Abstract
The genetic structure of populations of the tick Amblyomma ovale from five distinct areas of the Brazilian Atlantic rainforest was evaluated via DNA sequencing and associated with the presence of domestic dogs acting as hosts at the edge of forest fragments. Ticks were collected from domestic dogs and from the environment between 2015 and 2017. Four collection areas were located in the surroundings and within the Serra do Mar State Park, São Paulo State (23°37′21"S, 45°24′43"W), where dogs were bimonthly monitored along 2 years using camera traps and GSM trackers. To determine the spatial limits of genetic structure, ticks collected upon dogs living near the Serra do Baturié, Ceará State (4°15′40"S, 38°55′54"W) were included as well. A total of 39 haplotypes of 16S rRNA and Cox 1 mitochondrial genes sequences were observed, with 27 of them coming from areas within the Serra do Mar State Park. No haplotype was shared between the Serra do Mar and the Serra do Baturié indicating isolation of tick populations at the scale of 2000 km. Although three different haplotype lineages of A. ovale occurred within the Serra do Mar State Park, no genetic structure was found across the study sites within this park, suggesting high tick gene flow across a range of 45 km. Monitoring data from domestic dogs and wild carnivores showed that these species share the same habitats at the forest edge, with dogs playing a likely limited role in tick dispersal. Our findings have important implications for understanding the genetic structure of wide spread A. ovale along Brazilian rainforest remnants, which can further be associated to tick-borne infectious agents, such as Rickettsia parkeri, and used for predicting future patterns of tick diversity in the Brazilian Atlantic rainforest.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
By definition, a host represents most of the ecological requirements for a parasite. This dependency often demands specific adaptations that can lead to population divergence and even speciation when parasites exploit different host types (De Meeus 2012; McCoy et al. 2013; Dupraz et al. 2016). The recent application of molecular techniques to parasitic organisms has provided insightful information on their biology and ecology, which has been useful to measure parasite population structuring and to approach the factors associated with divergence, such as spatial isolation and host type. However, understanding the relative importance of these factors in structuring populations can be particularly challenging when parasites have broad distributions and use several host species to complete the life cycle (Nadolny et al. 2015; Dupraz et al. 2016; Mazé-Guilmo et al. 2016). This is frequently the case for ticks, where two or more phylogenetically distant hosts may be used for blood feeding in immature and adult life stages. For example, hard ticks (Ixodidae family) often need a minimum of two distinct host species for complete development: commonly one host type for larval and nymphal stages and a different one for the adult phase (Ogrzewalska et al. 2012; Krawczak et al. 2016a; Martins et al. 2016). Still, specificity for a particular host can vary. Moreover, there is frequently a range of different potential host species that can be exploited during each developmental stage. This complex life cycle provides these ectoparasites the chance to maintain populations across different ecological conditions and to disperse at different spatial scales (Ogrzewalska et al. 2012; Krawczak et al. 2016b; Mazé-Guilmo et al. 2016).
The phylogenetic relationships within the Amblyomma genus has been widely discussed and revised over the last decades, especially the branch which includes Amblyomma ovale (Aragão and Fonseca 1961; Murgas et al. 2013). While the geographic distribution of this tick extends across the Neotropical region from central-northern Argentina into the Nearctic region of Mexico, with a few records from the United States and Caribbean (Guglielmone et al. 2003), the knowledge on its life cycle has still some significant gaps, mainly in relation to host use (Martins et al. 2012; Krawczak et al. 2016b). Although small rodents are considered to play an important role for larvae and nymphs of A. ovale under natural conditions (Szabó et al. 2013), wild birds have been reported to be parasitized in some regions of Brazil (Ogrzewalska et al. 2009; Luz and Faccini 2013; Ramos et al. 2015). In addition, Gallus gallus was found to be an adequate host for immature stages under laboratory conditions (Martins et al. 2012). These results suggest that birds may play a role in maintaining immature of this tick in natural populations and, as a consequence, may also disperse specimens across geographic locations (Ogrzewalska et al. 2009; Luz and Faccini 2013). In turn, adults of A. ovale are considered to prefer wild carnivores but may frequently use dogs, especially if they are in contact with forest habitats (Ogrzewalska et al. 2009; Sabatini et al. 2010; Martins et al. 2012; Szabó et al. 2013; Maturano et al. 2015).
