating DR5 expression [26], the mechanism of action is not fully understood.
In the present study, we demonstrate that gefitinib enhances TRAIL-induced apoptosis in human colon cancer cells via inhibition of Akt and ERK and death receptor upregulation. Furthermore, we found that death receptor upregulation is mediated by autophagy and JNK activation.
Materials and methods
Reagents
A solution of 50 mM gefitinib (purchased from Cayman Chemical, Ann Arbor, MI) was prepared in 100 % dimethyl sulfoxide, stored in small aliquots at −80 °C, and diluted in cell culture medium as needed. His-tagged recombinant human TRAIL (rhTRAIL) was produced and purified as described previously [27]. Dulbecco’s modified Eagle’s medium: Nutrient Mixture F-12 (DMEM/F12) medium, Iscove’s Modified Dubecco’s Medium (IMDM), and fetal bovine serum were obtained from Gibco (Gibco, Grand Island, NY, USA). Trypsin and PMSF were obtained from Amersco (Amersco LLC, OH, USA). PI, WST-1 and 3-MA were purchased from Sigma (Sigma Aldrich, St Louis, MO, USA). Incomplete protease inhibitor cocktail tablets were acquired from Roche (Roche, Welwyn Garden City, UK). The ECL Plus Western Blotting Detection Kit was purchased from Beijing Saizhi technology co., LTD (Beijing, China). Penicillin/streptomycin was obtained from the Tina Jin Hao Yang Biological Manufacture Co., Ltd. (Tianjin, China). Antibodies against DR4 and caspase-9 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies against PARP, JNK, p-JNK, ERK, p-ERK, Akt, p-Akt-308, p-Akt-473, caspase-3, LC3, CHOP and DR5 were purchased from Cell Signaling (Danvers, MA, USA). Antibodies against actin and caspase-8 were purchased from BD Biosciences (Franklin Lakes, NJ, USA). Caspase inhibitor and SP600125 were purchased from Beyotime Institute of Biotechnology (Shanghai, China). All of the other reagents were of analytical grade or better.
Cell lines and cell culture
Unless otherwise stated, all cell lines were purchased from the American Type Culture Collection and were maintained in the appropriate growth medium supplemented with 10 % fetal calf serum and penicillin/streptomycin at 37 °C and 5 % CO2. Colon adenocarcinoma HCT116 human colon cancer cells were cultured in IMDM medium. HT-29 cells were cultured in DMEM/F12 medium. HMECs and NCM460 were cultured in DMEM medium.
WST-1 assay
Cell viability was assessed using the WST-1 assay. In this assay, cells were treated with different concentrations of gefitinib or TRAIL for 24 h. The medium was then removed, and 10 µl/well of the WST-1 solution (Beyotime Biotechnology, Jiangsu, China) was added. After 4 h incubation, the absorbance was measured at 450 nm using a microplate reader (Bio-Rad). All experiments were performed in triplicate and repeated at least three times. Cell proliferation is expressed as the percentage of the control, which was set to 100 %.
Annexin V-FITC/PI flow cytometric analysis
Cell apoptosis was quantified by flow cytometry using Annexin V-FITC/propidium iodide (PI) staining. Cells were plated in 6-well plates (Costa) for 24 h, followed by treatment for another 24 h with various concentrations of gefitinib or TRAIL. Cells were then stained with Annexin V-FITC/PI (Keygen Biotechnology, Jiangsu, China) according to the manufacturer’s instructions. Washed cells were re-suspended in 500 µl PBS and incubated with 10 µl PI and 10 µl Annexin V-FITC for 15 min at room temperature in the dark. Approximately 20,000 cells were counted for each measurement.
Propidium iodide (PI) staining for DNA fragmentation
Using flow cytometry, cell death was quantified with propidium iodide (PI) staining for DNA fragmentation and content. For this analysis, cells were first pre-treated with gefitinib for 24 h and then exposed to TRAIL for 24 h. Floating and adherent cells were collected and fixed in 75 % ethanol, followed by ribonuclease (RNase) A treatment and PI staining. A total of 20,000 fluorescent events were acquired at 610 nm following excitation at 488 nm.
