Abstract
Strain OCN044T was isolated from the homogenised tissue and mucus of an apparently healthy Acropora cytherea coral fragment collected from the western reef terrace of Palmyra Atoll in the Northern Line Islands and was taxonomically evaluated with a polyphasic approach. The morphological and chemotaxonomic properties are consistent with characteristics of the genus Vibrio: Gram-stain-negative rods, oxidase- and catalase-positive, and motile by means of a polar flagellum. Strain OCN044T can be differentiated as a novel subspecies based on 21 differences among chemotaxonomic features (e.g., fatty acids percentages for C12:0 and C18:1 ω7c), enzymatic activities (e.g., DNase and cystine arylamidase), and carbon sources utilized (e.g., L-xylose and D-melezitose) from its nearest genetic relative. Phylogenetic analysis and genomic comparisons show close evolutionary relatedness to Vibrio tetraodonis A511T but the overall genomic relatedness indices identify strain OCN044T as a distinct subspecies. Based on a polyphasic characterisation, differences in genomic and taxonomic data, strain OCN044T represents a novel subspecies of V. tetraodonis A511T, for which the name Vibrio tetraodonis subsp. pristinus subsp. nov. is proposed. The type strain is OCN044T (= LMG 31895T = DSM 111778T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Members of the family Vibrionaceae, with over 190 accepted species, are Gram-stain-negative Gammaproteobacteria predominantly found in marine and estuarine environments (Pruzzo et al. 2005; Oliver et al. 2012; Jiang et al. 2021). A high abundance are detected in eutrophic aquatic ecosystems and colonised marine organisms (Thompson et al. 2004). Vibrio species are implicated as etiological agents of disease in coral, bivalves, shrimp, fish, and humans (Baker-Austin et al. 2018). Noteworthy examples of marine Vibrio pathogens and their hosts include Vibrio pectenicida and scallops (Pecten maximus) (Lambert et al. 1998), Vibrio ostreicida and Flat Oysters (Ostrea edulis) (Prado et al. 2005, 2014), Vibrio tasmaeniensis and Pacific oysters (Crassostrea gigas) (Gay et al. 2004), and Vibrio coralliilyticus and various coral, urchins, and bivalves (Ben-Haim et al. 2003; Sussman et al. 2008; Ushijima et al. 2014, 2016; Vezzulli et al. 2010; Estes et al. 2004; Balbi et al. 2019; Kesarcodi-Watson et al. 2009; Li et al. 2020; Nguyen et al. 2019; Richards et al. 2015). In contrast, some Vibrio species are considered mutualistic or commensal symbionts, like the bioluminescent Aliivibrio fischeri (Ruby 1996; McFall-Ngai et al. 2012). A. fisheri can colonise a specialised organ of the squid Euprymna scolopes, which facilitates counter-luminescent camouflage during the night. Genomic analysis of the recently described species Vibrio tetraodonis A511T, isolated from the marine pufferfish Sphoeroides spengleri, identified gene families and clusters within the strain that may confer advantages to the pufferfish host (Azevedo et al. 2021). Additional Vibrio strains have been pursued as potential probiotics; for example, growth of the microalga Chaetoceros muelleri is improved when co-cultured with Vibrio alginolyticus C7b (Gomez-Gil et al. 2002; Verschuere et al. 2000; Sawabe et al. 2003; Riquelme et al. 2001). Researchers have also begun to assess the roles and interactions of mutualistic/commensal Vibrio in the coral holobiont (Koenig et al. 2011, Arboleda and Reichardt, 2009, Kvennefors et al. 2010). Studies on mutualistic Vibrio are less frequent than pathogenesis research, due in part to stigmatisation of this genus as being commonly associated with disease, which highlights the need for and novelty of additional work on non-pathogenic Vibrio species.
Strain OCN044T was isolated from an apparently healthy Acropora cytherea colony and, based on partial 16S rRNA gene sequencing alone, strain OCN044T was originally and incorrectly designated V. nereis strain OCN044 (Ushijima et al. 2016). This work assessed the virulence of various V. coralliilyticus strains and used strain OCN044T as a negative control bacterium during infection trials, in which it did not induce obvious disease signs in apparently healthy Acropora cytherea or Montipora capitata coral fragments. Based on its isolation and inability to infect its host in laboratory trials, strain OCN044T may be a member of the coral microflora. Much of the research on Vibrio species related to coral focuses on disease and pays little attention to members of the non-diseased microbial community. The continued study of this and other non-pathogenic Vibrio species may provide insight into interspecies interactions within the coral holobiont. The results of a polyphasic approach to characterize strain OCN044T support the classification of the isolate as a novel subspecies of V. tetraodonis A511T, for which the name Vibrio tetraodonis subsp. pristinus subsp. nov. is proposed.
Materials and methods
Isolation, cultivation, and phenotypic characterisation
Strain OCN044T was isolated from a fragment of healthy A. cytherea coral harvested from the western reef terrace of Palmyra Atoll in the Northern Line Islands as previously described (Ushijima et al. 2016). Briefly, the fragment was crushed in autoclaved seawater and the resulting crushate was plated on glycerol seawater agar (4 g l− 1 tryptone, 2 g l− 1 yeast extract, and 2 g l− 1 glycerol in a liter of seawater) as previously described (Ushijima et al. 2016). The plated crushate was incubated at 29 °C overnight and colonies were purified on the same medium.