The Brazilian Atlantic rainforest is considered a hotspot of biodiversity, with one of the world’s most rich environments in terms of species (Ribeiro et al. 2009). It is also one of the most threatened biomes on the planet. Its original range stretched along the eastern Brazilian coast and covered 131,546,000 ha (Verdade and Campos 2004; Frigeri et al. 2014). In contrast, if taking into account forest fragments above 100 ha in size, only 8.5% of its original surface remains currently (Castilho et al. 2012). If forest patches above 3 ha are considered then this proportion increases to 12.5% (Schnell et al. 2013). Seventy percent of the Brazilian population currently lives among the remaining forest patches of this biome (IBGE 2017). To help conserving the remaining native areas, several protected areas were created. One of these is the Serra do Mar State Park (IUCN category II), located on the seacoast of São Paulo State, with 332,000 ha spanning across and around 25 cities (Gigliotti and Santos 2013). Currently, this is the largest continuous remaining fragment of the Brazilian Atlantic rainforest. Given the high level of deforestation and perturbation of this natural rainforest ecosystem, the capacity of forest patches to sustain large mammals such as wild carnivore populations is limited, and access of domestic animals (i. e. dogs) into the forest area has notably increased (Frigeri et al. 2014). As consequence, contact with parasites typically associated with wildlife is expected (Sabatini et al. 2010; Frigeri et al. 2014). Indeed, over the last decade, a Rickettsia parkeri strain Atlantic rainforest-causing rickettsiosis has emerged in eastern Brazil, where it is primarily transmitted by A. ovale ticks (Szabó et al. 2013; Krawczak et al. 2016a; Krawczak and Labruna 2018). This emergence could be directly linked to an increased human exposure to ticks brought via domestic dogs entering Atlantic rainforest patches, one of the natural habitats of A. ovale (Labruna et al. 2007; Spolidorio et al. 2010; Szabó et al. 2013). The use of different alternative hosts along the forest edge may also result in a divergence between wild and domestic parasite populations, leading to associated changes in parasite ecology and, in the case of ticks, in the frequency and circulation of associated pathogens.
To investigate gene flow between wild host populations and dogs within forest patches and among the forest edge, between 2015 and 2017 we collected specimens of A. ovale upon dogs and directly from the environment in five remnants of Atlantic rainforest. Four of these areas were located on the boundaries of the Serra do Mar State Park, where dogs were monitored bimonthly through camera traps and GPS/GSM trackers. A fifth population was used to estimate the degree of long-range gene flow and included the analysis of ticks collected upon dogs living in the edge of an Atlantic rain forest fragment in Guaramiranga municipality, another preserved area of Atlantic rainforest located at northern limit of the biome, near 2000 km away from the Serra do Mar State Park.
Methods
Study area
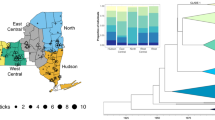
The study comprised four areas (A1–A4) within a section of the Serra do Mar State Park in the municipality of Caraguatatuba, along the northern coast of São Paulo State, in southeastern Brazil (Fig. 1). This municipality is 48,510 ha wide, with a population of 116,786 inhabitants living between the coast and the forest (IBGE 2017). Social and economic inequality is significant, and the use of the forest by the local population reflects this situation. As an external location to the Serra do Mar State Park, we included ticks collected in Serra do Baturié (A5), located in a Brazilian Atlantic rainforest fragment in Ceará State, northeast Brazil (Fig. 2). A straight line between Areas 1–4 and Area 5 is 2325 km long.
Geographic distribution of Amblyomma ovale haplotypes (H1–H39) and the haplotype network indicating the mutational steps among the different haplotype lineages. Different colors refer to different haplotype lineages and the relative size of the pie chart is proportional to the frequency of each lineage within the site
Within the Serra do Mar State Park, samples were obtained from four different areas (Fig. 1), three (A1–A3) at the park edge, where people and dogs live in close contact with the forest, and one completely within the park (A4). A1 was emplaced within a poor community characterized by subsistence plantations and hunting, 15 km away from A2. This last area was located into a low-income community as well yet without economic exploitation of the forest and separated from A3 by 15 km. A3 was represented by a middle-income residential zone, where inhabitants use the forest for leisure activities. Finally, A4 was located inside the limits of the park, in an area where dogs supposedly had no access, and the human dwellers correspond exclusively to the Park staff; this area was 3 km away from the following area (A2).
The areas where ticks were collected directly from vegetation were selected according to the following criteria: topographic gradient (from sedimentary plains to low and high slope mountain), altitude below 300 m, and accessible trails. The presence of dogs with access to forest edges was also an inclusion criterion for the selected areas.