Reverse transcription polymerase chain reaction (RT-PCR)
DR4 and DR5 mRNAs were detected using RT-PCR as follows: Total RNA from the treated cells was extracted using the Trizol reagent according to the supplier’s instructions. Two micrograms of total RNA was converted into cDNA using M-MLV reverse transcriptase (Promega) and then amplified using Ex-Taq polymerase (Takara). Total RNA was amplified by PCR with primers described previously: DR4 sense 5′-CTGAGCAACGCAGACTCGCTGTCCAC-3′, DR4 anti-sense 5′-TCCAAGGAC ACGGCAGAGCCTGTGCCAT-3′, DR5 sense 5′-AAGACCCTTGTGCTCGTT GTC-3′, DR5 anti-sense 5′-GACACATTCGATGTCACTCCA-3′, glyceraldehyde-3- phosphate dehydrogenase (GAPDH) sense, 5′-GTCTTCACCACCATGGAG-3′, and GAPDH antisense 5′-CCACCCTGTTGCTGTAGC-3′. Reactions were run at 94 °C for 2 min; 94 °C for 35 cycles of 30 s each; 50 °C for 30 s; and 72 °C for 45 s with an extension at 72 °C for 10 min. PCR products were run on a 1.5 % agarose gel and then stained with ethidium bromide. Stained bands were visualized under UV light and photographed.
Flow cytometric analysis of the expression of cell surface DR4 and DR5
Cells were treated with different concentrations of gefitinib for 24 h, collected and washed with cold 2 % FBS/PBS, and then labeled with anti-DR4 or anti-DR5 antibody for 20 min on ice. The secondary antibody used was phycoerythrin (PE)- conjugated goat anti-rabbit Ab, which was incubated for 20 min on ice in the dark. Cells were then washed in cold 2 % FBS/PBS and resuspended in 300 µl 2 % FBS/PBS and immediately analysed by flow cytometry. All the antibodies were diluted with 2 % FBS/PBS.
Transfection with siRNA
High purity controls (scrambled RNA), along with DR4 and DR5 siRNA oligos, were obtained from GenePharma (Shanghai, China). The targeting sequences of the siRNA constructs were: DR4 siRNA, 5′-AAGAACCAGCAGAGGUCACAA-3′; DR5 siRNA, 5′-AAGACCCUUGUGCUCGUUGUC-3′; CHOP siRNA, 5′-UUCUUGGUCGUCUCCAGUGUU-3′. Transfection of these siRNA duplexes was performed in 6-well plates using the Lipofectamine RNAiMAX (GenePharma, Shanghai, China) following the manufacturer’s instructions. After 24 h of transfection, cells were treated with gefitinib for 24 h. Whole cell extracts were prepared for western blot protein analyses.
Western blotting
Cells were lysed in lysis buffer (50 mM Tris/acetate, pH 7.4, 1 mM EDTA, 0.5 % Triton X-100, 150 mM sodium chloride, 0.1 mM PMSF, and Roche incomplete protease inhibitor cocktail). Protein concentrations were measured using the Bradford method [28]. Equal amounts of protein were separated by 12 % sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a nitrocellulose membrane, blotted with each antibody, and then detected by using the enhanced chemiluminescence (ECL) reagent.
Statistical analysis
Data were analyzed using the student’s t test for comparison between groups. Significance was defined as P values less than 0.05 or 0.01.
