Abstract
The strain designated NCCP-602T was isolated from tannery effluent, and displayed aerobic, gram-positive, rod-shaped cells that were characterized by oxidase negative, catalase positive, and non-motile features. The most favourable growth conditions were observed at a temperature of 30°C, pH 7.0, and NaCl concentration of 1% (w/v). It tolerated heavy metals at high concentrations of chromium (3600 ppm), copper (3300 ppm), cadmium (3000 ppm), arsenic (1200 ppm) and lead (1500 ppm). The results of phylogenetic analysis, derived from sequences of the 16S rRNA gene, indicated the position of strain NCCP-602T within genus Brevibacterium and showed that it was closely related to Brevibacterium ammoniilyticum JCM 17537T. Strain NCCP-602 T formed a robust branch that was clearly separate from closely related taxa. A comparison of 16S rRNA gene sequence similarity and dDDH values between the closely related type strains and strain NCCP-602T provided additional evidence supporting the classification of strain NCCP-602T as a distinct novel genospecies. The polar lipid profile included diphosphatidylglycerol, glycolipid, phospholipids and amino lipids. MK-7 and MK-8 were found as the respiratory quinones, while anteiso-C15:0, iso-C15:0, iso-C16:0, iso-C17:0, and anteiso-C17:0 were identified as the predominant cellular fatty acids (> 10%). Considering the convergence of phylogenetic, phenotypic, chemotaxonomic, and genotypic traits, it is suggested that strain NCCP-602 T be classified as a distinct species Brevibacterium metallidurans sp. nov. within genus Brevibacterium with type strain NCCP-602T (JCM 18882T = CGMCC1.62055T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breed laid the foundation for the identification of the genus Brevibacterium in 1953, with the inaugural type species designated as Brevibacterium linens. This bacterium was first isolated from surface-ripened cheese and was characterized as a gram-positive microorganism that is nonspore-forming, nonbranching, and possessing short rods, along with a notably high G+C content. The genus name “Brevibacterium” originates from the fusion of the Latin term “breviation” (signifying short) with the Greek term “bakteria” (meaning rod), which captures its unique morphological characteristics. Subsequently, genus Brevibacterium was expanded to include a variety of species with diverse morphological, physiological, and biochemical traits (Breed 1953). In a subsequent revision, the genus description was refined to encompass only those species that demonstrated chemotaxonomic and morphological traits in alignment with the characteristics of type species Brevibacterium linens (Collins et al. 1983). The venerable genus Brevibacterium stands as the exclusive representative of the family Brevibacteriaceae, residing within the order Micrococcales and the distinguished class Actinobacteria (Forquin-Gomez et al. 2014). However, the genus exhibits remarkable diversity in physiological, biochemical, and chemical characteristics, with a total of 37 species reported with valid published names https://lpsn.dsmz.de/genus/brevibacterium. Several novel species within genus Brevibacterium (sensu stricto), including B. spongiae (Huang et al. 2022; Zhang et al. 2023b), B. limosum, B. atlanticum (Pei et al. 2021), and B. rongguiense (Deng et al. 2020; Pei et al. 2020), have been found in recent reports. The members of this genus are frequently identified on human skin and are prominently found in cheese and dairy products (Collins et al. 1983; Roux and Raoult 2009; Wauters et al. 2004). In diverse environments, sporadic occurrences of Brevibacterium strains, including those characterized by elevated salinity levels (Bhadra et al. 2008) and poultry (Pascual and Collins 1999), have been detected. Brevibacterium picturae and Brevibacterium celere were isolated by Heyrman et al. (2004) and Ivanova et al. (2004) during examination of a ruined mural artwork and a brown alga’s deteriorated thallus.
The genus members have generic recognition in biotechnological industry patents, encompassing applications in antibiotic production, wastewater treatment, and the art of making cheese. Moreover, in addition to indigenous Agrobacterium and Corynebacterium strains, specific native Brevibacterium strains found in heavy metal-contaminated soils in Kuwait have demonstrated impressive resilience to both hydrocarbon and heavy metal contamination (Ali et al. 2012).Additionally, it has been demonstrated that certain strains of Brevibacterium linens can grow when provided with n-alkanes as carbon substrates. Leveraging these metabolic capabilities for remediation of crude oil pollution is another evident application. The influence of specific pollutants, such as cadmium sulfate or sodium arsenate, on enhancing this metabolic pathway remains uncertain; however, there is a positive indication of an increased rate of direct crude oil consumption. These findings underscore the potential of certain native Brevibacteriaceae species as valuable resources for bioremediation of complex organic–inorganic pollutants. Moreover, in addition to detoxifying pentachlorophenol, B. casei was also utilized for synthesis of gold and silver nanoparticles (Kalishwaralal et al. 2010; Ng et al. 2010; Verma and Singh 2013). Brevibacterium metallicus (Román-Ponce et al., 2018) and Brevibacillus parabrevis (Wani et al., 2023) exhibited tolerance to high concentrations of heavy metals, showed markable potential for bioremediation by accumulating high concentrations of copper, cadmium and zinc.
In this study, strain NCCP-602T was isolated from tannery effluent to characterize heavy metal tolerant potentially novel bacteria from Pakistan and its functional genomics for potential application in bioremediation. This study evaluated its potential application in bioremediation, detailed genomic functional analysis for heavy metal tolerance, and xenobiotic degradation-encoding genes of strain NCCP-602T, which were found to effectively tolerate high concentrations of heavy metals. Through polyphasic taxonomic analysis, strain NCCP-602T was identified as a novel heavy metal-tolerant species. This study focused on the strain NCCP-602T, which was initially isolated from a sample of tannery effluent. The primary objective was to establish its taxonomic classification by thoroughly examining its physiological, biochemical genotypic, and phylogenetic attributes. The taxonomic position of strain NCCP-602T was unequivocally confirmed through this comprehensive investigation, which confirmed its status as a novel species within genus Brevibacterium; for which the name Brevibacterium metallidurans sp. nov. was proposed.
