Abstract
The brown dog tick, Rhipicephalus sanguineus sensu lato, is a ubiquitous and taxonomically controversial pest of dogs with immense veterinary and public health significance. Genetic analyses of specimens from various geographical origins reveal intraspecific diversity within the taxon. Little information is available on the genetic characteristics of R. sanguineus s.l. in Nigeria, West Africa. In this study, 460 bp of the mitochondrial 16S rDNA gene of R. sanguineus s.l. collected from dogs in different ecological zones of Nigeria was amplified, sequenced and characterized. Phylogenetic and pairwise analyses were used to compare the sequences generated in this study to each other and to sequences in GenBank. The sequences in this study were highly similar (>98%) to each other and clustered with sequences of the R. sanguineus s.l. tropical lineage in GenBank. None of the sequences in this study clustered with the ‘southeastern Europe’ or temperate lineage. The mean intraspecific divergence among R. sanguineus s.l. in this study was 1.7% (range: 0−8.0%). Furthermore, the sequences in this study showed mean divergence of 1.5% (0−10%), 5.0% (3.8−13.9%) and 9.7% (6.9−19.8%) from sequences of the tropical, southeastern Europe and temperate lineages, respectively. Interestingly, sequences in this study showed a mean divergence of 9.3% (1.0−17.8%) from the Rhipicephalus sp. morphotype 4 (GenBank acc. nr. KC243850) earlier identified from cattle in Nigeria, suggesting diversity in this taxon in Nigeria. Further studies are needed to elucidate the veterinary and public health significance of R. sanguineus s.l. in Nigeria taking into cognizance the existence of intraspecific variation in vector competence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The brown dog tick, Rhipicephalus sanguineus s.l., is renowned for its widespread distribution and the number of pathogens it can transmit (Dantas-Torres 2010; Gray et al. 2013; Zemtsova et al. 2016; Jones et al. 2017). Therefore, this tick has continued to attract attention globally not because of its association with dogs but as a vector of pathogens of public health significance (Moraes-Filho et al. 2015; Chitimia-Dobler et al. 2017). Although the work of Nava et al. (2018) has established the name R. sanguineus sensu stricto (s.s.) for tick populations in the western Mediterranean region and for the so-called temperate lineage of R. sanguineus sensu lato (s.l.) in the southern hemisphere, the designation R. sanguineus s.l. is adopted for the ticks in this study.
Based on available biological, genetic and molecular evidences, R. sanguineus s.l. is a group of at least 17 morphologically closely related species as follows: Rhipicephalus aurantiacus Neumann; Rhipicephalus bergeoni Morel and Balis, Rhipicephalus boueti Morel; Rhipicephalus camicasi Morel, Mouchet and Rodhain; Rhipicephalus guilhoni Morel and Vassiliades; Rhipicephalus leporis Pomerantzev; Rhipicephalus moucheti Morel; Rhipicephalus pumilio Schulze; Rhipicephalus pusillus Gil Collado; Rhipicephalus ramachandrai Dhanda; Rhipicephalus rossicus Yakimov and Kol-Yakimova; R. sanguineus sensu stricto (s.s.); Rhipicephalus schulzei Olenev; Rhipicephalus sulcatus Neumann; Rhipicephalus tetracornus Kitaoka and Suzuki; Rhipicephalus turanicus Pomerantzev; and Rhipicephalus ziemanni Neumann. A new addition to this list is Rhipicephalus afranicus Bakkes (Bakkes et al. 2020). In view of the medical and veterinary importance of R. sanguineus s.l., there is the need for accurate identification. However, there are limitations with the morphological identification of the brown dog tick occasioned by the intraspecific variability of morphological traits and close similarity to those in related species with the result that they are often misidentified (Farid 1996; Ioffe-Uspenskiy et al. 1997; Oliveira et al. 2005).