Results
Gefitinib enhances TRAIL-induced apoptosis in HCT116 cells
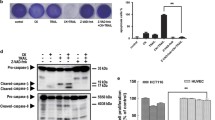
We initially investigated whether gefitinib could enhance TRAIL-induced apoptosis in TRAIL-sensitive HCT116 human colon cancer cells. Cell viability were tested by WST-1 assay. As shown in Fig. 1a, HCT116 cells were found to be moderately sensitive to TRAIL and gefitinib. However, in combination, they decreased cell viability significantly. PI staining demonstrated that gefitinib or TRAIL alone induced 26 or 19 % apoptosis, respectively. However, their combination enhanced apoptosis to 98 % (Fig. 1b). This finding was supported by Annexin V/PI staining as shown in Fig. 1c, where essentially the same levels of apoptosis could be attained with gefitinib or TRAIL alone or in combination (Fig. 1c). The degree of apoptosis was then evaluated by immunoblotting for apoptotic markers. As shown in Fig. 1d, HCT116 cells treated with gefitinib and TRAIL exhibited a markedly greater level of cleaved caspase-8, -9, and -3, along with the caspase-3 substrate poly(ADP-ribose) polymerase (PARP). Pre-treatment with the cell-permeable pan caspase inhibitor, z-VAD-fmk, effectively blocked apoptosis induced by this treatment from 98 to 22 % (Fig. 1b), indicating that gefitinib enhanced TRAIL-induced apoptosis of HCT116 cells in a caspase-dependent manner. In addition, our results showed that gefitinib significantly potentiated TRAIL-induced apoptosis in HCT116 cells, but not in immortalized human microvascular endotheilal cell lines (HMEC) (Fig. 1e) and NCM460 human normal colon cells (Fig. 1f), indicating that the combination of gefitinib and TRAIL preferentially target cancer cells.
Gefitinib enhances TRAIL-induced apoptosis in HCT116 cells. HCT116 cells, HMECs or NCM460 cells were co-treated with or without 50 µM gefitinib and/or TRAIL (25 ng/ml) for 24 h. Cell viability was analyzed using the WST-1 assay (a, e, or f), and cell apoptosis was teste by flow cytometry followed by PI staining (b) or Annexin V/PI staining (c) and western blot (d). The values represent the mean ± s.d. (n = 3). *P < 0.05, ** P < 0.01 (Student’s t test, two-tailed)
Additive effect of gefitinib and TRAIL on TRAIL-resistant HT-29 cells
We also investigated whether gefitinib could sensitize TRAIL-resistant HT-29 cancer cells to TRAIL. The WST-1 assay results showed that HT-29 cells were resistant to TRAIL and sensitive to gefitinib. The combination of TRAIL and gefitinib exerted an enhanced effect (Fig. 2a). Whereas TRAIL or gefitinib treatment alone induced 43 or 7.5 % apoptosis, respectively, their combination increased apoptosis to 72 % (Fig. 2b). Pre-treatment with the cell-permeable pan caspase inhibitor, z-VAD-fmk, effectively blocked apoptosis that was induced by this treatment, from 72 to 33 %. Similar results were obtained using Annexin V/PI staining (Fig. 2c). Consistent with these findings, combination treatment efficiently initiated caspase-3 processing and the cleavage of PARP (Fig. 2d).
Additive effect of gefitinib and TRAIL on TRAIL-resistant HT-29 cells. HT-29 cells were treated with or without 50 µM gefitinib and/or TRAIL (25 ng/ml) for 24 h. Cell viability was analyzed using the WST-1 assay (a) and cell apoptosis was investigated by flow cytometry PI staining (b) or Annexin V/PI staining (c) and western blot (d). Values represent the mean ± s.d. (n = 3). *P < 0.05, ** P < 0.01 as compared with control (Student’s t test, two-tailed)
Cell surface death receptor induction by gefitinib is required for the synergistic effect
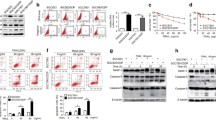
For mechanistic insight into gefitinib-enhanced TRAIL-induced apoptosis, we examined the effect of gefitinib on the expression of TRAIL death receptors. Our results showed that DR4 and DR5 were upregulated by gefitinib in a dose-dependent manner in both HCT116 and HT-29 cells (Fig. 3a). However, gefitinib did not impact either DR4 or DR5 mRNA expression levels, indicating post-transcription modulation (Fig. 3b). In support of this, we found that the translational inhibitor cycloheximide (CHX) blocked gefitinib-induced DR5 and DR4 expression (Fig. 3c). Interestingly, gefitinib increased cell surface DR4 and DR5 expression significantly in HCT116 cells, but only modestly in HT-29 cells (Fig. 3d). This might account for the synergistic effect or addictive effect observed with HCT116 cells or HT-29 cells.