Materials and Methods
Strain NCCP-602T was isolated from samples collected from tannery effluent in Sambrial, Sialkot, Pakistan. The aerobic recovery of strain NCCP-602T was achieved using a dilution plate method on tryptic soy agar (TSA, Difco), which contained varying concentrations of Pb2+, Cr2+, Cd2+, As2+, and Cu2+. Individual colonies of strain NCCP-602T were subsequently transferred to fresh plates for purification, after which 16S rRNA gene sequencing was used for identification of the purified colonies. For routine cultivation, TSA medium was used to grow the isolate at 30 °C, while for prolonged preservation, the mixture was stored at −80°C in tryptic soy broth (TSB; Difco) supplemented with 70% (v/v) glycerol.
To meet the essential criteria for characterizing novel aerobic bacterial taxa, comprehensive polyphasic characterization experiments were conducted on the isolated strain. The reference strains from closely related taxa, including Brevibacterium ammoniilyticum JCM 17537T, Brevibacterium celere JCM 13521T, Brevibacterium casei JCM 2594T, and B. linens JCM 1327T, were obtained from the Japan Collection of Microorganisms (JCM), Japan. Unless specified otherwise, the characterization experiments were conducted at a temperature of 30°C under the same laboratory conditions along with the reference strains.
Heavy metal tolerance
The ability of newly discovered strain NCCP-602T to endure increased levels of heavy metals (Pb2+, Cu2+, As2+,Cd2+ and Cr2+) was assessed via cultivation on TSA enriched with heavy metals at various concentrations over a period of five-days. In TSA, concentrations of Pb2+, As2+, Cr2+, Cd2+ and Cu2+ were carefully regulated within the range of 300‒3000 ppm by employing specific salts of Pb (NO3)2, CuSO4·5H2O, NaH2AsO4 Cd(NO3) and K2Cr2O7.
Antibiotic resistance
ATB-Vet Strep (BioMerieux, France) was used to investigate antibiotic resistance. Strain NCCP-602T and reference strains were inoculated to ATB-Vet strep according to the manufacturer’s instructions, and then incubated for 48 h at 37°C. After this incubation period, growth was visually assessed (Guérin-Faublée et al. 1993).
Morphological and physiological characterization
The strain growth was assessed on a range of agar media (Difco, US), which included Luria–Bertani (LB) agar, nutrient agar (NA), tryptic soy agar (TSA), and brain heart infusion agar (BHI) (Ventosa et al. 1982)., Colony morphology of strain NCCP-602T was examined on TSA after two days of incubation. For detailed cell morphology, a scanning electron microscope (EVO MA10-W) was used following established procedures using cells grown in TSB for 24 h. After immersion in a 2.5% glutaraldehyde solution for 3 h, the cultured cells were sequentially dehydrated in ethanol at concentrations of 30%, 50%, 70%, 90%, and 100% for 15 min each. Subsequently, the samples were subjected to one-hour drying process in a vacuum dryer. Next, a suitable area was selected, and pieces measuring 1 × 1 cm were cut. Finally, gold was sprayed for optimal observation under a scanning electron microscope (EVO MA10-W) (Abbas et al. 2015a). Gram staining was performed using a commercial Gram staining kit (Solarbio, Beijing) following the manufacturer’s instructions. Growth temperatures were assessed by growing strains on TSA media and incubated at different temperature conditions (4, 20, 25, 30, 37, 45, 50, 55, or 60°C) for 5 days. To investigate the range of pH for cell growth, TSA was modified to various pH values ranging from 4 to 10. Specific buffer systems, such as 0.1 M citric acid/0.1 M sodium citrate (pH 4.0‒5.0), 0.1 M KH2PO4/0.1 M NaOH (pH 6.0–8.0), and 0.1 M Na2CO3/0.1 M NaHCO3 (pH 9–10), were used (Xu et al. 2005) to achieve the desired pH of the media. NaCl tolerance was assess by growing cells on TSA supplemented with different NaCl concentrations in the range of 0‒15% w/v and incubation for a period of 5 days. Motility of the strains was determined using TSA as a semisolid medium (Tambalo et al. 2010). Oxidase activity was analysed by utilizing an oxidase reagent; catalase activity, 3‒10% hydrogen peroxide; degradation of tweezers (1% each of Tweens 20, 40, 60, and 80); and starch degradation (0.1% soluble starch) following conventional procedures as outlined in previous descriptions by Odds (1981). The physiological and biochemical properties of the strains were determined using API 50CH, API 20E, and API 20NE galleries (bioMerieux, France), whereas the API ZYM strip (bioMerieux, France) was utilized to analyse enzyme production API Kits were used following the manufacturer’s instructions. Biolog Universal Growth (BUG) medium was used for cultivation of the strains in Biolog experiments and BIOLOG test was performed using a BIOLOG GEN III microplate system (Inc. Hayward, CA, USA) in accordance with the manufacturer’s recommendations.
Phylogenetic analysis
Following the protocol described previously (Ahmed et al. 2007), 16S rRNA gene of strain NCCP-602T was amplified through PCR amplification using universal forward and reverse primers, namely, 9F (5’-GAGTTTGATCCTGGCTCAG-3’) and 1510R (5’-GGCTACCTTGTTACGA-3’). PCR product was sequenced using 16S rRNA gene-specific universal primers by commercial service of Macrogen (http://dna.macrogen.com/eng), Korea. The obtained sequences were assembled using BioEdit software, and the strain was identified using the EzTaxon server (http://eztaxon-e.ezbiocloud.net). The sequences of 16S rRNA gene of closely related reference strains were obtained via Ez-Taxon server database and aligned through CLUSTAL W following the Tamura–Nei model (Tamura and Nei 1993) for calculating evolutionary distances. MEGA 11 software was used to construct phylogenetic trees via the maximum-parsimony, neighbor-joining, and maximum-likelihood methods in accordance with protocols detailed conducted by Ahmed et al. (2014). The robustness of associations was evaluated through 1000 iterations of the bootstrap technique involving resampling of the tree topology (Kumar et al. 2016).