Molecular tools have been extensively employed as an adjunct to the classical methods in order to accurately identify and characterize members of the R. sanguineus taxon. Genetic markers such as the mitochondrial 16 and 12S of the ribosomal DNA (rDNA) and to a lesser extent the nuclear markers such as the internal transcribed spacer-2 (ITS2) region have been used to achieve rapid and reliable species identification of ticks within the R. sanguineus group (Black and Piesman 1994; Mangold et al. 1998; Nava et al. 2012; Dantas-Torres et al. 2013; Chitimia-Dobler et al. 2017; Almeida et al. 2017). Prior to the reports of Nava et al. (2018), at least two distinct lineages—‘temperate’ and ‘tropical’, based on molecular studies—have been considered under the taxon R. sanguineus (Moraes-Filho et al. 2011; Nava et al. 2012; Sanches et al. 2016; Jones et al. 2017). However, it has now been established that the so-called temperate lineage of R. sanguineus s.l. in the western Mediterranean region and the southern hemisphere are bona fide R. sanguineus s.s. (Nava et al. 2018), thus clarifying the debate on the taxonomy of this taxon.
It has been postulated that the introduction of the temperate species into tropical areas and vice versa do occur frequently, but particular requirements of each tick species possibly related to climatic factors such as annual temperature extremes might have precluded their establishment in the new climes (Moraes-Filho et al. 2011; Laatamna et al. 2020).
The brown dog tick is believed to have originated from Africa before it was able to spread throughout the world along with humans and their dogs (Balashov 1994; Walker et al. 2000; Guglielmone et al. 2003). Till date, there are comparatively few genetics studies on this tick species on the African continent, particularly on tick specimens from Nigeria, West Africa. Therefore, the aim of this study was to identify and characterize R. sanguineus s.l. infesting dogs in Nigeria using the 16S rDNA mitochondrial gene marker in order to augment the sketchy molecular data from the continent.
Materials and methods
Study location
Nigeria is a West African country located between 4–14°N and 2–15°E, and has a total area of about 925,796 km2. It shares international land borders with Republic of Benin to the west, Chad and Cameroon to the east and Niger in the north with coastlines on the Gulf of Guinea in the south and Lake Chad to the northeast. The country is covered by three types of vegetation: forests in the south, savannahs in the central and northern part, and Sahel at the northern fringes (Oguntunde et al. 2011). Relatedly, there are four climate types; the tropical monsoon over the south, tropical savanna in the central and northern regions, Sahel over the northern fringes and an alpine climate on the highlands and plateaus of Nigeria which are over 1520 m above sea level (Adedoyin 1989). Wide variation in annual precipitation is recorded over Nigeria ranging from as low as 450 mm in the Sahel to over 4000 mm in the coastal regions. Equally variable is the mean annual temperature that ranges from 21 °C in the alpine climate to as high as 40 °C in the Sahel climate (Oguntunde et al. 2011; Ogungbenro and Morakinyo 2014).
Ticks sampling
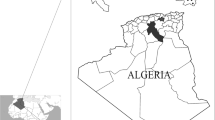
Ticks were collected from owned dogs in the Federal Capital Territory (FCT) Abuja and 11 states located in the various agro-climatic zones of Nigeria between June 2018 and September 2019 (Fig. 1). Ticks were removed using fine forceps from naturally infested dogs presented to veterinary clinics regardless of their clinical condition. Ticks were placed in labelled tubes containing 70% ethanol and transported to the Entomology Research Laboratory, National Veterinary Research Institute (NVRI), Vom, Nigeria.
Morphological identification of ticks
Ticks were identified morphologically under a stereomicroscope using standard keys (Walker et al. 2000; Dantas-Torres et al. 2013). Ticks were determined as R. sanguineus s.l. based on the following morphological characteristics. The male ticks were identified based on the long comma-shaped, dorsally tapering spiracles with an end tail that is less than half as wide as the adjacent festoon. The females were identified based on the presences of a U-shaped genital opening with triangular or crescent-shaped sclerites that slope slightly outward and are well separated from each other (Dantas-Torres et al. 2013). In total, 5515 ticks were morphologically identified as R. sanguineus s.l. and were included in this study.