Morphological, physiological, and culture-based characterisation of strain OCN044T and the related species V. tetraodonis A511T and Vibrio aquimaris DSM 109633T were carried out as previously described with some modifications (Beurmann et al. 2017; Azevedo et al. 2021; Franco et al. 2020). V. tetraodonis A511T and Vibrio aquimaris DSM 109633T were provided by the Collection of Aquatic Microorganisms (CAIM) in Mazatlan, Sinaloa, Mexico, and the Belgian Co-ordinated Collection of Micro-Organisms (BCCM) LMG collection in Ghent, Belgium, respectively. Strain properties were determined using lysogeny broth (LB) medium (Sigma-Aldrich, St. Louis, MO), glycerol artificial seawater medium buffered with 50 mM Tris base to pH 8.3 (GASW-Tris) and solidified with 1.5% agar (Ushijima and Häse, 2018), and Thiosulfate Citrate Bile Salts Sucrose (TCBS) agar (HiMedia, West Chester, PA), which was prepared according to the manufacturer’s instructions and supplemented with NaCl up to a 3% final concentration. Strains were routinely cultured at 28 °C in GASW-Tris, solidified with 1.5% agar as needed or shaken at 150 RPM overnight unless otherwise noted. Cell morphology was examined by transmission electron microscopy (TEM). Cells from an overnight culture of strain OCN044T grown in GASW-Tris at 28 °C were deposited on Formvar-coated grids, fixed with 1% uranyl acetate, and imaged on a Hitachi HT770 TEM at 100 kV. Swimming and swarming motility were determined on GASW-Tris plates supplemented with 0.26% or 1.5% agar, respectively; plates were incubated at 28 °C for 48 h and cultures were evaluated for movement outward from the inoculation site. Swimming was additionally assessed via the hanging drop method. Anaerobic growth was assessed on GASW-Tris plates incubated at 28 °C for 48 h using the GasPak System according to the manufacturer’s instructions (BD, Franklin Lakes, NJ). The pH range supporting growth was determined with GASW medium without TRIS base adjusted to pH 4.0–10.0 in increments of 0.5 using the following buffers: pH 4.0–6.0, citrate/Na2HPO4; pH 6.0–8.0, phosphate buffer; pH 9.0–10.0, glycine/NaOH (McCauley et al. 2015). The pH was adjusted prior to sterilisation, verified after sterilisation prior to inoculation, and verified again after incubation. The temperature range supporting growth was assessed on GASW-Tris medium incubated from 0 to 45 °C in 5 °C increments. Tolerance to NaCl was determined on LB medium buffered to pH 8.3 with 50 mM Tris base and supplemented with 0–10% NaCl in 0.5% increments from 0 to 2% and 1% increments from 3 to 10%.
Carbon source utilisation and enzyme activity tests were carried out using the API 50 CH, API ZYM, and API 20E kits according to the manufacturer’s instructions (BioMérieux, Marcy-l’Étoile, France), the only modification was the final concentration of the suspension medium for these kits was adjusted to 3% NaCl. Utilisation of citrate was determined on Simmon’s citrate medium (BD) supplemented with 3% NaCl according to the manufacturer’s instructions. Catalase activity was assessed with the addition of 3% H2O2 to colonies grown on GASW medium, with the formation of bubbles interpreted as a positive result. Determination of oxidase activity was carried out with BD BBL DrySlide tests according to the manufacturer’s instructions (BD). Nitrate and nitrite reduction tests were conducted as previously described [36], with the medium supplemented with 3% NaCl. SIM, MR-VP, and urease media (BD) were prepared with 3% NaCl and the tests were conducted according to the manufacturer’s instructions after two days of incubation at 28 °C. Substrate degradation activity was assessed with GASW-Tris supplemented with 1 % (w/v) Tweens 20, 40, 60, and 80, and clearance zones around colonies were determined after two days of incubation at 28 °C. Gelatinase activity was assessed with gelatinase medium (BD) supplemented with 3% NaCl after two days of incubation at 28 °C according to the manufacturer’s instructions. DNase activity was determined with DNase test agar with methyl green (BD) supplemented with 3% NaCl after two days of incubation at 28 °C.
Antibiotic susceptibility testing
Antibiotic susceptibility was determined by disc diffusion assays on Mueller-Hinton agar (BD) plates supplemented with 3% NaCl. After inoculation, plates were incubated at 28 °C for 18 h and zones of inhibition were recorded. Antibiotics were purchased from Thermo Fisher Scientific (Waltham, MA, USA), Hardy Diagnostics (Santa Maria, CA, USA), and Sigma-Aldrich and used in the following doses: ampicillin (Amp; 10 µg), penicillin G (Pn; 10 units), carbenicillin (Carb; 100 µg), vancomycin (Vanc; 30 µg), erythromycin (Ery; 15 µg), tetracycline (Tet; 30 µg), oxytetracycline (Oxy; 30 µg), streptomycin (Strep; 30 µg), spectinomycin (Spec; 100 µg), trimethoprim (Trim; 5 µg), gentamycin (Gm; 10 µg), chloramphenicol (Cm; 30 µg), kanamycin (Kan; 30 µg), neomycin (Neo; 30 µg), and vibriostatic agent 0129 (150 µg).
Chemotaxonomic characterisation
Cellular fatty acid analysis was carried out at the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) using strain OCN044T, V. tetraodonis A511T, and V. aquimaris DSM 109633T cells harvested during the exponential growth phase in GASW at 28 °C. Fatty acid methyl esters (FAMEs) were separated and identified according to the MIDI Sherlock Microbial Identification System, by Microbial ID (MIDI, Microbial ID, Newark, DE), and the published profiles of other related strains were used for comparison.
Phylogenetic analysis and genomic characterisation
Genomic DNA was isolated from strain OCN044T and V. tetraodonis A511T via phenol chloroform extraction and used in a PCR with the primers 8F (5’- AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’- TACGGYTACCTTGTTACGACTT − 3’) to amplify a fragment of the 16S rRNA gene (Aebischer et al. 2006; Sambrook 2001). The amplified product was purified with the Wizard SV Gel and PCR Clean-up System (Promega, Madison, WI) and sequenced by Sanger Sequencing in the Biotechnology Center at Southern Oregon University using the same primers. The 16S rRNA gene sequences of strain OCN044T and 44 related strains were aligned using the MUSCLE algorithm, and a maximum likelihood phylogenetic tree was generated using the MEGAX software package (Kumar et al. 2018).
Multilocus sequence analysis (MLSA) was conducted as previously described (Jiang et al. 2021). Briefly, coding sequences of the ftsZ, gapA, gyrB, mreB, pyrH, recA, rpoA, and topA genes were retrieved from the genome of strain OCN044T (Loughran et al. 2020) and from those of 44 related strains (Table S2), aligned and concatenated using MEGAX, and a maximum likelihood phylogenetic tree was generated.
