Abstract
A Gram-staining negative, aerobic, motile, and short rods strain, designated SYSU M60028T, was isolated from a Pearl River sediment sample in Guangzhou, Guangdong, China. The isolate could be able to grow at pH 6.0–8.0 (optimum, pH 7.0), 25–37 °C (optimum, 28 °C) and in the presence of 0–2% (w/v) NaCl (optimum, 0% NaCl). The cellular polar lipids of this strain were phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, phosphatidylcholine, one unidentified aminolipid and three unidentified lipids. The respiratory quinone of SYSU M60028T was found to be Q-10. The major fatty acids (> 5% of total) were summed feature 8, C16:0, and C18:1 ω7c 11-methy1. The genomic DNA G + C content was 69.9%. Phylogenetic analyses based on 16S rRNA gene sequences and core genes indicated that strain SYSU M60028T belonged to the genus Alsobacter and had the highest sequences similarities to Alsobacter metallidurans SK200a-9T (96.87%) and Alsobacter soli SH9T (96.87%). Based on the phenotypic, genotypic, and phylogenetic data, strain SYSU M0028T should be considered to represent a novel species of the genus Alsobacter, for which the name Alsobacter ponti sp. nov. is proposed. The type strain is SYSU M60028T (= CGMCC 1.19341T = KCTC 92046T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Alsobacter of the family Alsobacteraceae, class Alphaproteobacteria was first proposed by Bao et al. (2014). At the time of writing, the genus Alsobacter comprises two species with validly published names. Species of this genus have been reported from a garden soil sample (Bao et al. 2014) and a paddy soil sample (Sun et al. 2018). According to the former studies, Alsobacter species are Gram-staining negative, aerobic and rod-shape, containing Q-10 as the respiratory ubiquinone (Bao et al. 2014; Sun et al. 2018). In this study, we describe a new isolate which represents a novel species of this genus based on the analysis of morphological, phenotypic, genotypic, and phylogenetic characteristics.
Estuaries are the interaction zones between oceans and terrestrials, whose eco-environment health are essential to the sustainable development of human habitation and social economy. In order to investigate the cultivable bacteria community in the Pearl River estuary, surface sediment samples were collected from Pearl River estuary, Guangzhou, Guangdong, China. During this study, one novel isolate SYSU M60028T was cultivated from a sediment sample.
Material and method
Sample collection and isolation
The surface sediment samples (10 cm) were collected at the depth of 3 m from the Pearl River, Guangdong, China, and transferred by a sterile homogeneous bag. The temperature and pH of the samples were 28 °C and pH 7.5, respectively. The sediment samples were diluted 103- fold with sterile distilled water and 0.2 mL final suspension was plated on Reasoner's 2A (R2A; BD) agar (Yeast Extract 0.5 g, Proteose Peptone 0.5 g, Glucose 0.5 g, Soluble Starch 0.5 g, Casamino Acids 0.5 g, Sodium Pyruvate 0.3 g, K2HPO4 0.3 g, MgSO4-7H2O 0.05 g, Distilled Water 1000 ml, Agar 18 g, pH 7.0). The isolation plates were incubated at 28℃ for 7 days. Single colonies were selected and purified on the same medium. The purified strains were preserved as glycerol suspensions (20%, v/v) at − 80 °C for further use. Among the purified isolates, one off-white strain SYSU M60028T showed low 16S rRNA sequences similarities to Alsobacter metallidurans (96.87%) and Alsobacter soli (96.87%), and was selected for further novel species characterization. The basal growth conditions of the strain SYSU M60028T for all experiments were maintained at pH 7.0 and 28℃, unless otherwise stated.