Dog sampling and collection of free-living ticks
We sampled a cohort of 20 dogs, including individuals older than one year only, and born and raised in the region as described elsewhere (Pinter et al. 2008). In each of areas A1–A3, 20 dogs were sampled every two months, comprising a total of 60 dogs sampled on six occasions between June 2015 and April 2016. During each visit, dogs were examined for the presence of ticks.
Free-living ticks were collected in A4, inside the Serra do Mar State Park, where dogs supposedly had no access. To reach this area, a 3-km trail was walked in the direction of the forest core in April 2017. The ticks found on the vegetation were collected along the trail using drag sampling (Arzua and Brescovit 2006).
All adult ticks collected were preserved in 70% ethanol and taken to the Laboratory of Parasitic Diseases of the School of Veterinary Medicine and Animal Science of the University of São Paulo, where they were morphologically identified to species level following Barros-Battesti et al. (2006). Collecting procedures were previously approved by the “Instituto Chico Mendes” (ICMBio-SISBio permit 42793), “Companhia Ambiental do Estado de São Paulo” (CETESB – SP) and by the Ethics Committee of the School of Veterinary Medicine of the University de São Paulo (certificate number 6441270114).
Telemetry data
Six Global System for Mobile Communication (GSM) GPS-TK102B (CU-TEK®) trackers were used to track dog movements, two in each of areas A1–A3. The trackers were placed on the collars of dogs allowed to move freely and that were thought to use the forest (unrestricted dogs). Each device remained attached to the dog for a 1-week period, after which the devices were installed on other selected dogs from the same area. The devices were monitored weekly from October to December 2015. During the monitoring period, the devices were programmed to send a SMS message with the geographic coordinates every 30 min, from 6 pm to 8 am. This time was selected because this was the period when dogs were unrestricted and could possibly use the forest.
Camera trap records
From December 2015 to August 2016, six HD Auto-Focus Nature View camera traps (BUSHNELL®) were installed along Serra do Mar State Park trails used by GPS-tracked dogs and used to record animal usage along these paths. Two cameras were installed in each of areas A1–A3, one on the forest edge and other at least 300 m further into the forest. Every 25 days, batteries were changed and videos and photographs were downloaded. In addition, photographic record obtained by two camera traps installed in A2 was provided by other researchers working in the same area and period (unpublished data by Mora et al., 2017).
Analyses
DNA isolation and PCR amplification
Genomic DNA of ticks was isolated using the DNeasy Tissue kit (Qiagen, Valencia, CA, USA) following manufacturer’s instructions. Each tick was processed individually and two regions of mitochondrial DNA (16S rRNA and cox1 genes) were amplified by PCR (see AI in Supplementary materials for details) (Black and Piesman 1994; Simon et al. 1994). The amplified products were sequenced in both directions (forward and reverse) using the Sanger method (Eurofins Genomics, Courtaboeuf, France).
Genetic analyses
To ensure sequence quality and confirm species identifications, we manually verified the sequence chromatographs and checked sequence similarity to other sequences deposited in GenBank using BLASTN (Basic Local Alignment Search Tool-Nucleotide). The sequences of both genes were then aligned, edited manually using MEGA software (v.6.0.5), and concatenated for each individual (Tamura et al. 2013). Two files containing the alignments for each gene fragment were also created. For the three files, haplotypes were defined using DNAsp v.5.10.01 (Librado and Rozas 2009). Then, we performed the genetic analyses on each file.
A median-joining algorithm (MJ), based on parsimony criteria (Bandelt et al. 1999) using Network v.5.0.0.0 (fluxusengineering.com) was employed to visualize the frequency and spatial distribution of haplotypes. Genetic diversity (haplotype and nucleotide) within each A. ovale population was estimated using ARLEQUIN software v.3.5.1.2. (Excoffier et al. 1992). To assess the genetic neutrality of gene sequences, we used Tajima’s D and Fu’s Fs (Tajima 1989; Fu 1997), with 1000 random permutations, as implemented in ARLEQUIN.
The partition of genetic variance among sampled sites was estimated using an Analysis of Molecular Variance (AMOVA) with ARLEQUIN (Excoffier et al. 1992). To determine how genetic diversity was structured without a geographic constraint a priori, we also performed a Spatial Analysis of Molecular Variance (SAMOVA). This analysis identifies the spatial scale of a genetically homogeneous population, estimating the number of K clusters based on the maximized intergroup genetic variance–Fct index (Dupanloup et al. 2002).