A whole genome-based taxonomic analysis was performed as follows. Coding regions in all genomes were predicted and annotated with Prokka (version 1.14.5) set to default parameters (Seemann 2014). Phylogenetic analysis of the predicted amino acid sequences was conducted using PhyloPhlAN (version 3.0) with all parameters set to default except the “--diversity” parameter, which was set to “low” (Asnicar et al. 2020). A custom workflow was run in PhyloPhlAN using Diamond to map amino acid sequences and reference marker gene sets (Buchfink et al. 2015), MAFFT to align individual marker genes (Katoh and Standley, 2013), and trimAl to trim gaps and phylogenetically uninformative sites and to concatenate trimmed alignments (Capella-Gutiérrez et al. 2009). The concatenated alignment produced by the PhyloPhlAN pipeline was used to construct a maximum likelihood phylogeny of Vibrio species using IQ-Tree (version 2.0.3) (Minh et al. 2020). A total of 380 core conserved marker genes shared between 43 Vibrio genomes and 1 outgroup were identified by PhyloPhlAN, and the final concatenated alignment of trimmed alignments consisted of 90,492 characters. Model testing of the aligned and concatenated amino acid sequences was first performed using IQ-Tree with the “-m TESTONLY” option and the best-fit model “LG + F + G4” was used for phylogenetic inference. A maximum likelihood phylogenetic tree was constructed with IQ-Tree using the following parameters: −m LG + F + G4 -alrt 1000 -bb 1000 -T 24, with IQ-Tree run on the Pegasus high-performance computing cluster at The George Washington University. The resulting maximum likelihood phylogenomic tree was drawn using the ETE3 toolkit and a custom Python script (version 3.1.1), and edited in Inkscape and Adobe Illustrator (Huerta-Cepas et al. 2016).
DNA-DNA Hybridisation (isDDH) was conducted in silico using GGDC 2.1 with the default parameters (Meier-Kolthoff et al. 2013). ANIb and FastANI tools were run with default parameters to calculate the pairwise average nucleotide identities (ANI) between eight closely related Vibrio genomes used for phenotypic comparison.
Results and discussion
Isolation and ecology
In the summer of 2011, our group embarked on a research trip to Palmyra Atoll in the Northern Line Islands focused on identifying pathogens involved in a tissue loss disease affecting Acroporids. It was necessary to isolate bacteria from healthy coral to use as controls in infection trials on island because we were quite limited in our bacterial importation abilities. The vast majority of colonies isolated from apparently healthy Acropora cytherea fragments demonstrated a fast-growing swarming phenotype and were incapable of growing on TCBS agar; this morphology was absent from both diseased Acropora cytherea fragments and seawater. One such isolate was utilized as the bacterial control in infection trials. Due to restrictions on the number of bacterial isolates we were allowed to export from Palmyra Atoll, we returned with only pathogenic isolates and the strain utilized as the bacterial control in infection trials, which is denoted strain OCN044T (Ushijima et al. 2016). A comparison of the 16S sequence of strain OCN044T with a subset of metagenomic sequencing projects conducted on members of the Acropora genus and other coral and uploaded to NCBI, indicated at least eight instances of 100% sequence identity of 16S sequences derived from coral with the strain OCN044T 16S (Table S1). No such sequences with 100% identity to the strain OCN044T 16S were present in seawater or other collected material nearby in sequencing projects where the metadata was sufficient to determine the nature of individual sets of reads. These data are consistent with the potential of strain OCN044T associating with coral as a component of the holobiont.
Morphology, physiology, and biochemical analyses
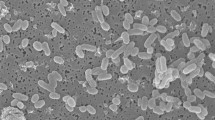
Strain OCN044T exhibited phenotypic and chemotaxonomic characteristics consistent with the genus Vibrio: Gram-stain-negative, rod-shaped cells, each with a single polar flagellum (Fig. 1), capable of swimming and swarming motility, anaerobic growth, and presence of the major fatty acids C12:0, C14:0, C16:0, summed feature 3 (C16:1ω7c and/or C16:1ω6c), C18:1ω7c, and/or C18:1ω6c, which are consistent with the genus Vibrio (Lambert et al. 1983). Growth of cream coloured, smooth, raised, opaque colonies with entire margins occurred from 10 to 37 °C, 1–6% NaCl, and at pH 6.5-9.0. Optimal growth was observed from 25 to 30 °C, 1–3% NaCl, and pH 7.5–8.5. Prolonged growth on GASW plates can result in colonies changing to a light-yellow colour but no diffusible pigment is observed, and strain OCN044T does not grow on TCBS agar even with NaCl supplementation (1–3% NaCl final concentration). The inability to grow on TCBS agar is shared with the related strain V. pectenicida CAIM 594T but is in contrast to the production of green colonies by V. tetraodonis A511T and Vibrio aquimaris DSM 109633T on the medium (Lambert et al. 1998). Strain OCN044T was also unable to grow in 0.5 or 8% NaCl, which differentiates it from related species including V. tetraodonis A511T, Vibrio aquimaris DSM 109633T, V. pectenicida DSM 19584T, and Vibrio caribbeanicus ATCC BAA-2122T (Hoffmann et al. 2012). Further, only strain OCN044T, V. pectenicida DSM 19584T, and V. ostreicida DSM 21433T (Prado et al. 2014) swarm while other related type strains do not display this motility. Notable characteristics that differentiate strain OCN044T from closely related Vibrio species are shown in Table 1.
In biochemical assessments, strain OCN044T is positive for oxidase, catalase, gelatinease, DNase, alkaline phosphatase, esterase (C4), esterase lipase (C8), lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, alpha-chymotrypsin, acid phosphatase, napthol ASBI phosphohydrolase, alpha glucosidase, and N-acetyl-beta-glucosamindase activites, ferments glucose, hydrolyses tween 40, 60, and 80, utilises glycerol, D-glucose, D-fructose, D-mannose, N-acetylglucosamine, D-maltose, D-trehalose, amidon (starch), and glycogen as carbon sources, is negative for both nitrate and nitrite reduction, urease activity, and is not observed to produce indole or acetoin (Table 1 and Table S2). Only strain OCN044T demonstrated cystine arylamidase activity among the type strains tested and used for comparison. Complete carbon source utilisation and enzyme activity profiles are presented in Tables S3&S4 in the Supplementary Material. Strain OCN044T is susceptible to erythromycin, tetracycline, spectinomycin, trimethoprim, chloramphenicol, and vibriostatic agent 0129.