Phenotypic characterization
Cultural characteristics were performed on Luria–Bertani (LB) agar, R2A agar, tryptic soy agar (TSA; Difco), and marine agar 2216E (MA; BD Difco) at 28 °C for 5 days. The microscopic morphological characteristics of the strain SYSU M60028T were observed by transmission electron microscope (JEM–1400FLASH, JEOL, JAPAN). The Gram-reaction was performed by the standard Gram reaction and was confirmed by the non-staining method using 3% KOH (Buck 1982). Growth temperature (4, 10, 15, 20, 25, 28, 30, 37, 45, 55, 60 and 65 °C), pH range (pH 4.0–10.0, at intervals of 1 unit, adjusted by using the buffer system) (Xu et al. 2005), and NaCl tolerance (0, 1, 2, 3, 4, 6, 9, 12 and 15%, w/v) were tested on R2A agar. Oxidase and catalase activities were determined by assessing the oxidation of 1% (w/v) tetramethyl-p-phenylenediamine (Kovacs 1956) and the formation of bubble on addition of 3% H2O2, respectively. Tweens (20, 40, 60, and 80) degradation, H2S production, milk coagulation and peptonization, cellulose, starch, gelatin hydrolysis, anaerobic growth, and urease activity were tested according to the methods previously described (Gonzalez et al. 1978; Smibert Krieg 1994). Other physiological and biochemical characteristics of strain SYSU M60028T were identified by GEN III Microplates system (Biolog, USA), API 20NE and API ZYM kits (bioMérieux) according to the manufacturer’s instructions.
Chemotaxonomy characterisation
Biomass for chemical and molecular studies of strain SYSU M60028T was obtained on R2A agar during the logarithmic growth period (3 days). For polar lipids, strain SYSU M60028T was extracted and identified by two-dimensional TLC on silica gel G60 plates (Merck) as previously described (Collins Jones 1980; Minnikin 1979). The respiratory ubiquinone was extracted from lyophilized cells as described by Collins et al. (1977), purified, and analysed by HPLC as described by Tamaoka et al. (1983). The cellular fatty acid methyl esters were extracted and analysed according to the standard protocol of the Microbial Identification System (Sherlock version 6.1; MIDI database: TSBA6) (Athalye et al. 1985).
Phylogenetic analysis
Genomic DNA extraction and PCR amplification of 16S rRNA gene were carried out according to the method described by Li et al. (2007). The obtained sequences were compared with the corresponding validly published species in the EzBioCloud database (Yoon et al. 2017). The 16S rRNA gene sequences of close related type strains were aligned using CLUSTAL X software package (Thompson et al. 1997) and were used for constructing the phylogenetic trees by MEGA X software (Kumar et al. 2018). Algorithms based on neighbour-joining (Saitou Nei 1987), maximum-likelihood (Felsenstein 1981), and maximum-parsimony (Fitch 1971) methods were used for generation of the phylogenetic trees. Kimura’s two-parameter model was used to calculate evolutionary distance matrices of the phylogenetic trees (Kimura 1980). Stability of the phylogenetic trees was evaluated by bootstrap analysis with 1000 replications (Felsenstein 1985). Chelatococcus reniformis B2974T was used as an outgroup.
Genome sequencing, annotation, and analysis
The whole genome sequencing of strain SYSU M60028T was performed by using Illumina NovaSeq platform at the Guangdong Microbial Culture Collection Center (GDMCC, Guangdong, China). Reads of each data sets were filtered, and high quality paired-end reads were assembled using readfq (version 10), SOAP denovo (version 2.04), and SPAdes (Bankevich et al. 2012; Li et al. 2008, 2010). The quality of the genomes was examined using CheckM v.1.0.7 (Parks et al. 2015). The COG database (Galperin et al. 2014) and KEGG database (Kanehisa Sato 2020; Kanehisa et al. 2022) were used to annotate the genome sequences. The secondary metabolite biosynthetic gene clusters were annotated by antiSMASH (Blin et al. 2021). The Average nucleotide identity (ANI) was calculated using JSpecies (http://jspecies.ribohost.com/jspeciesws/) (Richter et al. 2016). The digital DNA-DNA hybridization analysis was performed by using the DSMZ Genome-to-Genome Distance Calculator platform (http://ggdc.dsmz.de/distcalc2.php) (Goris et al. 2007). The phylogenomic tree was constructed according to the method described by Jiao et al. (2021). The multiple sequence alignments (MSAs) were generated by GTDB-Tk software (Chaumeil et al. 2019). The Maximum-likelihood phylogenetic tree was calculated by IQ-Tree with parameters (alrt 1000 bb 1000 nt AUTO) (Nguyen et al. 2015). The best-fit model (LG + F + R4) determined by ModelFinder was well supported by Akaike information criterion (AIC), corrected AIC (AICc), and Bayesian information criterion (Kalyaanamoorthy et al. 2017). The phylogenomic tree was visualized using the online Interactive Tree of Life program version 4.2 finally (Letunic Bork 2016).