To evaluate genetic divergences between A. ovale populations from Ceará and São Paulo States, two unrooted Neighbor-Joining trees were constructed for 16S rRNA and cox1 genes, and percentages of genetic identities were calculated upon the resulting alignments using Geneious R9 (Kearse et al. 2012).
Results
Tick sampling
Nineteen, 23 and 18 dogs were monitored in A1, A2, and A3, respectively. A total of 190 A. ovale specimens were collected directly from these animals: 155 in A1, 18 in A2, and 17 in A3. All ticks from A2 and A3, and a subset of 20 adult ticks from A1, were submitted to molecular analyses. A total of 13 adult A. ovale were collected from the environment in A4 and were also used in molecular analyses. In A5, a total of 32 individuals of A. ovale were collected from dogs living in the surroundings of Serra do Baturié in Ceará.
Telemetry data
A total of 12 dogs were monitored, four per area (A1–A3). Of these, two dogs were recorded inside the forest (in A1 and A3) (Fig. 1). The other two dogs seemed to roam only around the urban areas. At times, when the dogs were in the GSM shaded areas, it was not possible to register their position. Telemetry devices were previously tested on edge and core areas of the forest. In both areas the devices failed to transmit data in determined points (probably by the density of vegetation), therefore avoiding an accurate calculation of overall use of forest areas by tagged dogs.
Photographic records
During the monitoring period, camera traps recorded 2855 photographs and 156 videos. Thirty-eight of these files registered medium or large mammals (≥ 1 kg); 22 recorded the following species of carnivores: Eira barbara, Nasua nasua, Leopardus pardalis, Leopardus sp., Puma concolor and Canis lupus familiaris. Cameras installed at the forest edge (cameras E1 and E3, Fig. 1) recorded dogs in all studied areas. In A1, records of dogs were also obtained by the camera installed into the forest (camera I1, Fig. 1). People were also recorded walking in the forest alongside their dogs in A1 and A3, particularly in the morning for A1 (AM acronym, Fig. 1) and during the weekends or holidays for A3. Leopardus pardalis, Leopardus sp., and E. barbara were recorded by the camera installed at the forest edge (camera E3, Fig. 1) only 100 m away from houses, where dogs were also recorded, demonstrating an overlap of ranges in different periods: wild carnivores at night and dogs during the day. Puma concolor was also recorded in A1 and A2 by cameras installed inside the forest (cameras I1 and I2, Fig. 1).
Genetic analyses
A total of 97 adult A. ovale ticks were submitted to molecular analyses, from which we obtained 87 partial sequences of the 16S rRNA gene (275 bp), 78 of the cox1 gene (436 bp), and concatenated arrangements (711 bp) for 70 ticks (Table 1). The cox1 gene fragment yielded higher polymorphism than the 16S rRNA gene, with 54 and 22 variable sites, respectively (Table 2). Haplotype and nucleotide diversity were high within all tick populations (Table 3).
A total of 39 unique haplotypes were observed when both mitochondrial gene sequences were combined. Of these, 27 were observed in samples from A1–A4. None of the 12 observed haplotypes from A5 were shared with the Caraguatatuba samples (A1–A4). However, haplotype sharing was observed among the Caraguatatuba populations (A1–A4).
The Network showed the presence of four well-supported haplotype lineages, where the lineage from A5 was completely distinct from those of A1–A4 (Fig. 2). The lineages from A1–A4 were separated from the A5 lineage by more than 40 mutational steps (Fig. 2). The spatial distribution of haplotypes from the three lineages in A1–A4 was completely mixed (Fig. 2).
No deviation from neutrality was found at the two genes examined for either Tajima’s D or Fu’s F test (p > 0.05) suggesting no evidence of selection on the genes or recent population expansion (Table 3).
In agreement with network, AMOVA results showed that within Caraguatatuba, only 0.17% of the variation could be attributed to the interpopulation level; no barrier to gene flow seems to occur among these populations (Table 4). When A5 is included this value rises to 85.95% of the molecular variation.
The spatial analysis (SAMOVA) also indicated strong geographic isolation between Caraguatatuba (A1–A4) and Ceará (A5) samples (FCT = 0.87335), coinciding with the AMOVA results. Again, no significant differences were found among A1–A4 suggesting no small scale geographic structure.