Cell surface death receptors induction by gefitinib is required for the synergistic effect of gefitinib and TRAIL. HCT116 and HT-29 cells were treated with gefitinib at the indicated doses. DR4 and DR5 proteins were analyzed by western blotting (a), the mRNA expression were analyzed by RT-PCR (b), the cell surface DR4 and DR5 expression was tested by flow cytometry (d). HCT-116 cells were pretreated with 5 µM cycloheximide for 30 min before incubation with 50 µM gefitinib for 24 h. Whole-cell extracts were subjected to western blot analysis using anti-DR5 and anti-DR4 antibodies (c). HCT116 cells were transfected with DR4 and DR5 siRNA and then exposed to 50 μM gefitinib for 24 h. Whole-cell extracts were prepared and analyzed by western blotting (e) and the cell surface DR4 and DR5 expression was tested by flow cytometry (f). Control siRNA or DR4/DR5 siRNA transfected HCT116 cells were treated with or without 50 μM gefitinib and/or TRAIL (25 ng/ml) for 24 h. Cell viability was analyzed using the WST-1 assay (g) and the expression of caspase-3, -8 and PARP was assessed by western blotting (h)
To validate the role of cell surface death receptor expression on the enhanced effect from gefitinib on TRAIL-induced apoptosis in HCT116 cells, we used siRNAs specific for DR4 and DR5 (Fig. 3e). As shown in Fig. 3f, siRNAs for DR4 and DR5 reduced the cell surface expression of both death receptors. The WST-1 assay revealed that silencing of DR4 and DR5 increased the percentage of cell viability (Fig. 3g) and the cleavage of caspase-3, -8, and PARP significantly (Fig. 3h), suggesting that cell surface expression of death receptors indeed played a crucial role in the synergistic effect of gefitinib and TRAIL in HCT116 cells.
Gefitinib-induced death receptors upregulation is mediated by autophagy
Next, we were interested in assessing the mechanism by which gefitinib induced the expression of DR4 and DR5. Because gefitinib can induce autophagy in tumor cells [29], we investigated whether the observed upregulation of death receptors was related to autophagy. Our results showed that gefitinib produced a dose-dependent increase in LC3-II and decrease in p62 (Fig. 4a) that paralleled death receptor upregulation (Fig. 4a). Pre-treatment with autophagy inhibitor 3-methyladenine (3-MA, 5 mM) partially inhibited LC3-II upregulation (Fig. 4b) and death receptor upregulation (Fig. 4b), indicating that autophagy was associated with death receptor induction. In addition, 3-MA also substantially inhibited cell surface expression of death receptors (Fig. 4c) and reduced the extent of gefitinib/TRAIL-induced apoptosis in HCT116 cells from 95 to 69 % (Fig. 4d).
Gefitinib-induced death receptor upregulation is mediated by autophagy. HCT116 were treated with 50 μM gefitinib for the indicated hours. Protein expression was analyzed by western blotting (a). HCT116 cells were pre-treated with or without 3-MA for 1 h, then co-treated with gefitinib as the giving does for 24 h. Protein expression were analyzed by western blotting (b) and the cell surface DR4 and DR5 expression was tested by flow cytometry (c). HCT116 cells were pre-treated with 5 mM of 3-MA for 1 h then treated with or without gefitinib and/or TRAIL for 24 h. Cells were stained with PI and analyzed by flow cytometry (d)
Gefitinib-induced death receptors expression via JNK activation
Because Akt, ERK and JNK can mediate death receptor upregulation, we investigated whether these kinases played a role in gefitinib-induced DR4 and DR5 expression. Our results showed that gefitinib decreased Akt and ERK activation (Fig. 5a) and led to phosphorylation of JNK (Fig. 5b). In addition, gefitinib-induced DR4 and DR5 upregulation paralleled the activation of JNK (Fig. 5b). Pre-treatment with JNK inhibitor SP600125 (10 or 20 µM) significantly reduced JNK phosphorylation, as well as death receptor upregulation (Fig. 5c, d). Moreover, SP600125 reduced cell apoptosis induced by gefitinib and TRAIL from 90 to 51 % (Fig. 5e). We also found that pre-treatment with SP600125 significantly inhibited upregulation of JNK-modulated CHOP, the upstream transcription regulator of DR4 and DR5 (Fig. 5c). However, we saw no robust decrease in CHOP siRNA transfected HCT116 cells (Fig. 5f).