Chemotaxonomic characteristics
The predominant cellular fatty acids of strain OCN044T are typical for the genus Vibrio (including C12:0 (3.6%), C14:0 (6.4%), C16:0 (17.6%), summed feature 3 [C16:1ω7c and/or C16:1ω6c] (41.9%), C18:1ω7c (14.0%), and/or C18:1ω6c (3.0%)) (Table S5) (Lambert et al. 1983). The FAME profiles of closely related species are presented in Table S6 in the Supplementary Material, and notable differences in fatty acid content percentages are observed with C12:0, C12:0 3OH, C16:0, C18:1ω7c, and C16:1ω7c and/or C16:1ω6c.
Phylogenetic analysis and genomic characteristics
Prior sequencing of the strain OCN044T genome facilitated an assessment of the relevant features for existing within the coral holobiont (Loughran et al. 2020). Microbes on coral may quorum sense (QS) to maintain a competitive advantage during antagonistic interactions and nutrient source competition (Golberg et al. 2013). This strain potentially has the components of several QS systems (Table S7) but, like its close relative V. tetraodonis A511T, lacks luxI and luxM homologs, which could indicate alternative systems that do not rely on AI-1 or AHLs. Iron is a critical component of Vibrio metabolism, as well as a major limiting factor in oligotrophic marine environments (e.g., coral reefs), and strain OCN044T harbours genes for this purpose (Johnson 2013). Changes in seawater due to runoff and freshwater input can influence the osmoregulation of microbes, which can lead to changes in the microbial composition of the coral holobiont. Previous work indicates that, as salinity is modulated to alter osmotic conditions, coral communities can shift toward increasing levels of Vibrios (Röthig et al. 2016); strain OCN044T contains genes that would aid in this manner of osmoregulation to maintain its place in the holobiont. Coral-associated microbes have been found to exhibit chemotaxis as a means of establishing and maintaining themselves as part of the holobiont (Tout et al. 2015), and strain OCN044T has 34 proteins with potential MCP signal transduction domains as well as additional genes to facilitate chemotaxis. Together, these genomic components could promote strain OCN044T to persist as a constituent of the coral holobiont.
Analysis of the 16S rRNA placed strain OCN044T in the genus Vibrio and clusters with V. aquimaris DSM 109633T and V. tetraodonis A511T (Fig. S1 in the Supplementary Material). Comparison of the strain OCN044T 16S rRNA sequence indicates 98.56% identity with V. tetraodonis A511T, which is below the 98.65% same-species identity threshold (Kim et al. 2014). Within the complete genome of V. aquimaris DSM 109633T, nine copies of the 16S rRNA sequence are present and comparison of the strain OCN044T 16S rRNA sequence indicates 98.84–99.25% identity, which is slightly above the 98.65% same-species identity threshold. A multilocus sequence analysis (MLSA) was used to assess the relationship between strain OCN044T and other Vibrio species (Ushijima et al. 2016; Jiang et al. 2021). This analysis placed strain OCN044T next to V. tetraodonis A511T and in a cluster with V. aquimaris DSM 109633T and V. pectenicida CAIM 594T (Fig. S2 in the Supplementary Material). Grouping with V. pectenicida CAIM 594T in the MLSA indicates that strain OCN044T belongs within the eponymous Pectenicida clade. The G + C content of the genome of strain OCN044T is 42.40%, which is in line with reported values for related strains (Table 1) and falls within the 38–51 mol% typically observed for this genus (Farmer et al. 2005).
For a more robust analysis of phylogeny, a whole genome-based taxonomic analysis was performed. The resulting phylogenomic tree placed strain OCN044T as sister to V. tetraodonis A511T with a 100% bootstrap value (Fig. 2). This result contrasts with the 16S tree but is consistent with the MLSA tree where V. tetraodonis A511T is a closer relative to strain OCN044T than V. aquimaris DSM 109633T, which is not unexpected given the higher reliability of species relatedness in MLSA analyses than 16S phylogenies in the family Vibrionaceae. The grouping of strains near strain OCN044T in Fig. 2, further informed by the results of the MLSA in Figure S2, indicated that V. tetraodonis A511T, V. aquimaris DSM 109633T, V. pectenicida DSM 19584T, V. caribbeanicus ATCC BAA-2122T, V. ostreicida DSM 21433T, V. coralliilyticus ATCC BAA-450T, Vibrio neptunius LMG 20536T represented the most closely related type strains and served as the basis of comparison throughout this study.
A maximum likelihood phylogenomic tree using 381 conserved core genes (90,623 characters) shared between the 43 Vibrio species and the outgroup, Photobacterium damselae subsp. damselae strain ATCC 33539T. Ultrafast bootstrap (UFBoot) values were determined and, though not shown, are 100% at each node (Hoang et al. 2018). Accession numbers of genomes are shown in parentheses. Bar, number of substitutions per nucleotide position
To assess genomic similarity, in silico DNA-DNA hybridisation (isDDH) was conducted and ANI values compared. The isDDH values ranged from 18.1 to 63.8% between strain OCN044T and the other related type strains used for comparison, which are below the 70% cutoff for members of different species (Table 2) (Goris et al. 2007). The ANI values between the eight closely related Vibrio genomes named above (Table 3) were used for genomic comparison (Jain et al. 2018; Richter et al. 2015). When compared to strain OCN044T, an average ANI value of 95.3% was recorded for V. tetraodonis A511T, which falls within the threshold range of 95–96% commonly used to delineate species (Varghese et al. 2015; Jain et al. 2018; Chun et al. 2018; Ciufo et al. 2018; Kim et al. 2014).