Genome features
Whole-genome sequencing of strain NCCP-602T was performed using Illumina NovaSeq platform, employing commercial service of Sangon Biotech Co., Ltd., in Shanghai, China. Trimmomatic software (version 0.38) was used to filter out low-quality data and remove barcodes from the original reads (Bolger et al. 2014). The assembly of clean data was carried out using SPAdes software (version 3.11.1) (Bankevich et al. 2012). Contigs with lengths exceeding 500 bp were chosen for the prediction of genes via Prodigal software (version 2.6.3) (Besemer et al. 2001). We utilized the COG classifier (version 1.0.5, accessible at https://github.com/moshi4/COGclassifier) to annotate the protein-coding sequences of strain NCCP-602T alongside closely related type strains. The genomes of NCCP-602T were examined for resistance and virulence factors by searching the databases of the Pathosystems Resource Integration Center (PATRIC) and the Comprehensive Antibiotic Resistance Database (CARD) available at their respective websites: https://www.patricbrc.org and https://card.mcmaster.ca. Additionally, the annotation of enzymes related to carbohydrate metabolism and carbon metabolism was conducted using the HMM search tool (version 10) (Johnson et al. 2010) from the dbCAN2 database (Zhang et al. 2018). Using CheckM, the completeness and contamination of the genome assembly were assessed (Parks et al. 2015), resulting in a completeness of 98.97% and a contamination of 5.93%. For construction of phylogenomic tree, draft genome of 23 related type strains belonging to genus Brevibacterium were obtained from NCBI (http://www.ncbi.nlm.nih.gov/) database. The phylogenomic tree was inferred using EasyCGTree with default options. Evolutionary relationships were inferred using amino acid sequences of the bac120 gene set as default parameter in FastTree version 4.0 (Zhang et al. 2023a). The Type Strain Genome Server (TYGS) was utilized to construct a phylogenomic tree based on genomic sequences, including those of NCCP-602T and closely related type strains (Meier-Kolthoff and Göker 2019). The average nucleotide identity calculation was used to calculate the genomic distance between the NCCP-602T strain and related strains (Yoon et al. 2017). The calculation of digital DNA‒DNA hybridization (dDDH) value involved the application of formula 2 using genome‒to-genome distance calculator (GGDC, v. 3.0) (Meier-Kolthoff et al. 2022).
Gene network/pathway analysis
The genome sequence was annotated with KEGG (Kyoto Encyclopedia of Genes and Genomes) enzyme database. Each gene was assigned to a KEGG pathway. Physiological features of strain NCCP-602T were demonstrated by biochemical pathway maps constructed by result of individual analysis of KEGG pathways.
Chemotaxonomic analysis
For chemotaxonomic analysis, strain NCCP-602T was grown on TSA for 24 h to obtain necessary biomass. Subsequently, isoprenoid quinones were extracted from 100 to 150 mg of lyophilized cells. The method outlined by Minnikin et al. (1984) was followed for purification of quinones using TLC. The analytical method involved HPLC, employing a Cosmosil column (4.6 × 150 mm; Nacalai Tesque; reversed-phase silica gel; 5C18) and a mobile phase comprising methanol: 2-propanol (2:1) (Collins et al. 1977; Tamaoka 1986), which was used for determination of quinone. Quinone peaks were identified at a UV wavelength of 270 nm using Shimadzu equipment (Kyoto, Japan). Polar lipids were investigated through two-dimensional TLC (silica gel 60, Merck) plates (10 × 10 cm).Lyophilized cells (10 mg) were used to analyse amino acids in cell wall peptidoglycan. The cells were hydrolysed with 6 N HCl at 100°C for 18 h. Thin-layer chromatography (80:26:4:10) was performed on an HPTLC cellulose plate (10 × 10 cm, 1.05787.001 Merck, Germany) with a solvent combination of methanol-distilled water and 6 N HCl-pyridine for the analysis of whole-cell hydrolysate. To aid in identification, a TLC plate was also concurrently run with a standard solution of diaminopimelic acid (0.01 M, including meso-a2pm, DD-, and LL-) (Fig. 1).
Whole-cell fatty acids analysis was performed by growing strain NCCP-602T and reference strains on TSA media. Fatty acid methyl esters were prepared following the procedures described by Sasser (1990), andcellular fatty acids analysis was conducted using a gas chromatograph (model 6890; Hewlett Packard), with identification performed using the MIDI Sherlock version 4.5 system and the MIDI database TSGA40 4.10.
Results
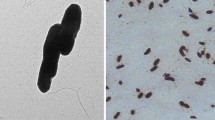
Following incubation at 30°C for 24‒48 h, strain NCCP-602T exhibited small, smooth, whitish colonies with glossy surfaces, a convex and spherical shape, entire margins, and a somewhat adhesive texture. The cells exhibited aerobic, nonmotile, and gram-positive staining and were rod-shaped (1.577‒0.448 µm diam.) (Fig. 2). Cells growth was monitored over a temperature range of 20‒42°C, revealing optimal growth between 30°C and 37°C within a span of 3 days. Strain NCCP-602T demonstrated no growth at pH 5.0 but exhibited robust growth within the pH range from 6.0 to 10.0, with a pH ranging from 7 to 8 for optimal growth.
Heavy metal tolerance
Genomic analysis of strain NCCP-602 T revealed an array of genes dedicated to maintaining heavy metal homeostasis. The copC gene has been shown to confer resistance to copper. The periplasmic proteins encoded by the copC gene exhibit a high affinity for copper atoms. The periplasmic accumulation of copper is regulated by four genes: copA, copC, copB and copD (Banci et al. 2003). The genome of strain NCCP-602T revealed the presence of the corA gene, which is responsible for conferring resistance to cobalt and magnesium (Table S5). The corA gene encodes for the transport of Mg2⁺ ions. The corA gene, responsible for Mg2⁺ transport, has been reported to exhibit a wide phylogenetic distribution among bacterial species (Groisman et al. 2013; Hmiel et al. 1989, 1986).