Gefitinib-induced death receptors expression is mediated by JNK activation. HCT116 were treated with gefitinib at the indicated doses. Proteins were analyzed by western blotting (a, b). HCT116 cells were pre-treated with or without 10 or 20 μM SP600125 and then treated with gefitinib in the giving doses. The expression of proteins were analyzed by western blotting (c) and the cell surface DR4 and DR5 expression was tested by flow cytometry (d). HCT116 cells were transfected with CHOP siRNA and then exposed to 50 μM gefitinib for 24 h. Whole-cell extracts were prepared and analyzed by western blotting (e). HCT116 cells were pre-treated with or without SP600125 and then co-treated with or without gefitinib and/or TRAIL for 24 h. The cells were stained with PI and were analyzed by flow cytometry (f)
Discussion
Previous studies had demonstrated that gefitinib could enhance TRAIL-induced apoptosis in non-small cell lung cancer cells, human bladder cells and esophageal squamous cell carcinoma [22–24, 26], but not in human colon cancer cells. Moreover, the mechanism of action by gefitinib had not been investigated. Here, we found that gefitinib could enhance TRAIL-induced apoptosis in human colon cancer HCT116 cells, as well as in TRAIL-resistant HT-29 cells. The combination of gefitinib and TRAIL synergistically enhanced HCT116 cell apoptosis, whereas it had only an additive effect on HT-29 cells. The degree of enhanced apoptosis was related to the cell surface expression of death receptors DR4 and DR5 in both cell lines, more significantly in HCT116 cells. Therefore, we hypothesized that death receptor trafficking pathways were different in HCT116 and HT-29 cells. Further studies are now underway to investigate this thoroughly. In contrast, gefitinib and TRAIL together had no effect on HMECs, indicating that their combination preferentially targets cancer cells and spares normal cells.
To determine whether the cell surface upregulation of death receptors by gefitinib is essential to sensitizing colon cancer cells to TRAIL, we silenced DR4 and DR5 using specific siRNAs in HCT116 cells. Our results showed that DR4 and DR5 siRNAs remarkably decreased cell surface expression of these death receptors, and significantly decreased subsequent potentiation of gefitinib on TRAIL-induced apoptosis. Based on these findings, we concluded that cell surface expression of death receptors by gefitinib is crucial to the synergistic effect of gefitinib and TRAIL.
Death receptor expression is tightly regulated by transcriptional and post- transcriptional mechanisms. Here we found that gefitinib did not alter mRNA expression of either death receptor, indicating that gefitinib might upregulate these death receptors by post-transcriptional mechanism. To confirm the hypothesis, the translation inhibitor cycloheximide was used. The results showed that cycloheximide blocked gefitinib-induced DR5 and DR4 expression.
Autophagy, a highly conserved and regulated cellular process employed by living cells to degrade proteins and cell organelles, has been reported to be induced following treatment with EGFR-TKIs in NSCLC and breast cancer cell lines [29, 30]. Therefore, we next asked whether autophagy could be related to gefitinib-induced death receptor upregulation. Our results showed that pre-treatment with autophagy inhibitor 3-MA substantially suppressed cell surface expression of death receptors and significantly reduced the extent of gefitinib/TRAIL-induced apoptosis. This finding allowed us to conclude that gefitinib treatment, at least in part, could account for the increase in DR4 and DR5 expression.