Nucleotide sequence identity for both 16S rRNA and cox1 haplotypes observed in Serra do Baturié (Ceará) population (A5) ranged between 95.6–100%. Identities for the same genes within Serra do Mar State Park populations (A1–A4) varied identically. Remarkably, comparisons between Serra do Baturié and Serra do Mar State Park populations showed nucleotide identities lower than 95.6%, reaching 92.0% in some cases (see Fig. 3 A2 Supplementary materials for details.).
Discussion
In Brazil, A. ovale has a unique life history. This species is one of the most widely distributed ticks parasitizing carnivores in their adult stages and associated to rodents and other small warm-blooded vertebrates in their immature stages. However, in the state of São Paulo (southeastern Brazil), A. ovale is not found above 300 m of altitude (Barbieri et al. 2015). These characters make this tick a parasite highly dependent on their hosts for interpopulational dispersal and consequent gene flow in subtropical areas. In this study, we found that A. ovale has high intra-population genetic diversity and high gene flow at a scale of tens of kilometres (within Caraguatatuba City), but can show strong structure at higher spatial scales (Ceará State to Sao Paulo State).
Evolutionary distances measured among haplotypes within populations were small, suggesting that tick diversification has (relatively) occurred locally with some potential mixing of different populations at some point in time. Additionally, population densities are high and stable enough to maintain this diversity over time. This type of population stability is also supported by the neutrality tests, which do not suggest recent extreme shifts in population demography. Moreover, comparisons of nucleotide identities between Serra do Baturié and Serra do Mar State Park A. ovale (< 95.0%) suggest that this species could be split into two putative sibling species.
Monitoring data from the dogs (telemetry and camera trap records) showed that these domestic animals only rarely enter the inner parts of large forest fragments, in opposition to what has been previously reported by other authors (Butler and Bingham 2000; Galetti and Sazima 2006). Despite a low sample size of dog tracks, this data and camera traps suggest that these animals are mainly restricted to forest edges and go deeply into the forest only when accompanied by their owners. In contrast, camera trap records clearly demonstrated the presence of wild carnivores in anthropized areas, including those areas where dogs were recorded. Thus, A. ovale ticks are likely brought to the forest edge by wild carnivores and rodents, and then use dogs as an alternative host, without the establishment of distinct populations exploiting exclusively domestic hosts. Indeed, this hypothesis is supported by a lack of genetic structure between areas A1–A3 (where only dogs were sampled) and A4 (where ticks were sampled from the vegetation). The SAMOVA did not detected evidence of genetic structuring within populations, a fact that is not unexpected if distinct host-associated populations exist in sympatry. This hypothesis is also the most parsimonious explanation of high genetic difference between Caraguatatuba and Ceará populations. Thus, it is likely that wild animals with limited dispersal distances due to high forest fragmentation are responsible for tick dispersal.
In contrast to what has been suggested for several Amblyomma species inhabiting Atlantic rainforest ecosystems (Ogrzewalska et al. 2011, 2012, 2015), the complete isolation of tick populations from Serra do Mar State Park and Serra do Baturié suggest that birds have an irrelevant role in the spread of larvae and nymphs of A. ovale. However, this assumption should be confirmed in a larger scale study, with a higher number of sites and complementary molecular techniques.
Due to its steep and relatively narrow coastal plain, Caraguatatuba was forgotten during the Brazilian economic cycles, and for that reason, some Atlantic rainforest fragments remained well preserved (Mouret 2017). As a consequence, Serra do Mar State Park is currently one of the largest ecological reserves for native Atlantic fauna in Brazil. In addition to this fact, high vagility of wild carnivores may explain haplotype diversity (and expected high effective population size) and high gene flow of A. ovale in the region. However, more subtle genetic differences among populations within the Serra do Mar State Park cannot be completely discarded until an analysis using polymorphic genetic markers such as single-nucleotide polymorphism (SNP) or microsatellites are applied. A diversity of factors may change across these areas that could alter the genetic composition of tick populations, including the presence and abundance of different host species. The effect of these factors on the genetic structure of A. ovale and its evolutionary trajectory will depend on the flexibility of the tick to successfully use different environments and host species (De Meeus 2012; McCoy et al. 2013; Dupraz et al. 2016). What is clear from our work is that forest fragmentation can play a major role in isolating tick populations and may have direct implications for the circulation of tick-borne disease agents, such as R. parkeri. Future work on this tick using more accurate genetic markers is now required for determining finer scale structure among contiguous forest regions, the relation to host and habitat type, and whether the extreme isolation observed at a larger spatial scale (> 2000 km) corresponds to a general pattern of this tick species when sampling isolated forest fragments. This data will become utterly important if we are to understand the epidemiology of tick-borne pathogens and the evolutionary trajectory of tick diversity.