Result and discussion
Phenotypic characteristics
Cells of strain SYSU M60028T were Gram-staining negative, aerobic, and had the ability to move by flagella (Fig. S4). Colonies were circular, punctiform and off-white in colour. Cells were short rods with size measuring 0.6–0.7 × 0.9–1.0 μm (Fig. S4). The isolate could be able to grow at pH 6.0–8.0 (optimum, pH 7.0), 25–37 °C (optimum, 28 °C) and in the presence of 0–2% (w/v) NaCl (optimum, 0% NaCl). Strain SYSU M60028T was positive for oxidase activity, α-glucosidase, H2S production, nitrate reduction and trypsin, but negative for milk coagulation and peptonization, Tweens (20, 40, 60, and 80), or catalase activity. The characteristics distinguishing strain SYSU M60028T from closely related strains are listed in Table 1. The detailed physiological characteristics of strain SYSU M60028T are given in the species descriptions below.
Chemotaxonomical characteristics
The respiratory quinone of strain SYSU M60028T was identified as ubiquinone Q-10. The major cellular fatty acids (> 5%) detected of strain SYSU M60028T were summed feature 8 (C18:1ω7c/ C18:1ω6c, 55.3%), C16:0 (9.0%), and C18:1 ω7c 11-methy1 (5.8%). The cellular polar lipids were phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, phosphatidylcholine, one unidentified aminolipid and three unidentified lipids (Fig. S5). The detailed fatty acid profile of strain SYSU M60028T was provided in Table S4.
Phylogenic and phylogenomic analysis
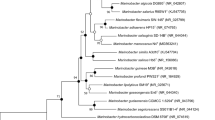
The 16S rRNA gene sequences of strain SYSU M60028T was submitted to GenBank with accession number ON881906. On pairwise comparison with the 16S rRNA gene sequences available in the EzBioCloud database, strain SYSU M60028T showed highest 16S rRNA gene sequences similarities to Alsobacter metallidurans SK200a-9T (96.87%) and Alsobacter soli SH9T (96.87%). The neighbour-joining tree based on 16S rRNA gene sequences showed that strain SYSU M60028T formed a distant clade with Alsobacter metallidurans SK200a-9T and Alsobacter soli SH9T (Fig. 1). An apparently stable lineage was also supported by maximum-parsimony and maximum-likelihood phylogenetic trees (Fig. S1, Fig. S2). Phylogenomic tree (Fig. S3) based on the concatenated alignment of 120 marker genes provided further evidence of this topological structure. Based on the 16S rRNA gene sequences identity and the phylogenetic relationship, Alsobacter metallidurans SK200a-9T and Alsobacter soli SH9T were used as reference type strains.