In the present study, proteins involved in zinc homeostasis, including Zn-dependent hydrogenase, Zn-binding hydrogenase, and neutral zinc metallopeptidase, were reported. The chrA gene, which is responsible for chromium resistance through encoding of a chromate-transporting protein, has been identified. The presence of Cr (IV) induces the expression of the chromate transporter gene chrA (He et al. 2010). Genome analysis revealed that the resistance of strain NCCP-602T to Mn (II) is mediated by the MnmtR and MnmA genes, which regulate the Mn (II) transport system.
The arsenic resistance of strain NCCP-602T is attributed to the presence of an arsenic efflux pump membrane protein and a metalloregulator belonging to the ArsR/SmtB family of transcription factors, which encode mechanisms for arsenic resistance. The presence of an arsenic operon, as well as genes conferring tolerance to copper and chromium, within the genome of strain NCCP-602T potentially contributes to its survival in natural wastewater environments (Flores et al. 2022).
Antibiotic susceptibility testing revealed that strain NCCP-602T was resistant to various antibiotic classes, including cephalothin, cefoperazone (a cephalosporin), tetracycline (tetracycline), lincomycin (lincosamide), oxacillin, penicillin (ampicillin), amoxicillin, amoxicillin-clavulanic acid (abeta lactam-beta lactamase inhibitor), colistin (polymyxin), cotrimethoprim, sulfonylens (sulfonamide), flumequin, oxolinic acid, enrofloxacin (quinones), nitrofurantoin (nitrofuran) and fucoxanthin (xanthophylls), however, it was found sensitive to gentamicin (aminoglycoside), rifamcin (rifampicin), spectinomycin (aminocyclitol), erythromycin (macrolides) and chloramphenicol (phenicol). Furthermore, the phenotype analysis indicated the presence of 49 antibiotic resistance genes, including antibiotic resistance efflux pump complexes. These genetic elements are responsible for conferring resistance to multiple antibiotic classes through molecular mechanisms (Fig. S7).
Biochemical characterization
Strain NCCP-602T shared numerous phenotypic traits with closely related strains; however, this strain also showed unique physiological and biochemical traits that distinguish it from the reference species. Table 1 provides a comprehensive overview of the distinct physiological and biochemical characteristics of strain NCCP-602T. Notable distinctions include its ability to utilize citrate, voges-proskauer, release N2 from nitrate, no nitrate reduction to NO2, trisodium citrate, gelatin hydrolysis, malic acid, glucose, N-acetyl-glucosamine, potassium gluconate, phenylacetic acid, and adipic acid (API 20E, API 20NE). The strain demonstrated high enzyme activity for α-glucosidase and acid phosphatase (API ZYM); use of glycerol, arabinose, glucose, maltose, melezitose, and L-arabitol as sources of carbon (API 50 CH); resistance against lincomycin, streptomycin, apramycin, mutactimycin, cephalothin, and kanamycin; and sensitivity against chloramphenicol, tetracycline and erythromycin, setting it apart from closely related species such as B. ammoniilyticum JCM 17537T, B. celere JCM 13521T and B. casei JCM 2594T. Strain NCCP-602T did not degrade Tween 20, 40, 60, or 80, and starch and can utilize L-alanine, citrate, inosine, and D-serine as carbon sources. Strain NCCP-602T was found to be positive for nalidixic acid, aztrename, sodium bromate and potassium tellurite (Biolog).
Furthermore, strain NCCP-602T exhibited notable resistance to typically harmful concentrations of heavy metals and can survive in TSA media supplemented with As (1200), Pb (1500 ppm), Cd (3000 ppm), Cu (3300 ppm), and Cr (3600 ppm). Compared to previously documented heavy metal-tolerant bacteria, strain NCCP-602T displays a relatively greater tolerance to these harmful concentrations of heavy metals, especially when contrasted with numerous other bacterial strains (Abbas et al. 2014, 2015b; Elahi et al. 2019; Mustapha and Halimoon 2015).
Phylogenetic analysis
Highest similarity (98.5%) of 16S rRNA gene sequence of strain NCCP-602 T was found with Brevibacterium ammoniilyticum (GenBank accession no. JF937067), while it displayed less than 98.5% similarity with that of other Brevibacterium species. Results of phylogenetic analysis indicated that strain NCCP-602T clustered within a specific clade alongside Brevibacterium casei and B. ammoniilyticum as its closest neighbours (Figure S2). Phylogenetic trees (Figures S3 and S4) constructed following maximum-likelihood and maximum-parsimony algorithms showed positioning of strain NCCP-602T within the same clade as Brevibacterium casei and B. ammoniilyticum suggesting a close relationship, and a robust bootstrap support was further observed in the neighbour-joining tree. The bac120 tree also indicated that strain NCCP-602T clustered with Brevibacterium ammoniilyticum KACC15558T (GCF-038429965.1) in the clade (Fig. 1). Determining the relatedness values of digital DNA‒DNA hybridization between closely related reference strains and the NCCP-602T strain revealed 64.7% relatedness for Brevibacterium ammoniilyticum JCM 17537T, 23.3% for B. celere JCM 13522T, and 31.1% for B. casei JCM 2594T. The calculated values fall below the defined threshold of 70%, which is desired for classification of the strain as a distinct new species in the genus (Wayne et al. 1987). The determined G+C content of 67.6 mol% in genomic DNA of strain NCCP-602T supports its taxonomic affiliation within Brevibacterium genus. This G+C content is observed to be consistent with that found in closely related strains, including Brevibacterium ammonylyticum JCM 17537T, B. celere JCM 13521T, and B. casei JCM 2594T. Although biochemical data presented in Table 1 reveal certain distinctions from the closely related reference strains, it is worth noting that strain NCCP-602T shares numerous characteristics with B. ammonilyticum JCM 17537T and B. celere JCM 13521T. The cumulative evidence strongly suggested that strain NCCP-602T constitutes a distinct and novel species within Brevibacterium genus.