References
Aragão HB, Fonseca F (1961) Notas de Ixodologia: IX. O complexo ovale do gênero Amblyomma. Memórias do Instituto Oswaldo Cruz 59:131–148. https://doi.org/10.1590/S0074-02761961000200002
Arzua M, Brescovit AD (2006) Métodos de coleta e preservação para identificação. In: Barros-Battesti DM, Arzua M, Bechara GH (eds) Carrapatos de importância médico-veterinária da região neotropical: um guia ilustrado para identificação de espécies, 1st edn. Vox/ICTTD-3/Butantan, São Paulo, pp 183–189
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48. https://doi.org/10.1093/oxfordjournals.molbev.a026036
Barbieri JM, Da Rocha CMBM, Bruhn FRP, Cardoso DL, Pinter A, Labruna MB (2015) Altitudinal assessment of Amblyomma aureolatum and Amblyomma ovale (Acari: Ixodidae), vectors of spotted fever group rickettsiosis in the State of São Paulo, Brazil. J Med Entomol 52:1170–1174. https://doi.org/10.1093/jme/tjv073
Barros-Battesti DM, Arzua M, Bechara. GH (2006) Carrapatos de importância médico veterinária da região neotropical: um guia ilustrado para identificação de espécies, 1st edn. São Paulo
Black WC, Piesman J (1994) Phylogeny of hard- and soft-tick taxa (Acari: Ixodida) based on mitochondrial 16S rDNA sequences. Proc Natl Acad Sci USA 91:10034–10038
Butler JR, Bingham J (2000) Demography and dog-human relationships of the dog population in Zimbabwean communal lands. Vet Rec 147:442–446. https://doi.org/10.1136/VR.147.16.442
Castilho CS, Marins-Sá LG, Benedet RC, Freitas TRO (2012) Genetic structure and conservation of Mountain Lions in the South-Brazilian Atlantic Rain Forest. Genet Mol Biol 35:65–73
De Meeus T (2012) Initiation à la génétique des populations naturelles applications aux parasites et à leurs vecteurs, IRD edn. Marseille
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581. https://doi.org/10.1046/j.1365-294X.2002.01650.x
Dupraz M, Toty C, Noël V, Estrada-Peňa A, González-Solís J, Boulinier T, Dujardin JP, McCoy KD (2016) Linking morphometric and genetic divergence with host use in the tick complex, Ornithodoros capensis sensu lato. Infect Genet Evol 46:12–22. https://doi.org/10.1016/j.meegid.2016.10.005
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Frigeri E, Cassano CR, Pardini R (2014) Domestic dog invasion in an agroforestry mosaic in southern Bahia, Brazil. Trop Conserv Sci 7:508–528. https://doi.org/10.1177/194008291400700310
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Galetti M, Sazima I (2006) Impact of feral dogs in an urban Atlantic forest fragment in southeastern Brazil. Natureza Conservação 4:146–151
Gigliotti C, Santos MJ (2013) A Expansão Urbana de Caraguatatuba (1950–2010): uma Análise das Transformações Sócio Espaciais. Caminhos de Geografia 14. http://www.seer.ufu.br/index.php/caminhosdegeografia/article/view/17794. Accessed 10 July 2017
Guglielmone AA, Estrada-Peña A, Keirans JE, Robbins RG (2003) Ticks (Acari: Ixodida) of the neotropical zoogeographic region. Atlanta Houten, International Consortium on Ticks and Tick-borne Diseases, p 173
IBGE (2017) IBGE | Brasil em Síntese | São Paulo | Caraguatatuba | Panorama [WWW Document]. https://cidades.ibge.gov.br/brasil/sp/caraguatatuba/panorama. Accessed 10 July 17
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Krawczak SF, Labruna MB (2018) The rice rat Euryoryzomys russatus, a competent amplifying host of Rickettsia parkeri strain Atlantic rainforest for the tick Amblyomma ovale. Ticks Tick Borne Dis 9:1133–1136. https://doi.org/10.1016/j.ttbdis.2018.04.013
Krawczak FS, Agostinho WC, Polo G, Moraes-Filho J, Labruna MB (2016a) Comparative evaluation of Amblyomma ovale ticks infected and noninfected by Rickettsia sp. strain Atlantic rainforest, the agent of an emerging rickettsiosis in Brazil. Ticks Tick Borne Dis 7:502–507. https://doi.org/10.1016/j.ttbdis.2016.02.007