Molecular characterization of Rhipicephalus sanguineus ticks
DNA extraction
In total 120 representative tick samples were selected from the morphologically identified R. sanguineus s.l. and included for the molecular studies. Five flat or partially engorged adult females and males each were selected from each of the study areas for DNA extraction. The ticks were removed from the 70% ethanol and washed in three changes of phosphate buffered saline (PBS), before DNA extraction. Genomic DNA was extracted using the QIAmp DNA Mini Tissue extraction kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. Eluted DNA were preserved at – 20 °C until PCR analysis.
PCR amplification and sequencing of 16S mitochondrial gene (16S rDNA)
Published primers, 16S + 1 (5′-CTG CTCAAT GAT TTT TTA AAT TGC TGT GG-3′) and 16 S-1 (5′-CCG GTC TGA ACT CAG ATC AAG T-3′) were used to amplify the 460 bp of 16S rDNA of Ixodidae (Black and Piesman 1994). The reaction was carried out in 50 µL volume made of 1.25 U Taq DNA polymerase (Fermentas), 5 µL 10 × Taq reaction buffer (including 15 mM MgCl2), 5 µL PCR nucleotide Mix (0.2 mM each), 1.5 µL (1 µM final concentration) of each primer, and 5 µL template DNA; PCR grade water (BioConcept, Allschwil, Switzerland) was used to make up the volume. The cycling conditions were as follows: an initial denaturation step at 95 °C for 3 min was followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 51 °C for 1 min and extension at 72 °C for 2 min. Final extension was performed at 72 °C for 5 min. The PCR was carried out using the Applied Biosystem thermal cycler (GeneAmp 9700) in the Molecular Biology Unit, Parasitology Division, NVRI, Vom, Nigeria.
The resulting PCR products were electrophoresed on a 1.5% agarose gel stained with SafeView from Applied Biological Materials (Richmond, BC, Canada) to check the size of amplified fragments by comparison to 100 bp DNA ladder from New England Biolabs (Ipswich, MA, USA) under a blue light transilluminator (Cleaver Scientific, Rugby, UK). Amplified products of the expected size were sequenced at a commercial facility (Macrogen Europe, Amsterdam, The Netherlands). Sequences obtained were manually corrected by visual analysis of the electrophoregram using Bioedit v.7.0.5.3 software and compared with the GenBank database by using the BLAST program (https://www.ncbi.nlm.nih.gov/BLAST).
Phylogenetic and pairwise analyses
The 14 sequences of R. sanguineus s.l. obtained in this study were aligned with each other and with other sequences in the GenBank using the Clustal W. Partial sequences of 16S mitochondrial rDNA representing the tropical, temperate, and the ‘Southeastern Europe’ lineages of R. sanguineus s.l. and other Rhipicephalus spp. from GenBank were included in the phylogenetic analysis. Also, 16S rDNA sequences of Rhipicephalus sp. morphotype 4 (KC243849, KC243850) earlier identified from cattle in Nigeria and R. afranicus (MK158990), recently described as the afrotropical lineage of R. turanicus from South Africa, were included. In addition, mitochondrial 16S rDNA sequences of R. turanicus, R. guilhoni, R sulcatus, R. muhsamae, R. zumpti and R. simus from various countries in GenBank were used. Sequence of R. microplus (EU918176) from Argentina was used as an outgroup. A general time reversible model with gamma distribution was selected to make the phylogenetic trees. Phylogenetic analyses were performed with the Maximum-likelihood (ML) and Neighbor Joining (NJ) methods. The phylogenetic trees were tested using 1000 replicate bootstrap values.
Pairwise analysis was conducted to compare the 16S rDNA sequences of R. sanguineus s.l. obtained from the various ecological zones of Nigeria in this study to identify inter- and intraspecies variation. Furthermore, pairwise estimates were used to determine the percent sequence divergence for the 16S rDNA gene of samples from this study with 16 sequences retrieved from the GenBank.