Functional genomics
With a cumulative size of 3.9 Mbp and an N50 length of 236,461 Kbp, the definitive draft genome sequence of strain NCCP-602T included 83 contigs. The phylogenomic tree generated from the genomes of closely related strains clearly demonstrated that strain NCCP-602T constituted a unique clade within genus Brevibacterium (Figure S5), thereby reinforcing its phylogenetic placement. With the use of Prodigal software, the prediction of protein-coding genes yielded a total of 2621 genes. Supplementary Table S1 provides a comprehensive list of genomic features for strain NCCP-602T alongside the reference strains B. ammoniilyticum JCM 17537T, B. celere JCM 13521T, and B. casei JCM 2594T. Despite the significantly high average nucleotide identity (ANI) value (95.4%), digital DNA‒DNA hybridization (dDDH) values between strain NCCP-602T and other Brevibacterium members remained below the recognized threshold values (70%) critical for distinguishing novel prokaryotic species (Richter and Rosselló-Móra 2009) (Table S1). The extensive protein profile of strain NCCP-602T, as categorized in Clusters of Orthologous Groups (COGs) database, exhibited significant similarities to those of its three closely related strains, B. ammoniilyticum JCM 17537T, B. celere JCM 13521T, and B. casei JCM 2594T. The principal protein groups encompassed translation, transcription, coenzyme metabolism, and transport, as well as amino acid metabolism and transport. Notably, cell motility-related genes were present in both strain NCCP-602T and its related strains, although genes associated with flagella were notably absent (Table S2). Moreover, carbohydrate degradation proficiency of the strains was also evaluated within the genomes of closely related type strains and strain NCCP-602T. The presence of five families, namely, auxiliary activities (AAs), glycoside hydrolases (GHs), carbohydrate-binding modules (CBMs), glycosyltransferases (GTs), and carbohydrate esterases (CEs), across the scrutinized genomes is presented in Table S3. However, in case of strain NCCP-602T, certain subfamilies, including GH0, GH13_3, GH13_16, GH13_26, GH16_24, GH23, GH33, GH170, GT5, and GT35, were notably absent. Additionally, antiSMASH version 7.0.0 was used to predict clusters of secondary metabolite genes, and analysis revealed that strain NCCP-602T contains gene clusters associated with ectoine, hydrogen cyanide, and siderophores. These clusters serve a protective function against oxidative stress in bacteria (Table S4) (Giani and Martínez-Espinosa 2020; Graf et al. 2008; Saha et al. 2016). The orthologous genes of closely related strains and strain NCCP-602T were clustered using the OrthoVenn3 web server (Wang et al. 2015) with the OrthoFinder algorithm (Emms and Kelly 2019). Among 3298 orthologous gene clusters identified, 2254 were shared by the four genomes. A customized version of CGView (http://cgview.ca) was utilized to generate a circular genome map for strain NCCP-602T (Figure S6A). It is worth noting that genome of strain NCCP-602T harbored 11 unique gene clusters containing 26 proteins (Figure S6B).
Genetic elements involved in bioremediation and environmental protection
KEEG annotation of the genome of the NCCP-602T strain confirmed the presence of specific enzymes responsible for encoding the xenobiotic resistance and degradation capability (Table S6). The genes responsible for histidine degradation; Cr (VI) intracellular reduction; the degradation of lethal nitronates; and catechol degradation, ring cleavage and phthalate hydrolysis were identified. Several nitrate, sulphur, and phosphate reductase and transport genes in the NCCP-602T genome are presented in (Table S6). The NCCP-602T genome profile confirms that the NCCP-602T strain uses GABA as a nutritional source in the rhizome, resulting in the regulation of GABA levels in plants (Renault et al. 2011). Various genes responsible for tolerance of strain NCCP-602T to osmotic stress and temperature were identified from genome of the strain (Table S6). The presence of osmotic stress tolerance genes in the genetic elements of strain NCCP-602T indicates that this strain has the ability to survive in highly saline environments.
Chemotaxonomic analysis
Results of cellular fatty acid analysis depicted the following profile of strain NCCP-602T: anteiso-C15:0, iso-C15:0, iso-C16:0, C17:0, and anteiso-C17:0 (Table S7)., Strain NCCP-602T differs from other members of Brevibacterium genus by variations in quantities of specific components. Nevertheless, predominant components of its fatty acid composition closely mirror those identified in closely related reference strains. Strain NCCP-602T exhibited elevated concentrations of iso-C15:0, iso-C16:0, and iso-C17:0, constituting 18.65% of fatty acid composition in comparison to the reference strains (Table S7).
The distinctive amino acid meso-diaminopimelic acid was identified in cell wall peptidoglycan of strain NCCP-602T, which is a characteristic feature observed in Brevibacterium species. The primary component MK-7 was identified in the respiratory quinone, comprising 67% of its composition, with minor components such as MK-8 (33%). Results of whole-cell composition revealed the sugars such as glucose, ribose, and mannitol. Significantly, occurrence of meso-diaminopimelic acid, a notable quantity of iso-C-15:0, and prevailing occurrence of major respiratory quinone as MK-7 closely conforms to the distinctive characteristics observed in members of Brevibacterium genus.
Major components of polar lipid profile in strain NCCP-602T included diphosphatidylglycerol, unknown glycolipids, unknown phospholipids and amino lipids. The strain exhibited a distinctive lipid profile with negative responses to ninhydrin sprays but positive reactions to alpha-naphthol reagents. The glycolipid (GL) exhibited chromatographic mobility typical of a diglycosyldiacylglycerol (Figure S1). This polar lipid composition closely mirrors that of related species, such as Brevibacterium ammoniilyticum JCM 17537T (Kim et al. 2013), which has been analysed at same lab conditions. The polar lipid profile of strain NCCP-602T, which is similar to that of B. ammoniilyticum JCM 17537T, contains unknown amino phospholipids and phospholipids. Notably, although amino lipid (AL) are present in both NCCP-602T and the reference species (Figure S1), however, levels of these amino lipids were relatively low.
Based on comprehensive analysis of phenotypic, physiological, phylogenetic, and DNA‒DNA relatedness and the chemotaxonomic data, there is clear distinctions established between strain NCCP-602T and other closely related validly classified members within genus Brevibacterium. Therefore, results of these studies support the recognition of this strain as a novel species within genus Brevibacterium, for whichas the name Brevibacterium metallidurans sp. nov. has been proposed with NCCP-602T has been designated as the type strain of Brevibacterium metallidurans.