Krawczak FS, Munoz-Leal S, Guztzazky AC, Oliveira SV, Santos FCP, Angerami RN, Moraes-Filho J, de Souza JC, Labruna MB (2016b) Rickettsia sp. strain atlantic rainforest infection in a patient from a spotted fever-endemic area in southern Brazil. Am J Trop Med Hyg 95:551–553. https://doi.org/10.4269/ajtmh.16-0192
Labruna MB, Sanfilippo LF, Demetrio C, Menezes AC, Pinter A, Guglielmone AA, Silveira LF (2007) Ticks collected on birds in the state of Sao Paulo, Brazil. Exp Appl Acarol 43:147–160
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. https://doi.org/10.1093/bioinformatics/btp187
Luz HR, Faccini JLH (2013) Ticks on Brazilian birds: overview. In: Birds—evolution and behaviour, breeding strategies, migration and spread of disease, vol 1, pp 97–126. https://doi.org/10.1007/s10493-012-9572-7
Martins TF, Moura MM, Labruna MB (2012) Life-cycle and host preference of Amblyomma ovale (Acari: Ixodidae) under laboratory conditions. Exp Appl Acarol 56:151–158. https://doi.org/10.1007/s10493-011-9506-9
Martins TF, Peres MG, Costa FB, Bacchiega TS, Appolinario CM, Antunes JMA, de P, Vicente, Megid AF, Labruna J, Martins MB, Peres TF, Costa MG, Bacchiega FB, Appolinario TS, Antunes CM, JMA de P, Vicente, Megid AF, Labruna J, M.B (2016) Ticks infesting wild small rodents in three areas of the state of São Paulo, Brazil. Ciência Rural 46:871–875. https://doi.org/10.1590/0103-8478cr20150671
Maturano R, Faccini JH, Daemon E, Fazza PC, Bastos R (2015) Additional information about tick parasitism in Passeriformes birds in an Atlantic Forest in southeastern Brazil. Parasitol Res 114:4181–4193. https://doi.org/10.1007/s00436-015-4651-4
Mazé-Guilmo E, Blanchet S, Mccoy KD, Loot G (2016) Host dispersal as the driver of parasite genetic structure: a paradigm lost? Ecol Lett 19:336–347. https://doi.org/10.1111/ele.12564
McCoy KD, Léger E, Dietrich M (2013) Host specialization in ticks and transmission of tick-borne diseases: a review. Front Cell Infect Microbiol 3:1–12. https://doi.org/10.3389/fcimb.2013.00057
Mouret S (2017) Revolução Industrial no Brasil—Início, incentivo e consequências. https://www.estudopratico.com.br/revolucao-industrial-no-brasil/. Accessed 10 July 2017
Murgas IL, Castro AM, Bermúdez SE (2013) Current status of Amblyomma ovale (Acari: Ixodidae) in Panama. Ticks Tick Borne Dis 4:164–166. https://doi.org/10.1016/j.ttbdis.2012.09.002
Nadolny R, Gaff H, Carlsson J, Gauthier D (2015) Comparative population genetics of two invading ticks: evidence of the ecological mechanisms underlying tick range expansions. Infect Genet Evol 35:153–162. https://doi.org/10.1016/j.meegid.2015.08.009
Ogrzewalska M, Pacheco RC, Uezu A, Richtzenhain LJ, Ferreira F, Labruna MB (2009) Ticks (Acari: Ixodidae) infesting birds in an Atlantic rain forest region of Brazil. J Med Entomol 46:1225–1229. https://doi.org/10.1603/033.046.0534
Ogrzewalska M, Uezu A, Jenkins CN, Labruna MB (2011) Effect of forest fragmentation on tick infestations of birds and tick infection rates by Rickettsia in the Atlantic Forest of Brazil. EcoHealth 8:320–331. https://doi.org/10.1007/s10393-011-0726-6
Ogrzewalska M, Saraiva DG, Moraes-Filho J, Martins TF, Costa FB, Pinter A, Labruna MB (2012) Epidemiology of Brazilian spotted fever in the Atlantic Forest, state of São Paulo, Brazil. Parasitology 139:1283–1300. https://doi.org/10.1017/S0031182012000546
Ogrzewalska M, Literák I, Capek M, Sychra O, Calderón V, Rodríguez BC, Prudencio C, Martins TF, Labruna MB (2015) Bacteria of the genus Rickettsia in ticks (Acari: Ixodidae) collected from birds in Costa Rica. Ticks Tick Borne Dis 6:478–482. https://doi.org/10.1016/j.ttbdis.2015.03.016