Results
In this study, 5515 ticks collected from 472 dogs in 11 states and the Federal Capital Territory (FCT), of Nigeria were morphologically identified as R. sanguineus s.l. PCR amplification of the 16S rDNA fragment was successful in 123 of the 130 selected specimens from the morphologically identified R. sanguineus s.l. in this study. Sequences were obtained from 16 selected samples representing the Sahel (n=3), Savanna (n=7) and the Guinea (n=6) ecological zones. One sequence from Plateau state was identified as R. muhsamae and another from Ogun state was identified as R. (B.) decoloratus and these were not included in the analyses. In all, 14 sequences of R. sanguineus s.l. were included in this study. All sequences from this study were submitted to the GenBank database under accession numbers MW560380–MW560390 and MZ470353–MZ470357.
Phylogenetic and pairwise analyses of Rhipicephalus sanguineus s.l. in Nigeria
The phylogenetic analysis of the 16S rDNA sequences inferred by the ML (Fig. 2) and NJ (Fig. 3) methods generated trees that segregated the Nigerian tick sequences into distinct cluster with sequences of the tropical lineage of R. sanguineus s.l. from Mozambique, South Africa, Egypt, Colombia and Brazil in GenBank. The low bootstrap support seen in the phylogenetic trees can be due to sequencing errors which may be consistent within labs but not between labs. Although, the sequences from this study fall in the same clade, they are positioned in different positions suggesting some degree of diversity among the Nigerian sequences. Furthermore, representative 16S rDNA sequences of the temperate and the ‘Southeastern Europe’ of R. sanguineus s.l. lineages and those of R. turanicus in GenBank formed distinct clades. However, the 16S rDNA gene sequence of the newly described R. afranicus from South Africa referred to as the ‘afrotropical’ lineage of R. turanicus clustered with R. sulcatus from Zambia and Zimbabwe and were placed distantly from R. turanicus in both the ML and NJ generated trees (Figs. 2, 3). The sequences of Rhipicephalus sp. morphotype 4 (KC243849 and KC243850) earlier identified from cattle in Nigeria formed a distinct cluster separate from other sequences (Figs. 2, 3).
Phylogenetic tree of Rhipicephalus sanguineus s.l. inferred from 402 bp 16S rDNA using the ML method. The bootstrap consensus tree was inferred from 1000 replicates. Sequences obtained in this study are indicated with black dots. GenBank accession numbers and countries of sequences and lineages are indicated. Rhipicephalus microplus (EU918176) from Argentina was used as outgroup
Phylogenetic tree of Rhipicephalus sanguineus s.l. inferred from 402 bp 16S rDNA using the NJ method. The bootstrap consensus tree was inferred from 1000 replicates. Sequences obtained in this study are indicated with black dots. GenBank accession numbers and countries of sequences and lineages are indicated. Rhipicephalus microplus (EU918176) from Argentina was used as outgroup
The nucleotide differences and sequence divergence on pairwise comparisons of mitochondrial 16S rDNA gene are presented in Table 1. The 16S rDNA sequences were used to assess the genetic variation within local populations of samples from the various collection sites within Nigeria as well as sequences from different countries in GenBank. Most samples collected from dogs in this study demonstrated 100 % identity. The mean intraspecific divergences among R. sanguineus s.l. in this study was 1.7% (range: 0.0−8.0%). However, the pairwise distances of three sequences (PH5, MG717 and KW4) showed variation of 0.8, 1.2, and 4.9%, respectively, from other sequences from Nigeria (Table 1). The greatest within-population diversity in 16S rDNA sequences was observed in one of the sequence KW4. This sequence demonstrated 13.6, 13.7 and 19.8% divergence from the tropical, southeastern Europe and temperate lineages of R. sanguineus s.l. sequences in GenBank. Furthermore, it showed 10.5 and 19.5% divergence from sequences of R. afranicus and R. turanicus, respectively (Table 1). Interestingly, the Rhipicephalus sp. morphotype 4 (KC243850) earlier identified from cattle in Nigeria showed a mean divergence of 9.3% (1.0–17.8%) from sequences in this study. All the sequences in this study showed ≥ 8.6% divergence from sequence of R. turanicus in GenBank (KY413795, KF145150). Furthermore, the sequences in this study showed mean divergence of 1.5% (0.0−10%), 4.97% (3.8−13.9%) and 9.74% (6.9−19.8%) from sequences of the tropical, southeastern Europe and temperate lineages, respectively (Table 1).