Description of Brevibacterium metallidurans sp. nov.
Brevibacterium metallidurans (me.tal.li.du’rans. L. neut. n. metallum, metal; L. pres. part. durans, enduring; N.L. part. adj. metallidurans, enduring high metal concentrations).
The cells possess distinctive features characterized by their gram-positive, aerobic, non-motile, absence of spore formation, and with a rod-shaped morphology, (diameters ranging from 1.57 to 0.45 µm). On tryptic soy agar, colonies were circular and small, exhibited smooth and shining surfaces, had a convex structure, and presented a whitish appearance with entire margins and a somewhat sticky texture. After thriving across a broad range of pH values (6–10), cells exhibited optimal growth at pH 7–8. Strain is adaptable to a broad temperature range of 20–42°C, with optimal growth observed at 30–37°C. Additionally, strain has the capacity to withstand NaCl concentrations ranging from 0 to 15% (w/v), with optimal growth achieved at 1% NaCl. Remarkably, the strain also displays resistance to heavy metals, including arsenic (As), chromium (Cr), copper (Cu), lead (Pb), and cadmium (Cd), at high concentrations. Moreover, it can neither hydrolyse starch nor degrade tween 20, 40, 60 or 80, however it showed a positive reaction to α-glucosidase, Voges Proskauer, nitrate reduction to N2, assimilation of glucose, adipic acid, N-acetyl-glucosamine, trisodium citrate, acid phosphatase, potassium gluconate, hydrolysis of gelatin and malate, citrate utilization, arabinose, phenylacetic acid, maltose, and L-arabitol whereas negative reactions for melezitose and glycerol. The strains can use L-alanine, citrate, inosine, and D-serine as carbon sources and are positive for nalidixic acid, aztrename, sodium bromate, and potassium tellurite (Biolog). The predominant polar lipids include diphosphatidylglycerol, glycolipids, and a minor amount of amino lipids and unknown phospholipids. Major cellular fatty acids included anteiso-C15:0, iso-C15:0, iso-C16:0, and C17:0, with anteiso-C17:0. MK-7 and MK-8 are identified as dominant respiratory quinones. DNA G + C content was determined to be 67.6 mol%.
Strain NCCP-602T (JCM 18882T = CGMCC1.62055T) was isolated from wastewater collected from Sialkot, Sambrial, Pakistan. Accession number of 16S rRNA gene sequence was AB920787 submitted in DDBJ/EMBL/GenBank. The whole-genome shotgun sequencing project for strain NCCP-602T has been archived in GenBank under accession numbers BioProject: PRJDB17310, BioSample: SAMD00729978, and WGS: BAAAAF010000001-BAAAAF010000083.
Data availability
Accession number of 16S rRNA gene sequence of strain NCCP-602T was AB920787 submitted in DDBJ/EMBL/GenBank. The whole-genome shotgun sequencing project for strain NCCP-602T has been archived in GenBank under accession numbers BioProject: PRJDB17310, BioSample: SAMD00729978, and WGS: BAAAAF010000001-BAAAAF010000083 .
References
Abbas S, Ahmed I, Kudo T, Iida T, Ali GM, Fujiwara T, Ohkuma M (2014) Heavy metal-tolerant and psychrotolerant bacterium Acinetobacter pakistanensis sp. nov. isolated from a textile dyeing wastewater treatment pond. Pak J Agr Sci 51:595–608
Abbas S, Ahmed I, Iida T, Lee Y-J, Busse H-J, Fujiwara T, Ohkuma M (2015a) A heavy-metal tolerant novel bacterium, Alcaligenes pakistanensis sp. nov., isolated from industrial effluent in Pakistan. Antonie Van Leeuwenhoek 108:859–870. https://doi.org/10.1007/s10482-015-0540-1
Abbas S, Ahmed I, Kudo T, Iqbal M, Lee Y-J, Fujiwara T, Ohkuma M (2015b) A heavy metal tolerant novel bacterium, Bacillus malikii sp nov., isolated from tannery effluent wastewater. Antonie Van Leeuwenhoek Int J Gen Mol Microbiol 108:1319–1330. https://doi.org/10.1007/s10482-015-0584-2
Ahmed I, Sin Y, Paek J, Ehsan M, Hayat R, Iqbal M, Chang YH (2014) Description of Lysinibacillus pakistanensis. Int J Agric Biol 16:447–450
Ahmed I, Yokota A, Fujiwara T (2007) A novel highly boron tolerant bacterium, Bacillus boroniphilus sp. nov., isolated from soil, that requires boron for its growth. Extremophiles 11:217–224
Ali N, Dashti N, Al-Mailem D, Eliyas M, Radwan S (2012) Indigenous soil bacteria with the combined potential for hydrocarbon consumption and heavy metal resistance. Environ Sci Pollut Res 19:812–820. https://doi.org/10.1007/s11356-011-0624-z
Banci L, Bertini I, Ciofi-Baffoni S, Gonnelli L, Su X-C (2003) Structural basis for the function of the N-terminal domain of the ATPase CopA from Bacillus subtilis. J Biol Chem 278:50506–50513
Bankevich A et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. https://doi.org/10.1093/nar/29.12.2607
Bhadra B, Raghukumar C, Pindi PK, Shivaji S (2008) Brevibacterium oceani sp. nov., isolated from deep-sea sediment of the Chagos Trench, Indian Ocean. Int J Syst Evol Microbiol 58:57–60. https://doi.org/10.1099/ijs.0.64869-0
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Breed R (1953) The Brevibacteriaceae fam. nov. of order Eubacteriales. Rias Commun VI Congr Int Microbiol Roma 1:13–14
Collins MD, Farrow JAE, Goodfellow M, Minnikin DE (1983) Brevibacterium casei sp. nov. and Brevibacterium epidermidis sp. nov. Syst Appl Microbiol 4:388–395. https://doi.org/10.1016/S0723-2020(83)80023-X
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230. https://doi.org/10.1099/00221287-100-2-221
Deng T et al (2020) Brevibacterium rongguiense sp. nov., isolated from freshwater sediment. Int J Syst Evol Microbiol 70:5205–5210. https://doi.org/10.1099/ijsem.0.004379
Elahi A, Ajaz M, Rehman A, Vuilleumier S, Khan Z, Hussain SZ (2019) Isolation, characterization, and multiple heavy metal-resistant and hexavalent chromium-reducing Microbacterium testaceum B-HS2 from tannery effluent. J King Saud Univ—Sci 31:1437–1444. https://doi.org/10.1016/j.jksus.2019.02.007