On the basis of 16S rRNA sequence comparison alone, strain OCN044T and V. aquimaris DSM 109633T would be considered more related. However, the advent of genome sequencing has resulted in a transition from 16S rRNA-based phylogenies to whole-genome comparisons as the basis for taxonomy due to greater resolution from longer inputs (Parks et al. 2018; Murray et al. 2020; Hugenholtz et al. 2021). It has also been noted that the 16S rRNA gene is more conserved than the whole genome and does not provide sufficient resolution among species, whereas ANI and other whole-genome comparisons facilitate finer resolution among species (Varghese et al. 2015; Jain et al. 2018). Strain OCN044T and V. tetraodonis A511T display a 95.3% average ANI for the concordant pair and isDDH of 63.8%, whereas strain OCN044T and V. aquimaris DSM 109633T display a 94.5% average ANI for the concordant pair and isDDH of 58.9%. Based on these data, it is clear that strain OCN044T is more closely related to V. tetraodonis A511T than V. aquimaris DSM 109633T. While the ANI cutoff for different species is 95–96% and the isDDH cutoff is 70%, previous work has indicated that the isDDH cutoff for subspecies classification is 79–80% (Meier-Kolthoff et al. 2014). Additional work has indicated that conspecific genomes have ANI values of ≥ 97% (Van Rossum et al. 2020). Our data demonstrate that strain OCN044T is below the species and subspecies cutoff values for isDDH when compared to V. tetraodonis A511T, but is also not conspecific with V. tetraodonis A511T. Because 95.3% ANI is slightly higher than the 95% lower bound for species demarcation, we define OCN044T as a subspecies rather than a novel species to remain within accepted genomic parameters. When considered with the other results, these data fulfil the currently accepted genomic criteria to indicate that strain OCN044T is sufficiently genetically divergent to be considered to represent a novel subspecies.
Strain OCN044T may be distinguished from phylogenetically related strains by its cystine arylamidase activity and its inability to grow at 8% NaCl. Strain OCN044T differs from the most closely related strain, V. tetraodonis A511T, in growth on TCBS agar in 0.5% NaCl, or at 37 °C; swarming motility; in gelatinase, DNase, arginine dihydrolase, beta galactosidase, and N-acetyl-beta-glucosaminidase activities; and in the utilization of L-xylose, amygdalin, arbutin, salicin, D-lactose, inulin, D-melezitose, gentiobiose, and D-turanose. Additionally, while V. tetraodonis A511T was isolated from a pufferfish and is thought to act as a potentially beneficial component of its microbiome (Azevedo et al. 2021), strain OCN044T was isolated from coral and identical 16S sequences are found in metagenomic projects assessing the coral holobiont, which indicates the potential for these two strains to associate with different hosts. The preceding summary of characteristics and genomic analyses support the inclusion of strain OCN044T in the genus Vibrio and demonstrate that it exists as a distinct and novel subspecies, for which the name Vibrio tetraodonis subsp. pristinus is proposed.
Description of Vibrio tetraodonis subsp. pristinus subsp. nov.
Vibrio tetraodonis subsp. pristinus (pris.tin’us L. nom. n. pristinus, meaning pristine; referring to the pristine nature of the reefs surrounding Palmyra Atoll, the location from which the strain was isolated).
The cells are Gram-stain-negative, non-spore-forming rods, motile by a single polar flagellum with swimming and swarming activity. Colonies are cream-colored and opaque on GASW-Tris agar, with growth at 10–37 °C, 1–6% NaCl, and at pH 5.5–10; they do not grow on TCBS agar. Luminescence is not observed. Facultative anaerobe that ferments glucose but not mannose, inositol, sorbitol, rhamnose, sucrose, melibiose, amygdalin, or arabinose. Positive for oxidase and catalase activity, does not reduce nitrate or nitrite, does not utilise citrate, and is positive for gelatinase activity. Does not produce acetoin, indole, hydrogen sulfide, and does not demonstrate β-galactosidase, arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase, urease, or tryptophan deaminase activity. Tween 40, 60, and 80 are hydrolysed but Tween 20 is not. Positive for gelatinase, DNase, alkaline phosphatase, esterase (C4), esterase lipase (C8), lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, alpha-chymotrypsin, acid phosphatase, napthol ASBI phosphohydrolase, alpha glucosidase, and N-acetyl-beta-glucosamindase; and is negative for alpha and beta galactosidase, beta glucuronidase, beta glucosidase, alpha mannosidase, and alpha fucosidase. Utilised carbon sources include glycerol, D-glucose, D-fructose, D-mannose, N-acetylglucosamine, D-maltose, D-trehalose, amidon (starch), and glycogen; but not erythritol, D- or L-arabinose, D-ribose, D- or L-xylose, D-adonitol, Methyl-BD-Xylopyranoside, D-galactose, L-sorbose, L-rhamnose, dulcitol, inositol, D-mannitol, D-sorbitol, Methyl-ad-Mannopyranoside, Methyl-ad-Glucopyranoside, amygdalin, arbutin, esculin, salicin, D-cellobiose, D-lactose, D-meliboise, D-saccharose (sucrose), inulin, D-melezitose, D-raffinose, xylitol, gentiobiose, D-turanose, D-lyxose, D-tagatose, D-fuccose, L-fuccose, D-arabitol, L-arabitol, potassium gluconate, potassium 2-ketogluconate, potassium 5-ketogluconate.
Dominant fatty acids are C16:0 (17.6%), C18:1 ω7c (14.0%), and C16:1 ω7c and/or C16:1 ω6c (41.9%). Susceptible to erythromycin, tetracycline, spectinomycin, trimethoprim, chloramphenicol, and vibriostatic agent 0129, but resistant to ampicillin, penicillin G, carbenicillin, vancomycin, oxytetracycline, streptomycin, gentamycin, kanamycin, and neomycin.
The type strain, OCN044T (= LMG 31895T = DSM 111778T), was isolated from the tissues of a fragment of apparently healthy A. cytherea coral off the western reef terrace of Palmyra Atoll in the Northern Line Islands. The DNA G + C content of the type strain is 42.40%.
Emended description of Vibrio tetraodonis subsp. tetraodonis subsp. nov.