Discussion
The results of the phylogenetic analyses showed that the 16S rDNA sequences of R. sanguineus s.l. in Nigeria in this study clustered in one clade along with members of the tropical lineage from other tropical climes, similar to previous reports (Moraes-Filho et al. 2011; Zemtsova et al. 2016; Jones et al. 2017). This confirms that there is a minimal variability in genetic sequences of the R. sanguineus s.l. in Nigeria. However, this finding differed from the earlier results of a study involving ticks from north Africa where sequences were grouped into two distinct clades (Chitimia-Dobler et al. 2017). None of the 16S rDNA sequences in this study clustered with the so called southeastern Europe lineages where some sequences of R. sanguineus s.l. from Egypt were placed in the study by Chitimia-Dobler et al. (2017). This difference may be related to climatic differences, as the sequences that grouped with the southeastern Europe lineage were from Egypt which is located north of the Sahara and experience more temperate climate compared to countries within the sub-Sahara region. Therefore, unlike the situation in north Africa where at least two lineages of R. sanguineus s.l. were living in sympatry, only the tropical lineage was identified in Nigeria in this study. Results of the phylogenetic analyses were corroborated by the genetic analysis which further confirmed a high genetic convergence among populations of R. sanguineus s.l. collected from dogs in this study with a mean intraspecific variation of 1.7% (0.0−8.0%). Furthermore, no clear pattern exists in the sequence data from the various agro-ecological zones, suggesting significant dispersal and population mixing within R. sanguineus s.l. in Nigeria. However, the diversity displayed in the pairwise analysis may suggest the possibility of the existence of more than one haplotype under the taxon R. sanguineus s.l. in Nigeria.
Rhipicephalus sp. morphotype 4, earlier identified from cattle in Nigeria, was grouped separately from other Rhipicephalus spp. included in this study and showed marked divergence of > 6% from sequences of R. sanguineus s.l. in GenBank. Thus, lending credence to the earlier suggestions for the re-evaluation of R. sanguineus s.l. from the Afrotropical Region (Sanches et al. 2016; Zemtsova et al. 2016) and the possibility to name Rhipicephalus sp. morphotype 4 as distinct species (Almeida et al. 2017). The differential ability of R. sanguineus s.l. to transmit pathogens to humans, pets, livestock and wildlife has been demonstrated, hence the need to conduct studies that will not only assist to clarify its taxonomy (Gray et al. 2013; Dantas-Torres et al. 2013; Almeida et al. 2017) but also to determine their vector capacity for various pathogens of veterinary and public health significance in Nigeria. Equally, results of such studies will assist in the formulation of effective disease surveillance and control measures. Furthermore, similar to biological studies, the role of the animal host from which Rhipicephalus sp. were removed in the genetic analyses should be investigated. This may clarify some of the inconsistencies recorded for various R. sanguineus s.l. collected from different animal hosts.