Although autophagy is known to regulate key processes associated with TRAIL resistance [31], the molecular mechanism of autophagy-mediated TRAIL resistance remains unknown [32, 33]. Recently, TRAIL-resistant breast cancer cells were reported to have a high basal level of autophagosomes that sequestered DR4 and DR5, which may contribute to the deficiency of these molecules on the cell surface [34]. This suggests that basal autophagosomes and drug-induced-autophagosomes might function differently in TRAIL receptor turnover. The precise role of autophagy in TRAIL and its receptor turnover remain unclear, and further studies are required to resolve this issue.
Because the inhibition of autophagy only partially decreased the effect of gefitinib on DR4 and DR5 expression, another mechanism of action aside from autophagy is likely also in play. Because Akt, ERK and JNK can mediate death receptor upregulation [11, 35, 36], we investigated whether these kinases could be that missing link. Our results demonstrated that gefitinib only provoked JNK activation, and that gefitinib-induced death receptor upregulation required the activation of JNK.
In addition to death receptor upregulation, the down-regulation of ERK and Akt by gefitinib may also lead to enhancement of TRAIL-induced apoptosis. Therefore, we concluded that gefitinib treatment of cancer cells results in the enhancement of TRAIL-induced apoptosis. Gefitinib effectively enhanced TRAIL-induced apoptosis by inhibiting Akt and ERK activation and up-regulating autophagy- and JNK- mediated death receptors expression (Fig. 6). Our findings contribute significantly to the understanding of the anticancer activity of gefitinib and warrant further evaluation for the combination of gefitinib and TRAIL as a potential therapeutic regimen against human colon cancer.
References
Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA et al (1995) Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 3(6):673–682
Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, Smith C, Smolak P, Goodwin RG, Rauch CT, Schuh JC, Lynch DH (1999) Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med 5(2):157–163. doi:10.1038/5517
Ashkenazi A, Pai RC, Fong S, Leung S, Lawrence DA, Marsters SA, Blackie C, Chang L, McMurtrey AE, Hebert A, DeForge L, Koumenis IL, Lewis D, Harris L, Bussiere J, Koeppen H, Shahrokh Z, Schwall RH (1999) Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest 104(2):155–162. doi:10.1172/JCI6926
Pan G, Ni J, Wei YF, Yu G, Gentz R, Dixit VM (1997) An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 277(5327):815–818
Chaudhary PM, Eby M, Jasmin A, Bookwalter A, Murray J, Hood L (1997) Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-kappaB pathway. Immunity 7(6):821–830
Pan G, O’Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, Dixit VM (1997) The receptor for the cytotoxic ligand TRAIL. Science 276(5309):111–113
Kimberley FC, Screaton GR (2004) Following a TRAIL: update on a ligand and its five receptors. Cell Res 14(5):359–372. doi:10.1038/sj.cr.7290236
Plummer R, Attard G, Pacey S, Li L, Razak A, Perrett R, Barrett M, Judson I, Kaye S, Fox NL, Halpern W, Corey A, Calvert H, de Bono J (2007) Phase 1 and pharmacokinetic study of lexatumumab in patients with advanced cancers. Clin Cancer Res 13(20):6187–6194. doi:10.1158/1078-0432.CCR-07-0950
Hotte SJ, Hirte HW, Chen EX, Siu LL, Le LH, Corey A, Iacobucci A, MacLean M, Lo L, Fox NL, Oza AM (2008) A phase 1 study of mapatumumab (fully human monoclonal antibody to TRAIL-R1) in patients with advanced solid malignancies. Clin Cancer Res 14(11):3450–3455. doi:10.1158/1078-0432.CCR-07-1416