Emms DM, Kelly S (2019) OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:238. https://doi.org/10.1186/s13059-019-1832-y
Flores A et al (2022) Genome mining, phylogenetic, and functional analysis of arsenic (As) resistance operons in Bacillus strains, isolated from As-rich hot spring microbial mats. Microbiol Res 264:127158. https://doi.org/10.1016/j.micres.2022.127158
Forquin-Gomez M-P, Weimer B, Sorieul L, Kalinowski J, Vallaeys T (2014) The Family Brevibacteriaceae. Springer, Berlin, pp 141–154. https://doi.org/10.1007/978-3-642-30138-4_169
Giani M, Martínez-Espinosa RM (2020) Carotenoids as a protection mechanism against oxidative stress in Haloferax mediterranei. Antioxidants (basel, Switzerland) 9:1060. https://doi.org/10.3390/antiox9111060
Graf R, Anzali S, Buenger J, Pfluecker F, Driller H (2008) The multifunctional role of ectoine as a natural cell protectant. Clin Dermatol 26:326–333. https://doi.org/10.1016/j.clindermatol.2008.01.002
Groisman EA, Hollands K, Kriner MA, Lee E-J, Park S-Y, Pontes MH (2013) Bacterial Mg2+ homeostasis, transport, and virulence. Annu Rev Genet 47:625–646
Guérin-Faublée V, Flandrois J-P, Richard YG (1993) Antimicrobial susceptibility testing of actinomyces pyogenes: Comparison of disk diffusion test and Api ATB Strep system with the Agar dilution method. Zentralblatt Für Bakteriologie 279:377–386. https://doi.org/10.1016/S0934-8840(11)80370-1
He M, Li X, Guo L, Miller SJ, Rensing C, Wang G (2010) Characterization and genomic analysis of chromate resistant and reducing Bacillus cereus strain SJ1. BMC Microbiol 10:221. https://doi.org/10.1186/1471-2180-10-221
Heyrman J, Verbeeren J, Schumann P, Devos J, Swings J, De Vos P (2004) Brevibacterium picturae sp. nov., isolated from a damaged mural painting at the Saint-Catherine chapel (Castle Herberstein, Austria). Int J Syst Evol Microbiol 54:1537–1541. https://doi.org/10.1099/ijs.0.63144-0
Hmiel S, Snavely M, Florer J, Maguire M, Miller C (1989) Magnesium transport in Salmonella typhimurium: genetic characterization and cloning of three magnesium transport loci. J Bacteriol 171:4742–4751
Hmiel S, Snavely M, Miller C, Maguire M (1986) Magnesium transport in Salmonella typhimurium: characterization of magnesium influx and cloning of a transport gene. J Bacteriol 168:1444–1450
Huang Y et al (2022) Phenotypic and genomic characteristics of Brevibacterium zhoupengii sp. nov., a novel halotolerant actinomycete isolated from bat feces. J Microbiol (seoul, Korea) 60:977–985. https://doi.org/10.1007/s12275-022-2134-8
Ivanova EP et al (2004) Brevibacterium celere sp. nov., isolated from degraded thallus of a brown alga. Int J Syst Evol Microbiol 54:2107–2111. https://doi.org/10.1099/ijs.0.02867-0
Johnson LS, Eddy SR, Portugaly E (2010) Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform 11:431. https://doi.org/10.1186/1471-2105-11-431
Kalishwaralal K, Deepak V, Ram Kumar Pandian S, Kottaisamy M, BarathManiKanth S, Kartikeyan B, Gurunathan S (2010) Biosynthesis of silver and gold nanoparticles using Brevibacterium casei. Colloids Surf, B 77:257–262. https://doi.org/10.1016/j.colsurfb.2010.02.007
Kim J, Srinivasan S, You T, Bang JJ, Park S, Lee S-S (2013) Brevibacterium ammoniilyticum sp. nov., an ammonia-degrading bacterium isolated from sludge of a wastewater treatment plant. Int J Syst Evol Microbiol 63:1111–1118. https://doi.org/10.1099/ijs.0.039305-0
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M (2022) TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res 50:D801–D807. https://doi.org/10.1093/nar/gkab902
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. https://doi.org/10.1038/s41467-019-10210-3
Minnikin D, O’donnell A, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett J (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Mustapha MU, Halimoon N (2015) Screening and isolation of heavy metal tolerant bacteria in industrial effluent. Procedia Environ Sci 30:33–37. https://doi.org/10.1016/j.proenv.2015.10.006
Ng TW, Cai Q, Wong C-K, Chow AT, Wong P-K (2010) Simultaneous chromate reduction and azo dye decolourization by Brevibacterium casei: Azo dye as electron donor for chromate reduction. J Hazard Mat 182:792–800. https://doi.org/10.1016/j.jhazmat.2010.06.106
Odds FC (1981) Biochemical tests for identification of medical bacteria. J Clinic Pathol 34:572. https://doi.org/10.1136/jcp.34.5.572-a
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055
Pascual C, Collins MD (1999) Brevibacterium avium sp. nov., isolated from poultry. Int J Syst Evol Microbiol 49:1527–1530. https://doi.org/10.1099/00207713-49-4-1527
Pei S, Niu S, Xie F, Wang W, Zhang S, Zhang G (2021) Brevibacterium limosum sp. nov., Brevibacterium pigmenatum sp. nov., and Brevibacterium atlanticum sp. nov., three novel dye decolorizing actinobacteria isolated from ocean sediments. J Microb (seoul, Korea) 59:898–910. https://doi.org/10.1007/s12275-021-1235-0
Pei S, Xie F, Niu S, Ma L, Zhang R, Zhang G (2020) Brevibacterium profundi sp. nov., isolated from deep-sea sediment of the Western Pacific Ocean. Int J Syst Evol Microbiol 70:5818–5823. https://doi.org/10.1099/ijsem.0.004483
Renault H et al (2011) GABA accumulation causes cell elongation defects and a decrease in expression of genes encoding secreted and cell wall-related proteins in Arabidopsis thaliana. Plant Cell Physiol 52:894–908
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131. https://doi.org/10.1073/pnas.0906412106
Román-Ponce B, Ramos-Garza J, Arroyo-Herrera I, Maldonado-Hernández J, Bahena-Osorio Y, Vásquez-Murrieta MS, Wang ET (2018) Mechanism of arsenic resistance in endophytic bacteria isolated from endemic plant of mine tailings and their arsenophore production. Arch Microbiol 200:883–895.