In public health perspective, R. sanguineus s.l. is closely associated with dogs and consequently humans (Walker et al. 2000; Gray et al. 2013; Jones et al. 2017). The brown dog tick has been incriminated as the putative vector of several zoonotic diseases (Dantas-Torres et al. 2011; Gray et al. 2013). In Nigeria, close contact between dogs and humans is common, and human infestation with the brown dog tick has been reported (Okoli et al. 2006). Apart from the traditional role of dogs for companionship, security, hunting and shepherding, dog meat serves as source of protein to humans (Kamani et al. 2013). In addition, dog breeding is a booming business in Nigeria leading to the mass importation of foreign breeds of dogs from various climatic regions into the country. As a result, there is high inter- and intra-country mobility of dogs for sale and mating purposes with the propensity of the introduction and spread of ectoparasites, especially ticks. Hence, the need for constant surveillance of ticks infecting dogs to determine the species present and their veterinary and public health implications. Expectedly, none of the R. sanguineus s.l. analyzed in this study clustered with the temperate lineage. A recent study on the continent has reported the occurrence of R. sanguineus s.s. from steppe and high plateau regions of Algeria, but the Sahara Desert might have acted as a biogeographic barrier to the incursion of this species into the Sub-Sahara region (Laatamna et al. 2020). Generally, our findings are in agreement with the ‘20 °C’ rule for the survival and colonization of the tropical and temperate lineages of R. sanguineus s.l. (Zemtsova et al. 2016). The mean annual temperate in Nigeria is > 20 °C which is the upper limit for the survival of the temperate lineage of R. sanguineus (Nava et al. 2012; Zemtsova et al. 2016). Thus, even if imported dogs were brought into Nigeria with the temperate lineage of R. sanguineus s.l., the high environmental temperature may preclude its survival and proliferation. Contrariwise, the tropical lineage of R. sanguineus s.l. has been reported in temperate regions (Inokuma et al. 1996; Jones et al. 2017). This observation may lend credence to the hypothesized origin of R. sanguineus s.l. in Afrotropical regions before spreading to other parts of the world (Balashov 1994; Walker et al. 2000; Guglielmone et al. 2003).
It is worth mentioning that there is a record of the morphological identification of R. afranicus on cattle in Plateau state, Nigeria, in 1948 and possibly a voucher specimen exists (Bakkes et al. 2020). However, there appears to be no other reports on this tick species in Nigeria since then. Interestingly, the 16 S rDNA sequence of R. afranicus (GenBank MK158990), the recently described Afrotropical lineage that was included in the phylogenetic analyses, clustered with sequences of R. sulcatus from Zambia (MN944866) and Zimbabwe (MN944868) similar to the report of the authors (Bakkes et al. 2020). They were more closely placed to the clusters of R. guilhoni from Nigeria (KC243851, KC243583) than that of R. turanicus, suggesting further taxonomic considerations (Gray et al. 2013; Nava et al. 2018). Taxonomic conclusions are limited when using 16 S only as it is not a good marker for detecting phylogenetic divergence deeper than the species level. Therefore, the use of 16 S sequences in this study resulted in low bootstrap support for assessing these relationships. Moreover, the phylogenetic placements of all species within the Afrotropical sanguineus complex is uncertain at this stage due to the lack of nuclear data and mitochondrial genomes.
In conclusion, the ability of R. sanguineus s.l. to adopt various strategies for survival has earned it the appellation as a ‘catholic’ tick (Dantas-Torres 2010), in addition to other descriptions such as The Pan-tropical dog tick, kennel tick or brown dog tick. There is no doubt to its versatility, broad host and climate range and the ability to vector pathogens of veterinary and public health importance. Added to these is the existence of diverse haplotypes under the taxon R. sanguineus manifested in varying biological and molecular studies. This tick will continue to engage the attention of the research community for some time, particularly the veterinary and public health practitioners. In Nigeria, efforts should be geared towards elucidating the epidemiology and vector competence of this tick for various pathogens of veterinary and public health significance, taking into cognizance the existence of intraspecific variations.