Camidge DR, Herbst RS, Gordon MS, Eckhardt SG, Kurzrock R, Durbin B, Ing J, Tohnya TM, Sager J, Ashkenazi A, Bray G, Mendelson D (2010) A phase I safety and pharmacokinetic study of the death receptor 5 agonistic antibody PRO95780 in patients with advanced malignancies. Clin Cancer Res 16(4):1256–1263. doi:10.1158/1078-0432.CCR-09-1267
Cheng H, Hong B, Zhou L, Allen JE, Tai G, Humphreys R, Dicker DT, Liu YY, El-Deiry WS (2012) Mitomycin C potentiates TRAIL-induced apoptosis through p53-independent upregulation of death receptors: evidence for the role of c-Jun N-terminal kinase activation. Cell Cycle 11(17):3312–3323. doi:10.4161/cc.21670
Dolloff NG, Mayes PA, Hart LS, Dicker DT, Humphreys R, El-Deiry WS (2011) Off-target lapatinib activity sensitizes colon cancer cells through TRAIL death receptor up-regulation. Science Transl Med 3(86):86ra50. doi:10.1126/scitranslmed.3001384
Baselga J, Averbuch SD (2000) ZD1839 (‘Iressa’) as an anticancer agent. Drugs 60(Suppl 1):33–40 discussion 41–32
Baselga J, Rischin D, Ranson M, Calvert H, Raymond E, Kieback DG, Kaye SB, Gianni L, Harris A, Bjork T, Averbuch SD, Feyereislova A, Swaisland H, Rojo F, Albanell J (2002) Phase I safety, pharmacokinetic, and pharmacodynamic trial of ZD1839, a selective oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with five selected solid tumor types. J Clin Oncol 20(21):4292–4302
Wakeling AE, Guy SP, Woodburn JR, Ashton SE, Curry BJ, Barker AJ, Gibson KH (2002) ZD1839 (Iressa): an orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res 62(20):5749–5754
Lorusso PM (2003) Phase I studies of ZD1839 in patients with common solid tumors. Semin Oncol 30 (Suppl 1):21–29. doi:10.1053/sonc.2003.50029
Schlessinger J (2000) Cell signaling by receptor tyrosine kinases. Cell 103(2):211–225
Sibilia M, Fleischmann A, Behrens A, Stingl L, Carroll J, Watt FM, Schlessinger J, Wagner EF (2000) The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell 102(2):211–220
Gschwind A, Fischer OM, Ullrich A (2004) The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer 4(5):361–370. doi:10.1038/nrc1360
Magne N, Fischel JL, Tiffon C, Formento P, Dubreuil A, Renee N, Formento JL, Francoual M, Ciccolini J, Etienne MC, Milano G (2003) Molecular mechanisms underlying the interaction between ZD1839 (‘Iressa’) and cisplatin/5-fluorouracil. Br J Cancer 89(3):585–592. doi:10.1038/sj.bjc.6601131
Tortora G, Caputo R, Damiano V, Melisi D, Bianco R, Fontanini G, Veneziani BM, De Placido S, Bianco AR, Ciardiello F (2003) Combination of a selective cyclooxygenase-2 inhibitor with epidermal growth factor receptor tyrosine kinase inhibitor ZD1839 and protein kinase A antisense causes cooperative antitumor and antiangiogenic effect. Clin Cancer Res 9(4):1566–1572
Xu F, Tian Y, Huang Y, Zhang LL, Guo ZZ, Huang JJ, Lin TY (2011) EGFR inhibitors sensitize non-small cell lung cancer cells to TRAIL-induced apoptosis. Chin J Cancer 30(10):701–711. doi:10.5732/cjc.011.10107
Shrader M, Pino MS, Lashinger L, Bar-Eli M, Adam L, Dinney CP, McConkey DJ (2007) Gefitinib reverses TRAIL resistance in human bladder cancer cell lines via inhibition of AKT-mediated X-linked inhibitor of apoptosis protein expression. Cancer Res 67(4):1430–1435. doi:10.1158/0008-5472.CAN-06-1224