Neighbour–joining phylogenetic tree based on 16S rRNA gene sequences showing the relationship between strain SYSU M60028T and their closest relatives. Bootstrap values (> 50%) based on 1000 replications are given at the nodes. Bar represents 0.01 substitutions per nucleotide position. Chelatococcus reniformis B2974T (KJ469373) was used as an outgroup
Genome analysis
The genome properties of strain SYSU M60028T included 56 contigs amounting to 4,727,519 bp. The G + C content based on DNA genomic was 69.9% (Table S1). The completeness and contamination of the genomic were 98.59% and 0.26%, respectively. The dDDH value comparing strain SYSU M60028T with Alsobacter soli SH9T and Alsobacter metallidurans SK200a-9T were 21.2% and 20.8% (Table S2), which were below the cutoff (70%) recommended as the criterion for interspecies identity (Wayne 1988). The ANI values comparing SYSU M60031T with Alsobacter soli SH9T and Alsobacter metallidurans SK200a-9T were 75.39% and 75.37% (Table S2), further suggesting the strain SYSU M60028T represents a novel species belonging to the genus Alsobacter.
According to the genome annotation results, the genome of the strain SYSU M60028T contained 4,264 protein-coding genes. The number of genes assigned to COG database and KEGG database were 3,304 and 3,732, respectively. COGs (Clusters of Orthologous Groups) categories distributions for the genes were presented in the Table S3. Amino acid transport and metabolism (COG category E) was the most abundant represented CDs (sequence coding for amino acids in protein) of strain SYSU M60028T, followed by general function prediction only (COG category R), function unknown (COG category S), and carbohydrate transport and metabolism (COG category G). According to the antiSMASH database, T3PKS, NRPS, NRPS-like and terpene gene clusters were four main clusters of the strain SYSU M60028T, which were responsible for the biosynthesis of secondary metabolites (Table S5). Strain SYSU M60028T had the potential to produce terpene, which offered new insight into biosynthesis of terpene in bacteria.
According to the annotation results of KEGG analysis, strain SYSU M60028T contained 14 genes which were relevant to the nitrogen cycle, including dissimilatory nitrate reduction, denitrification, and nitrification. Genome annotation results suggested that strain SYSU M60028T contained 4 genes (narGHI, nirK, norBC, and nosZ), which formed a complete denitrification pathway. Compared with type strains, Alsobacter soli SH9T also had the ability of denitrification, while Alsobacter metallidurans SK200a-9T did not have this complete pathway. This result was identified by the positive result of nitrate reduction test in API 20NE kit. Ntr C (a DNA-binding transcriptional regulator) was predicted in strain SYSU M60028T. The previous studies had indicated that Ntr C participated in nitrogenous compounds utilization, biological nitrogen fixation, the biosynthesis of biopolymer and maintenance on carbon–nitrogen balance (Hervas et al. 2008). Therefore, the strain might involve in nitrogen-transforming reactions and remove nitrogen, which meant this strain could play an important role in nitrogen cycle and reducing the eutrophication of nearshore waters (Falkowski et al. 2008). Strain SYSU M60028T also contained a complete assimilatory sulfur pathway (cysND, cysC, cysH, and cysJI), while Alsobacter metallidurans SK200a-9T and Alsobacter soli SH9T did not have this complete pathway. This result was consistent with the positive result of H2S production test and suggested the strain might reduce sulfate to sulfide, use sulfate as a terminal electron acceptor for the degradation of organic compounds, resulting in the production of sulphide, and kept the dynamics of sulfur cycle in balance (Muyzer et al. 2008). In addition, the genome contained 25 genes related to flagellar assembly. The diffusion of cells in semi-solid medium, and the flagellar observed by transmission electron microscope (Table S3) further supported the result of genome analysis.