Characteristics are as those given for the species description by Azevedo et al. (2021) with the following additions. Growth occurs at pH 6.0–10.0 with optimum growth between pH 7.0–9.0. Swimming but not swarming motility. Ferments glucose and Tween 40, 60, and 80 are hydrolysed but Tween 20 is not. Does not produce hydrogen sulfide, reduce nitrate or nitrite, or utilise citrate. Negative for DNase, gelatinase, urease, cystine arylamidase, alpha galactosidase, beta glucuronidase, beta glucosidase, N-acetyl-beta-glucosamindase, alpha mannosidase, and alpha fucosidase. Positive for alkaline phosphatase, esterase (C4), esterase lipase (C8), lipase (C14), leucine arylamidase, valine arylamidase, trypsin, alpha-chymotrypsin, acid phosphatase, napthol ASBI phosphohydrolase, beta galactosidase, and alpha glucosidase. Utilised carbon sources include glycerol, D-xylose, Methyl-BD-Xylopyranoside, D-glucose, D-fructose, D-mannose, Methyl-ad-Mannopyranoside, Methyl-ad-Glucopyranoside, N-acetylglucosamine, amygdalin, arbutin, salicin, D-maltose, D-lactose, D-trehalose, inulin, D-melezitose, amidon (starch), glycogen, gentiobiose, and D-turanose; but not erythritol, D- or L-arabinose, D-ribose, L-xylose, D-adonitol, D-galactose, L-sorbose, L-rhamnose, dulcitol, inositol, D-mannitol, D-sorbitol, esculin, D-cellobiose, D-meliboise, D-saccharose (sucrose), D-raffinose, xylitol, D-lyxose, D-tagatose, D-fuccose, L-fuccose, D-arabitol, L-arabitol, potassium gluconate, potassium 2-ketogluconate, potassium 5-ketogluconate.
Dominant fatty acids are C16:0 (18.7%), C18:1 ω7c (10.4%), and C16:1 ω7c and/or C16:1 ω6c (42.4%). Susceptible to erythromycin, gentamycin, kanamycin, trimethoprim, chloramphenicol, vancomycin, and vibriostatic agent 0129, but resistant to ampicillin, penicillin G, carbenicillin, tetracycline, oxytetracycline, streptomycin, spectinomycin, and neomycin. The type strain is A511T (= CBAS 712T = A511T).
References
Aebischer T, Fischer A, Walduck A, Schlötelburg C, Lindig M, Schreiber S, Meyer TF, Bereswill S, Göbel UB (2006) Vaccination prevents Helicobacter pylori-induced alterations of the gastric flora in mice. FEMS Immunol Med Microbiol 46:221–229
Arboleda M, Reichardt W (2009) Epizoic communities of prokaryotes on healthy and diseased scleractinian corals in Lingayen Gulf, Philippines. Microb Ecol 57:117–128
Asnicar F, Thomas AM, Beghini F, Mengoni C, Manara S, Manghi P, Zhu Q, Bolzan M, Cumbo F, May U, Sanders JG, Zolfo M, Kopylova E, Pasolli E, Knight R, Mirarab S, Huttenhower C, Segata N (2020) Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat Commun 11:2500
Azevedo GPR, Mattsson HK, Lopes GR, Vidal L, Campeão M, Tonon LAC, Garcia GD, Tschoeke DA, Silva BS, Otsuki K, Gomez-Gil B, Swings J, Thompson FL, Thompson CC (2021) Vibrio tetraodonis sp. nov.: genomic insights on the secondary metabolites repertoire. Arch Microbiol 203:399–404
Baker-Austin C, Oliver JD, Alam M, Ali A, Waldor MK, Qadri F, Martinez-Urtaza J (2018) Vibrio spp. infections. Nat Rev Dis Primers 4:1–19
Balbi T, Auguste M, Cortese K, Montagna M, Borello A, Pruzzo C, Vezzulli L, Canesi L (2019) Responses of Mytilus galloprovincialis to challenge with the emerging marine pathogen Vibrio coralliilyticus. Fish Shellfish Immunol 84:352–360
Ben-Haim Y, Thompson FL, Thompson CC, Cnockaert MC, Hoste B, Swings J, Rosenberg E (2003) Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol 53:309–315
Beurmann S, Ushijima B, Svoboda CM, Videau P, Smith AM, Donachie SP, Aeby GS, Callahan SM (2017) Pseudoalteromonas piratica sp. nov., a budding, prosthecate bacterium from diseased Montipora capitata, and emended description of the genus Pseudoalteromonas. Int J Syst Evol Microbiol 67:2683–2688
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, De Meyer S, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Ciufo S, Kannan S, Sharma S, Badretdin A, Clark K, Turner S, Brover S, Schoch CL, Kimchi A, DiCuccio M (2018) Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int J Syst Evol Microbiol 68:2386–2392
Estes RM, Friedman CS, Elston RA, Herwig RP (2004) Pathogenicity testing of shellfish hatchery bacterial isolates on Pacific oyster Crassostrea gigas larvae. Dis Aquat Organ 58:223–230
Farmer JJ, Janda JM, Brenner FW, Cameron DN, Birkhead KM (2005) Genus I. Vibrio Pacini 1854, 411AL. In: Garrity GM, Brenner DJ, Krieg NR, Staley JR (eds) Bergey’s manual of systematic bacteriology. Springer, New York, pp 494–546
Franco A, Rückert C, Blom J, Busche T, Reichert J, Schubert P, Goesmann A, Kalinowski J, Wilke T, Kämpfer P, Glaeser SP (2020) High diversity of Vibrio spp. associated with different ecological niches in a marine aquaria system and description of Vibrio aquimaris sp. nov. Syst Appl Microbiol 43:126123
Gay M, Berthe FC, Le Roux F (2004) Screening of Vibrio isolates to develop an experimental infection model in the Pacific oyster Crassostrea gigas. Dis Aquat Organ 59:49–56
Golberg K, Pavlov V, Marks RS, Kushmaro A (2013) Coral-associated bacteria, quorum sensing disrupters, and the regulation of biofouling. Biofouling 29:669–682
Gomez-Gil B, Roque A, Velasco-Blanco G (2002) Culture of Vibrio alginolyticus C7b, a potential probiotic bacterium, with the microalga Chaetoceros muelleri. Aquaculture 211:43–48
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS (2018) UFBoot2: Improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522
Hoffmann M, Monday SR, Allard MW, Strain EA, Whittaker P, Naum M, McCarthy PJ, Lopez JV, Fischer M, Brown EW (2012) Vibrio caribbeanicus sp. nov., isolated from the marine sponge Scleritoderma cyanea. Int J Syst Evol Microbiol 62:1736–1743
Huerta-Cepas J, Serra F, Bork P (2016) ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol 33:1635–1638
Hugenholtz P, Chuvochina M, Oren A, Parks DH, Soo RM (2021) Prokaryotic taxonomy and nomenclature in the age of big sequence data. ISME J 15:1879–1892
Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S (2018) High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114
Jiang C, Tanaka M, Nishikawa S, Mino S, Romalde JL, Thompson FL, Gomez-Gil B, Sawabe T (2021) Vibrio clade 3.0: New Vibrionaceae evolutionary units using genome-based approach. Curr Microbiol 79:10
Johnson CN (2013) Fitness factors in vibrios: a mini-review. Microb Ecol 65:826–851
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30:772–780
Kesarcodi-Watson A, Kaspar H, Lategan MJ, Gibson LF (2009) Challenge of New Zealand Greenshell mussel Perna canaliculus larvae using two Vibrio pathogens: a hatchery study. Dis Aquat Organ 86:15–20
Kim M, Oh H-S, Park S-C, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Koenig JE, Bourne DG, Curtis B, Dlutek M, Stokes HW, Doolittle WF, Boucher Y (2011) Coral-mucus-associated Vibrio integrons in the Great Barrier Reef: genomic hotspots for environmental adaptation. ISME J 5:962–972
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Kvennefors EC, Sampayo E, Ridgway T, Barnes AC, Hoegh-Guldberg O (2010) Bacterial communities of two ubiquitous Great Barrier Reef corals reveals both site- and species-specificity of common bacterial associates. PLoS ONE 5:e10401
Lambert MA, Hickman-Brennerm FW, Farmer JJ III, Moss CW (1983) Differentiation of Vibrionaceae species by their cellular fatty acid composition. Int J Syst Bacteriol 33:777–792
Lambert C, Nicolas JL, Cilia V, Corre S (1998) Vibrio pectenicida sp. nov., a pathogen of scallop (Pecten maximus) larvae. Int J Syst Bacteriol 48 Pt 2:481–487
Li R, Dang H, Huang Y, Quan Z, Jiang H, Zhang W, Ding J (2020) Vibrio coralliilyticus as an agent of red spotting disease in the sea urchin Strongylocentrotus intermedius. Aquac Rep 16:100244
Loughran RM, Esquivel AR, Deadmond MC, Koyack MJ, Paddock BE, O’Hanlon SM, Ushijima B, Saw JH, Videau P (2020) Draft genome sequence of Vibrio sp. strain OCN044, isolated from Palmyra Atoll, Northern Line Islands. Microbiol Resour Announc 9:e00042–e00020
McCauley EP, Haltli B, Kerr RG (2015) Description of Pseudobacteriovorax antillogorgiicola gen. nov., sp. nov., a bacterium isolated from the gorgonian octocoral Antillogorgia elisabethae, belonging to the family Pseudobacteriovoracaceae fam. nov., within the order Bdellovibrionales. Int J Syst Evol Microbiol 65:522–530
McFall-Ngai M, Heath-Heckman EA, Gillette AA, Peyer SM, Harvie EA (2012) The secret languages of coevolved symbioses: insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol 24:3–8
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf 14:60
Meier-Kolthoff JP, Hahnke RL, Petersen J, Scheuner C, Michael V, Fiebig A, Rohde C, Rohde M, Fartmann B, Goodwin LA, Chertkov O, Reddy TBK, Pati A, Ivanova NN, Markowitz V, Kyrpides NC, Woyke T, Göker M, Klenk H-P (2014) Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci 9:2
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R (2020) IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534
Murray AE, Freudenstein J, Gribaldo S, Hatzenpichler R, Hugenholtz P, Kämpfer P, Konstantinidis KT, Lane CE, Papke RT, Parks DH, Rossello-Mora R, Stott MB, Sutcliffe IC, Thrash JC, Venter SN, Whitman WB, Acinas SG, Amann RI, Anantharaman K, Armengaud J, Baker BJ, Barco RA, Bode HB, Boyd ES, Brady CL, Carini P, Chain PSG, Colman DR, DeAngelis KM, de los Rios MA, Estrada-de los Santos P, Dunlap CA, Eisen JA, Emerson D, Ettema TJG, Eveillard D, Girguis PR, Hentschel U, Hollibaugh JT, Hug LA, Inskeep WP, Ivanova EP, Klenk H-P, Li W-J, Lloyd KG, Löffler FE, Makhalanyane TP, Moser DP, Nunoura T, Palmer M, Parro V, Pedrós-Alió C, Probst AJ, Smits THM, Steen AD, Steenkamp ET, Spang A, Stewart FJ, Tiedje JM, Vandamme P, Wagner M, Wang F-P, Yarza P, Hedlund BP, Reysenbach (2020) A-L Roadmap for naming uncultivated Archaea and Bacteria. Nat Microbiol 5:987–994
Nguyen TV, Alfaro AC, Young T, Merien F (2019) Tissue-specific immune responses to Vibrio sp. infection in mussels (Perna canaliculus): A metabolomics approach. Aquaculture 500:118–125
Oliver JD, Pruzzo C, Vezzulli L, Kaper JB (2012) Vibrio species. In: Food microbiol, pp401–439
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, Hugenholtz P (2018) A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004
Prado S, Romalde JL, Montes J, Barja JL (2005) Pathogenic bacteria isolated from disease outbreaks in shellfish hatcheries. First description of Vibrio neptunius as an oyster pathogen. Dis Aquat Organ 67:209–215
Prado S, Dubert J, Romalde JL, Toranzo AE, Barja JL (2014) Vibrio ostreicida sp. nov., a new pathogen of bivalve larvae. Int J Syst Evol Microbiol 64:1641–1646
Pruzzo C, Huq A, Colwell RR, Donelli G (2005) Pathogenic Vibrio species in the marine and estuarine environment. In: Belkin S, Colwell RR (eds) Oceans and Health: Pathogens in the Marine Environment. Springer US, Boston, MA, pp 217–252
Richards GP, Watson MA, Needleman DS, Church KM, Häse CC (2015) Mortalities of Eastern and Pacific oyster larvae caused by the pathogens Vibrio coralliilyticus and Vibrio tubiashii. Appl Environ Microbiol 81:292–297
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J (2015) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931
Riquelme CE, Jorquera MA, Rojas AI, Avendaño RE, Reyes N (2001) Addition of inhibitor-producing bacteria to mass cultures of Argopecten purpuratus larvae (Lamarck, 1819). Aquaculture 192:111–119
Röthig T, Ochsenkühn MA, Roik A, van der Merwe R, Voolstra CR (2016) Long-term salinity tolerance is accompanied by major restructuring of the coral bacterial microbiome. Mol Ecol 25:1308–1323
Ruby EG (1996) Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol 50:591–624
Sambrook J (2001) Molecular cloning: a laboratory manual / Joseph Sambrook, David W. Russell. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y