References
Adedoyin JA (1989) Initiation of West African squall lines. Meteorol Atmos Phys 41:99–103. https://doi.org/10.1007/BF01043455
Almeidia C, Simões R, Coimbra-Dores MJ, Rosa F, Dias D (2017) Mitochondrial DNA analysis of Rhipicephalus sanguineus s.l. from the Western Iberian peninsula. Med Vet Entomol 31:167–177. https://doi.org/10.1111/mve.12222
Bakkes DK, Chitimia-Dobler L, Matloa D, Oosthuysen M, Mumcuoglu KY, Mans BJ, Matthee CA (2020) Integrative taxonomy and species delimitation of Rhipicephalus turanicus (Acari: Ixodida: Ixodidae). Internat J Parasitol. https://doi.org/10.1016/j.ijpara.2020.04.0050020-7519/2020
Balashov YS (1994) Importance of continental drift in the distribution and evolution of ixodid ticks. Entomol Rev 73:42–50
Black WC, Piesman J (1994) Phylogeny of hard and soft-tick taxa (Acari: Ixodida) based on mitochondrial 16S rDNA sequences. Proc Natl Acad Sci USA 91:10034–10038
Chitimia-Dobler L, Langguth J, Pfeffer M, Kattner S, Küpper T, Friese D, Dobler G, Guglielmone AA, Nava S (2017) Genetic analysis of Rhipicephalus sanguineus sensu lato ticks parasites of dogs in Africa north of the Sahara based on mitochondrial DNA sequences. Vet Parasitol 239:1–6. https://doi.org/10.1016/j.vetpar.2017.04.012
Dantas-Torres F (2010) Biology and ecology of the brown dog tick, Rhipicephalus sanguineus. Parasit Vectors 3:26. http://www.parasitesandvectors.com/content/3/1/26
Dantas-Torres F, Latrofa MS, Annoscia G, Gianelli A, Parisi A, Otranto D (2013) Morphological and genetic diversity of Rhipicephalus sanguineus sensu lato from the New and Old Worlds. Parasit Vectors 6:213. https://doi.org/10.1186/1756-3305-6-213
Dantas-Torres F, Latrofa MS, Otranto D (2011) Quantification of Leishmania infantum DNA in females, eggs and larvae of Rhipicephalus sanguineus. Parasites Vectors 4:56
Farid HA (1996) Morphological keys for the separation of the Rhipicephalus sanguineus group of ticks (Acarina: Ixodidae) in Egypt. J Egypt Soc Parasitol 26:453–460
Gray J, Dantas-Torres F, Estrada-Peña A, Levin M (2013) Systematics and ecology of the brown dog tick, Rhipicephalus sanguineus. Ticks Tick Borne Dis 4:171–180
Guglielmone AA, Estrada-Peña A, Keirans JE, Robbins RG (2003) Ticks (Acari: Ixodidae) of the Neotropical zoogeographic region. In: special publication international cons. Ticks Tick-Borne Dis-2. Atalanta, Hauten, p 173
Inokuma H, Tamura K, Onishi T (1996) Seasonal occurrence of Rhipicephalus sanguineus in Okayama prefecture, Japan and effect of temperature on development of the tick. J Vet Med Sci 58:225–228
Ioffe-Uspensky I, Mumcuoglu KY, Uspensky I, Galun R (1997) Rhipicephalus sanguineus and R. turanicus (Acari: Ixodidae): closely related species with different biological characteristics. J Med Entomol 34(1):74–81
Jones EO, Gruntmeir JM, Hamer SA, Little SE (2017) Temperate and tropical lineages of brown dog ticks in North America. Vet Parasitol Reg Stud Rep 7:58–61
Laatamna A, Oswald B, Chitimia-Dobler L, Bakkes DK (2020) Mitochondrial 16S rRNA gene analysis reveals occurrence of Rhipicephalus sanguineus sensu stricto from steppe and high plateaus regions Algeria. Parasitol Res 1199(7):2085–2091. https://doi.org/10.1007/s00436-020-06725-0
Kamani J, Baneth G, Mumcuoglu KY, Waziri NE, Eyal O, Guthmann Y, Harrus S (2013) Molecular detection and characterization of tick-borne pathogens in dogs and ticks from Nigeria. PLoS Negl Trop Dis 7(3):e2108. https://doi.org/10.1371/journal.pntd.0002108
Mangold AJ, Bargues MD, Mas-Coma S (1998) Mitochondrial 16S rDNA sequences and phylogenetic relationships of species of Rhipicephalus and other tick genera among Metastriata (Acari: Ixodidae). Parasitol Res 84:478–484