Conclusion
The results of the phylogenetic analysis showed the strain SYSU M60028T was a member of Alsobacter. Combining the morphological and chemotaxonomic characteristics, the strain SYSU M60028T could be distinguished from other Alsobacter species. For example, the proportions of fatty acids and polar lipid composition or the utilization of carbon and nitrogen source of strain SYSU M60028T are significantly different from those of the related species. Compared with Alsobacter metallidurans SK200a-9T (4630) and Alsobacter soli SH9T (4751), Strain SYSU M60028T contained the least number of protein-coding genes (4290). According to the annotation results of KEGG analysis, strain SYSU M60028T contained the complete assimilatory sulfur pathway (cysND, cysC, cysH, and cysJI), while Alsobacter metallidurans SK200a-9T and Alsobacter soli SH9T did not have this complete pathway. Strain SYSU M60028T contained 4 genes (narGHI, nirK, norBC, and nosZ), which were formed a complete denitrification pathway. Compared with type strains, Alsobacter soli SH9T also had the ability of denitrification, while Alsobacter metallidurans SK200a-9T did not have this complete pathway. Thus, based on above results, we propose the strain SYSU M60028T as a novel species of the genus Alsobacter, for which the name Alsobacter ponti sp. nov. is proposed.
Description of Alsobacter ponti sp. nov
Alsobacter ponti (pon’ti. L. gen. masc. n. ponti, of the sea).
Cells are Gram-staining negative, aerobic, motile, and short rods (0.6–0.7 × 0.9–1.0 μm). Colonies on R2A agar are off-white, circular, and smooth. Able to grow at pH 6.0–8.0 (optimum, pH 7.0), 25–37 °C (optimum, 28 °C) and in the presence of 0–2% (w/v) NaCl (optimum, 0% NaCl). Positive for oxidase activity, but negative for milk coagulation and peptonization, Tweens (20, 40, 60, and 80), or catalase activity. Positive for sterase (C4), esterase lipase (C8), α-chymotrypsin, trypsin, alkaline phosphatase, cystine arylamidase, valine arylamidase and leucine arylamidase. Negative for hydrolysis of urea, cellulose and gelatin, reduction of nitrates to nitrites, lipase (C14), acid phosphatase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, and α-mannosidase. The predominant respiratory quinone is Q-10. The cellular polar lipids are phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, phosphatidylcholine, one unidentified aminolipid and three unidentified lipids. The major fatty acids (> 5% of total) are summed feature 8, C16:0 and C18:1 ω7c 11-methy1. The G + C content of the genome DNA is 69.9%.
The type strain, SYSU M60028T (= CGMCC 1.19341T = KCTC 92046T) was isolated from a sediment sample collected from the Pearl River estuary, Guangdong, China. The 16S rRNA gene sequences of strain SYSU M60028T has been submitted to GenBank with accession number ON881906. The accession numbers of raw data and assembly genome are SRR23021099 and JANCLU000000000, respectively.
Data and materials availability
The 16S rRNA gene sequences of strain SYSU M60028T has been deposited in GenBank under the accession number ON881906. Whole Genome Shotgun project of strain SYSU M60028T has been deposited at DDBJ/ENA/GenBank under accession number JANCLU000000000. The genome raw data of strain SYSU M60028T has been deposited in GenBank under the accession number SRR23021099. Five supplementary tables and five supplementary figures are included on the online supplementary file.