Roux V, Raoult D (2009) Brevibacterium massiliense sp. nov., isolated from a human ankle discharge. Int J Syst Evol Microbiol 59:1960–1964. https://doi.org/10.1099/ijs.0.007864-0
Saha M, Sarkar S, Sarkar B, Sharma BK, Bhattacharjee S, Tribedi P (2016) Microbial siderophores and their potential applications: a review. Environ Sci Pollut Res Int 23:3984–3999. https://doi.org/10.1007/s11356-015-4294-0
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101. MIDI inc, Newark
Tamaoka J (1986) Analysis of bacterial menaquinone mixtures by reverse-phase high-performance liquid chromatography. In: Methods in Enzymology. Academic Press, Cambridge, pp 251–256. https://doi.org/10.1016/S0076-6879(86)23028-1
Tambalo DD, Del Bel KL, Bustard DE, Greenwood PR, Steedman AE, Hynes MF (2010) Regulation of flagellar, motility and chemotaxis genes in Rhizobium leguminosarum by the VisN/R-Rem cascade. Microbiology 156:1673–1685. https://doi.org/10.1099/mic.0.035386-0
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. https://doi.org/10.1093/oxfordjournals.molbev.a040023
Ventosa A, Quesada E, Rodriguez-Valera F, Ruiz-Berraquero F, Ramos-Cormenzana A (1982) Numerical taxonomy of moderately halophilic gram-negative rods. Microbiology 128:1959–1968
Verma T, Singh N (2013) Isolation and process parameter optimization of Brevibacterium casei for simultaneous bioremediation of hexavalent chromium and pentachlorophenol. J Basic Microbiol 53:277–290. https://doi.org/10.1002/jobm.201100542
Wang Y, Coleman-Derr D, Chen G, Gu YQ (2015) OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res 43:W78–W84. https://doi.org/10.1093/nar/gkv487
Wani PA, Rafi N, Wani U, Bilikis AH, Khan MSA (2023) Simultaneous bioremediation of heavy metals and biodegradation of hydrocarbons by metal resistant Brevibacillus parabrevis OZF5 improves plant growth promotion. Bioremed J 27(1):20–31.
Wauters G, Haase G, Vr A, Charlier J, Ml J, Van Broeck J, Delmée M (2004) Identification of a novel Brevibacterium species isolated from humans and description of Brevibacterium sanguinis sp. nov. J Clin Microbiol 42:2829–2832. https://doi.org/10.1128/jcm.42.6.2829-2832.2004
Wayne LG et al (1987) Report of the Ad Hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol 37:463–464. https://doi.org/10.1099/00207713-37-4-463
Xu P et al (2005) Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int J Syst Evol Microbiol 55:1149–1153. https://doi.org/10.1099/ijs.0.63407-0
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. https://doi.org/10.1007/s10482-017-0844-4
Zhang DF, He W, Shao Z, Ahmed I, Zhang Y, Li WJ, Zhao Z (2023a) EasyCGTree: a pipeline for prokaryotic phylogenomic analysis based on core gene sets. BMC Bioinform 24:390. https://doi.org/10.1186/s12859-023-05527-2
Zhang H et al (2018) dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101. https://doi.org/10.1093/nar/gky418
Zhang M, Song Q, Sang J, Li Z-Y (2023b) Brevibacterium spongiae sp. Nov., isolated from marine sponge Hymeniacidon sp. Int J Syst Evol Microbiol 73:005869. https://doi.org/10.1099/ijsem.0.005869
Acknowledgements
The authors gratefully acknowledge the kind cooperation of the Japan Collection of Microorganisms (JCM), Japan, for providing the reference strains.
Funding
This study was supported by fundings from Guangdong Provincial Science and Technology Plan Project (Grant No. 2022A0505020001), and the Higher Education Commission of Pakistan to S.M. throught the International Research Support Initative Program (IRSIP). This work was also partly supported by financial assistance from the PSDP-funded Project Research for Agricultural Development Project (RADP) under a subproject (Grant No. CS-55/RADP/PARC) entitled "Establishment of Microbial Bio-Resource Laboratories: National Culture Collection of Pakistan (NCCP)" from Pakistan Agricultural Research Council (PARC), Islamabad, Pakistan, and partly from the Japan Society for Promotion of Science (JSPS) and the Chinese Academy of Sciences President’s International Fellowship Initiative (Grant No. 2020VBA0020).
Author information
Authors and Affiliations
Contributions
Credit authorship contribution statement Conceptualization, IA, MA and SA; Methodology, SM, SA, HCW, and IA; Formal analysis, SA, SM and IA; Software, SM, SA, SZ and IA; Data curation, SM, SA and IA; Project administration, Resources and Funding acquisition: IA, WJL; Supervision, IA, WJL. MA; Writing – original draft, SM, and IA; Writing – review & editing, SM, SA, SZ, HCW, MX, MA, WJL and IA.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Manzoor, S., Abbas, S., Zulfiqar, S. et al. Functional genomics and taxonomic insights into heavy metal tolerant novel bacterium Brevibacterium metallidurans sp. nov. NCCP-602T isolated from tannery effluent in Pakistan. Antonie van Leeuwenhoek 117, 111 (2024). https://doi.org/10.1007/s10482-024-02006-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10482-024-02006-3