Moraes-Filho J, Krawczak FS, Costa FB, Soares JF, Labruna MB (2015) Comparative evaluation of the vector competence of four South American populations of the Rhipicephalus sanguineus group for the bacterium Ehrlichia canis, the agent of canine monocytic ehrlichiosis. PLoS ONE 10:e0139386
Moraes-Filho J, Marcili A, Nieri-Bastos FA, Richtzenhain LJ, Labruna MB (2011) Genetic analysis of ticks belonging to the Rhipicephalus sanguineus group in Latin America. Acta Trop 117:51–55. https://doi.org/10.1016/j.actatropica
Nava S, Beati L, Venzal JM, Labruna MB, Szabó MP, Petney T, Saracho-Bottero MN, Tarragona EL, Dantas-Torres F, Silva MMS, Mangold AJ, Guglielmone AA, Estrada-Peña A (2018) Rhipicephalus sanguineus (Latreille, 1806): neotype designation, morphological re-description of all parasitic stages and molecular characterization. Ticks Tick Borne Dis 9(6):1573–1585
Nava S, Mastropaolo M, Venzal JM, Mangold AJ, Guglielmone AA (2012) Mitochondrial DNA analysis of Rhipicephalus sanguineus sensu lato (Acari: Ixodidae) in the Southern cone of South America. Vet Parasitol 190:547–555. https://doi.org/10.1016/j.vetpar.2012.06.032
Ogungbenro SB, Morakinyo TE (2014) Rainfall distribution and change detection across climatic zones in Nigeria. Weather Clim Extrem 5–6:1–6
Oguntunde PG, Abiodun BJ, Lischeid G (2011) Rainfall trends in Nigeria, 1901–2000. J Hydrol 411:207–218
Okoli IC, Okoli CG, Opara M (2006) Environmental and multi-host infestation of the brown dog tick, Rhipicephalus sanguineus in Owerri, South-East Nigeria—a case report. Vet Arhiv 76:93–100
Oliveira PR, Bechara GH, Denardi SE, Saito KC, Nunes ET, Szabo´ MP, Mathias MI (2005) Comparison of the external morphology of Rhipicephalus sanguineus (Latreille, 1806) (Acari: Ixodidae) ticks from Brazil and Argentina. Vet Parasitol 129:139–147
Sanches GS, Evora PM, Mangold AJ, Jittapalapong S, Rodriguez-Mallon A, Guzman PE, Bechara GH, Carmarga-Mathias MI (2016) Molecular, biological, and morphometric comparisons between different geographic populations of Rhipicephalus sanguineus sensu lato. Vet Parasitol 215:78–87
Walker JB, Keirans JE, Horak IG (2000) The genus Rhipicephalus (Acari: Ixodidae): a guide to the brown ticks of the world. Cambridge University Press, Cambridge, p 643
Zemtsova GE, Apanaskevich DA, Reeves WK, Hahn M, Snellgrove A, Levin ML (2016) Phylogeography of Rhipicephalus sanguineus sensu lato and its relationships with climatic factors. Exp Appl Acarol 69:191–203. doi https://doi.org/10.1007/s10493-016-0035-4
Acknowledgements
The assistance of veterinarians and field officers in the various NVRI Outstation laboratories where sampling was conducted is highly appreciated. I am equally grateful to the staff of the Parasitology Divisions NVRI, Vom, for their technical assistance during the course of this work, and to J. González-Miguel of the Laboratory of Parasitology, Institute of Natural Resources and Agrobiology of Salamanca (IRNASA-CSIC), Salamanca, Spain, for designing figure 1.
Funding
This work did not receive any funds from government, private of public sources.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Ethical approval
Permission for this study was granted by the Animal Use and Care Committee (AUCC), National Veterinary Research Institute (NVRI) Vom, Nigeria. Oral consent was obtained from dog owners before tick collection.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kamani, J. Molecular identification and genetic analysis of Rhipicephalus sanguineus sensu lato of dogs in Nigeria, West Africa. Exp Appl Acarol 85, 277–289 (2021). https://doi.org/10.1007/s10493-021-00664-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10493-021-00664-w