Sawabe T, Setoguchi N, Inoue S, Tanaka R, Ootsubo M, Yoshimizu M, Ezura Y (2003) Acetic acid production of Vibrio halioticoli from alginate: a possible role for establishment of abalone–V. halioticoli association. Aquaculture 219:671–679
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069
Sussman M, Willis BL, Victor S, Bourne DG (2008) Coral pathogens identified for White Syndrome (WS) epizootics in the Indo-Pacific. PLoS ONE 3:e2393
Thompson FL, Li Y, Gomez-Gil B, Thompson CC, Hoste B, Vandemeulebroecke K, Rupp GS, Pereira A, De Bem MM, Sorgeloos P, Swings J (2003) Vibrio neptunius sp. nov., Vibrio brasiliensis sp. nov. and Vibrio xuii sp. nov., isolated from the marine aquaculture environment (bivalves, fish, rotifers and shrimps). Int J Syst Evol Microbiol 53:245–252
Thompson FL, Iida T, Swings J (2004) Biodiversity of Vibrios. Microbiol Mol Biol Rev 68:403
Tout J, Jeffries TC, Petrou K, Tyson GW, Webster NS, Garren M, Stocker R, Ralph PJ, Seymour JR (2015) Chemotaxis by natural populations of coral reef bacteria. ISME J 9:1764–1777
Ushijima B, Häse CC (2018) Influence of chemotaxis and swimming patterns on the virulence of the coral pathogen Vibrio coralliilyticus. J Bacteriol 200:e00791–e00717
Ushijima B, Videau P, Burger AH, Shore-Maggio A, Runyon CM, Sudek M, Aeby GS, Callahan SM (2014) Vibrio coralliilyticus strain OCN008 is an etiological agent of acute Montipora white syndrome. Appl Environ Microbiol 80:2102–2109
Ushijima B, Videau P, Poscablo D, Stengel JW, Beurmann S, Burger AH, Aeby GS, Callahan SM (2016) Mutation of the toxR or mshA genes from Vibrio coralliilyticus strain OCN014 reduces infection of the coral Acropora cytherea. Environ Microbiol 18:4055–4067
Van Rossum T, Ferretti P, Maistrenko OM, Bork P (2020) Diversity within species: interpreting strains in microbiomes. Nat Rev Microbiol 18:491–506
Varghese NJ, Mukherjee S, Ivanova N, Konstantinidis KT, Mavrommatis K, Kyrpides NC, Pati A (2015) Microbial species delineation using whole genome sequences. Nucleic Acids Res 43:6761–6771
Verschuere L, Rombaut G, Sorgeloos P, Verstraete W (2000) Probiotic bacteria as biological control agents in aquaculture. Microbiol Mol Biol Rev 64:655–671
Vezzulli L, Previati M, Pruzzo C, Marchese A, Bourne DG, Cerrano C (2010) Vibrio infections triggering mass mortality events in a warming Mediterranean Sea. Environ Microbiol 12:2007–2019
Acknowledgements
The authors thank Dr. Tina Carvalho in the Biological Electron Microscopy Facility at the University of Hawai‘i at Mānoa for assistance with electron microscopy, Nancy Shough (Southern Oregon University, SOU) for technical support, Lauren Millman (SOU) for administrative support, Charles Lein (Pierrepont School) for consultation on Latin use, Dalcione Reis for aid in strain acquisition, and Dan Loughran for continued physiological and emotional support. We gratefully acknowledge the computing resources provided on the High Performance Computing Cluster operated by Research Technology Services at the George Washington University.
Funding
This work was supported by startup funds from SOU to M.J.K. and P.V., from UNCW to B.U., and from GWU to J.H.S. M.O.G. was supported by an NSF South Dakota EPSCoR RII Track-1: Building on the 2020 Vision: Expanding Research, Education and Innovation in South Dakota, Award 1849206, and a Supporting Talent for Research Trajectories (START) grant award from Dakota State University.
Author information
Authors and Affiliations
Contributions
RML:investigation, writing—review and editing. Sarah A. Emsley: investigation, writing—original draft preparation, writing—review and editing. Tori Jefferson: investigation, writing—review and editing. Benjamin J. Wasson: investigation, writing—review and editing. Monica C. Deadmond: investigation, writing—review and editing. Taylor L. Knauss: investigation, writing—original draft preparation, writing—review and editing. Kaysa M. Pfannmuller: investigation, writing—review and editing. Katherine J. Lippert: investigation, writing—review and editing. Gregory Miller: resources, writing—review and editing. Lauren C. Cline: investigation, writing—review and editing. David K. Oline: investigation, resources, writing—review and editing, funding. Marc J. Koyack: resources, writing—review and editing, funding. Silvia Grant-Beurmann: investigation, resources, writing—review and editing. Michael O. Gaylor: investigation, resources, writing—review and editing, funding. Jimmy K. Saw: methodology, investigation, writing—review and editing, visualization, funding. Blake Ushijima: conceptualization, methodology, validation, investigation, resources, writing—review and editing, visualization, supervision, project administration, funding. Patrick Videau: conceptualization, methodology, validation, investigation, resources, writing—review and editing, visualization, supervision, project administration, funding.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Repositories
The strain collection identifiers for Vibrio tetraodonis subsp. pristinus subsp. nov. are LMG 31895 and DSM 111778. The 16S rRNA gene sequences for Vibrio tetraodonis subsp. pristinus subsp. nov. and V. tetraodonis A511T are deposited at DDBJ/ENA/GenBank under the accession numbers MW872696 and ON808596, respectively, and the draft genome of Vibrio tetraodonis subsp. pristinus subsp. nov. was deposited at DDBJ/ENA/GenBank under the accession number WWEU00000000.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Loughran, R.M., Emsley, S.A., Jefferson, T. et al. Vibrio tetraodonis subsp. pristinus subsp. nov., isolated from the coral Acropora cytherea at Palmyra Atoll, and creation and emended description of Vibrio tetraodonis subsp. tetraodonis subsp. nov. Antonie van Leeuwenhoek 115, 1215–1228 (2022). https://doi.org/10.1007/s10482-022-01766-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-022-01766-0