References
Athalye M, Noble WC, Minnikin DE (1985) Analysis of cellular fatty acids by gas chromatography as a tool in the identification of medically important Coryneform bacteria. J Appl Bacteriol 58:507–512
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single–cell sequencing. J Comput Biol 19:455–477
Bao Z, Sato Y, Fujimura R, Ohta H (2014) Alsobacter metallidurans gen. nov., sp. nov., a thallium-tolerant soil bacterium in the order Rhizobiales. Int J Syst Evol Microbiol 64:775–780
Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema Marnix H, Weber T (2021) antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49:W29–W35
Buck JD (1982) Nonstaining (KOH) method for determination of gram reactions of marine bacteria. Appl Environ Microbiol 44:992–993
Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH (2019) GTDB–Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36:1925–1927
Collins M, Jones D (1980) Lipids in the classification and identification of Coryneform bacteria containing peptidoglycans based on 2,4–diaminobutyric Acid. J Appl Microbiol 48:459–470
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in Actinomycetes and Corynebacteria. J Gen Microbiol 100:221–230
DE Minnikin CM, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47:87–95
Falkowski P, Fenchel T, Delong E (2008) The microbial engines that drive earth’s biogeochemical cycles. Science 320:1034–1039
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol 20:406–416
Galperin MY, Makarova KS, Wolf YI, Koonin EV (2014) Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269
Gonzalez C, Gutierrez C, Ramirez C (1978) Halobacterium vallismortis sp. nov. An amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24:710–715
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Hervas AB, Canosa I, Santero E (2008) Transcriptome analysis of Pseudomonas putida in response to nitrogen availability. J Bacteriol 190:416–420
Jiao JY, Fu L, Hua ZS, Liu L, Salam N, Liu PF, Lv AP, Wu G, Xian WD, Zhu QY, Zhou EM, Fang BZ, Oren A, Hedlund BP, Jiang HC, Knight R, Cheng L, Li WJ (2021) Insight into the function and evolution of the Wood-Ljungdahl pathway in Actinobacteria. ISME J 15:3005–3018
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589
Kanehisa M, Sato Y (2020) KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci 29:28–35
Kanehisa M, Sato Y, Kawashima M (2022) KEGG mapping tools for uncovering hidden features in biological data. Protein Sci 31:47–53
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kovacs N (1956) Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 178:703
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242-245
Li WJ, Xu P, Schumann P, Zhang YQ, Pukall R, Xu LH, Stackebrandt E, Jiang CL (2007) Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int J Syst Evol Microbiol 57:1424–1428
Li R, Li Y, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714
Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Li S, Yang H, Wang J, Wang J (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272
Muyzer G, Stams A (2008) The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol 6:441–454
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055
Richter M, Rosselló-Móra R, Glöckner FO, Peplies J (2016) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Smibert RM, Krieg NR (1994) Phenotypic characterization.In methods for general and molecular bacteriology. American Society For Microbiology, pp 611–651
Sun L, Liu H, Chen W, Huang K, Lyu W, Gao X (2018) Alsobacter soli sp. nov., a novel bacterium isolated from paddy soil, emended description of the genus Alsobacter and description of the family Alsobacteraceae fam. nov. Int J Syst Evol Microbiol 68:3902–3907
Tamaoka J, Katayama-Fujimura Y, Kuraishi H (1983) Analysis of bacterial menaquinone mixtures by high performance liquid chromatography. J Appl Bacteriol 54:31–36
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wayne LG (1988) International committee on systematic bacteriology announcement of the report of the ad hoc committee on reconciliation of approaches to bacterial systematics. J Appl Bacteriol 64:433–434
Xu P, Li WJ, Tang SK, Zhang YQ, Chen GZ, Chen HH, Xu LH, Jiang CL (2005) Naxibacter alkalitolerans gen. nov., sp nov., a novel member of the family 'Oxalobacteraceae’ isolated from China. Int J Syst Evol Microbiol 55:1149–1153
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole–genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Acknowledgements
The authors are grateful to Prof. Aharon Oren (The Hebrew University of Jerusalem, Israel) for helping with the etymology. This work was supported by the National Natural Science Foundation of China (No. 32200090), Innovation Group Project of Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) (Nos. 311021006, 311021004), and Guangdong Basic and Applied Basic Research Foundation, China (Nos. 2023A1515012270, 2022A1515010756).
Author information
Authors and Affiliations
Contributions
WJL designed research and project outline. QQD, JLL and SHL performed isolation, deposition and polyphasic taxonomy. PDW, XQL and YY performed genome analysis. QQD, ZWY and WJL drafted the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Deng, QQ., Luo, XQ., Li, SH. et al. Alsobacter ponti sp. nov., a novel denitrification and sulfate reduction bacterium isolated from Pearl River sediment. Antonie van Leeuwenhoek 116, 987–994 (2023). https://doi.org/10.1007/s10482-023-01861-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-023-01861-w