Teraishi F, Kagawa S, Watanabe T, Tango Y, Kawashima T, Umeoka T, Nisizaki M, Tanaka N, Fujiwara T (2005) ZD1839 (Gefitinib, ‘Iressa’), an epidermal growth factor receptor-tyrosine kinase inhibitor, enhances the anti-cancer effects of TRAIL in human esophageal squamous cell carcinoma. FEBS Lett 579 (19):4069–4075. doi:10.1016/j.febslet.2005.06.031
Yerbes R, Lopez-Rivas A, Reginato MJ, Palacios C (2012) Control of FLIP(L) expression and TRAIL resistance by the extracellular signal-regulated kinase1/2 pathway in breast epithelial cells. Cell Death Differ 19(12):1908–1916. doi:10.1038/cdd.2012.78
Yan D, Ge Y, Deng H, Chen W, An G (2015) Gefitinib upregulates death receptor 5 expression to mediate rmhTRAIL-induced apoptosis in Gefitinib-sensitive NSCLC cell line. Onco Targets Ther 8:1603–1610. doi:10.2147/OTT.S73731
Kim SH, Kim K, Kwagh JG, Dicker DT, Herlyn M, Rustgi AK, Chen Y, El-Deiry WS (2004) Death induction by recombinant native TRAIL and its prevention by a caspase 9 inhibitor in primary human esophageal epithelial cells. J Biol Chem 279(38):40044–40052. doi:10.1074/jbc.M404541200
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Han W, Pan H, Chen Y, Sun J, Wang Y, Li J, Ge W, Feng L, Lin X, Wang X, Jin H (2011) EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PloS One 6(6):e18691. doi:10.1371/journal.pone.0018691
Dragowska WH, Weppler SA, Wang JC, Wong LY, Kapanen AI, Rawji JS, Warburton C, Qadir MA, Donohue E, Roberge M, Gorski SM, Gelmon KA, Bally MB (2013) Induction of autophagy is an early response to gefitinib and a potential therapeutic target in breast cancer. PloS One 8(10):e76503. doi:10.1371/journal.pone.0076503
Han J, Hou W, Goldstein LA, Lu CS, Stolz DB, Yin XM, Rabinowich H (2008) Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells. J Biol Chem 283(28):19665–19677. doi:10.1074/jbc.M710169200
Hou W, Han J, Lu C, Goldstein LA, Rabinowich H (2008) Enhancement of tumor-TRAIL susceptibility by modulation of autophagy. Autophagy 4(7):940–943
Yuan BZ, Chapman J, Ding M, Wang J, Jiang B, Rojanasakul Y, Reynolds SH (2013) TRAIL and proteasome inhibitors combination induces a robust apoptosis in human malignant pleural mesothelioma cells through Mcl-1 and Akt protein cleavages. BMC Cancer 13(1):140. doi:10.1186/1471-2407-13-140
Di X, Zhang G, Zhang Y, Takeda K, Rosado LA, Zhang B (2013) Accumulation of autophagosomes in breast cancer cells induces TRAIL resistance through downregulation of surface expression of death receptors 4 and 5. Oncotarget 4(9):1349–1364
Thamkachy R, Kumar R, Rajasekharan KN, Sengupta S (2016) ERK mediated upregulation of death receptor 5 overcomes the lack of p53 functionality in the diaminothiazole DAT1 induced apoptosis in colon cancer models: efficiency of DAT1 in Ras-Raf mutated cells. Mol Cancer. doi:10.1186/s12943-016-0505-7
Shoeb M, Ramana KV, Srivastava SK (2013) Aldose reductase inhibition enhances TRAIL-induced human colon cancer cell apoptosis through AKT/FOXO3a-dependent upregulation of death receptors. Free Radical Bio Med 63:280–290. doi:10.1016/j.freeradbiomed.2013.05.039
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81302172) and the China Postdoctoral Science Foundation (2013M530959 & 2014T70270). We would also like to thank Prof. KH Mayo for critical reading and editing of this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Chen, L., Meng, Y., Guo, X. et al. Gefitinib enhances human colon cancer cells to TRAIL-induced apoptosis of via autophagy- and JNK-mediated death receptors upregulation. Apoptosis 21, 1291–1301 (2016). https://doi.org/10.1007/s10495-016-1287-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-016-1287-5