Pinter A, Horta MC, Pacheco RC, Moraes-Filho J, Labruna MB (2008) Serosurvey of Rickettsia spp. in dogs and humans from an endemic area for Brazilian spotted fever in the State of São Paulo, Brazil. Cadernos de Saúde Pública 24: 247–252. https://doi.org/10.1590/S0102-311X2008000200003
Ramos DG, Melo AL, Martins TF, Alves AS, Pacheco TA, Pinto LB, Pinho JB, Labruna MB, Dutra V, Aguiar DM, Pacheco RC (2015) Rickettsial infection in ticks from wild birds from Cerrado and the Pantanal region of Mato Grosso, midwestern Brazil. Ticks Tick Borne Dis 6:836–842. https://doi.org/10.1016/j.ttbdis.2015.07.013
Ribeiro MC, Metzger JP, Martensen AC, Ponzoni FJ, Hirota MM (2009) The Brazilian Atlantic Forest: how much is left, and how is the remaining forest distributed? Implications for conservation. Biol Conserv 142:1141–1153. https://doi.org/10.1016/j.biocon.2009.02.021
Sabatini GS, Pinter A, Nieri-Bastos FA, Marcili A, Labruna MB (2010) Survey of ticks (Acari: Ixodidae) and their rickettsia in an Atlantic rain forest reserve in the State of São Paulo, Brazil. J Med Entomol 47:913–916. https://doi.org/10.1603/ME10073
Schnell JK, Harris GM, Pimm SL, Russell GJ (2013) Quantitative analysis of forest fragmentation in the atlantic forest reveals more threatened bird species than the current red list. PLoS One 8:e65357. https://doi.org/10.1371/journal.pone.0065357
Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87:651–701. https://doi.org/10.1093/aesa/87.6.651
Spolidorio MG, Labruna MB, Mantovani E, Brandão PE, Richtzenhain LJ, Yoshinari NH (2010) Novel spotted fever group Rickettsiosis, Brazil. Emerg Infect Dis 16:521–523. https://doi.org/10.3201/eid1603.091338
Szabó MP, Nieri-Bastos FA, Spolidorio MG, Martins TF, Barbieri AM, Labruna MB (2013) In vitro isolation from Amblyomma ovale (Acari: Ixodidae) and ecological aspects of the Atlantic rainforest Rickettsia, the causative agent of a novel spotted fever rickettsiosis in Brazil. Parasitology 140: 719–728. https://doi.org/10.1017/S0031182012002065
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Verdade LM, Campos CB (2004) How much is a puma worth? economic compensation as an alternative for the conflict between wildlife conservation and livestock production in Brazil. Biota Neotrop 4:1–4. https://doi.org/10.1590/S1676-06032004000200014
Acknowledgements
We would like to thank Olivier Duron for helpful discussions. This work was not possible without financial support from FAPESP from processes 2014/648-3 and 2016/5355-0 (BEPE). Partial funding for this work came from the ANR Grant ESPEVEC (ANR blanc ANR-13-BSV7-0018-01) to KDM. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior–Brasil (CAPES)–Finance Code 001.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10493_2019_350_MOESM2_ESM.tif
Figure 3 Gray-scale heatmap matrixes calculated upon the alignments of 275 bp partial fragments of 16S rRNA (A) and cox1 (B) genes from Serra do Baturié (A5) and Serra do Mar State Park (A1–A4) populations of A. ovale. In each tree, Serra do Baturié sequences are denoted with orange and Serra do Mar State Park sequences with blue branches. Evolutionary relationships were inferred using Tamura-Nei genetic distance model. Support values obtained after 1000 bootstrap replications are indicated for main branches. Resulting trees were midpoint-rooted and transformed to cladrograms (TIF 12968 KB)
Rights and permissions
About this article
Cite this article
Fournier, G.F.S.R., Pinter, A., Santiago, R. et al. A high gene flow in populations of Amblyomma ovale ticks found in distinct fragments of Brazilian Atlantic rainforest. Exp Appl Acarol 77, 215–228 (2019). https://doi.org/10.1007/s10493-019-00350-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10493-019-00350-y