Abstract
A facultative anaerobic bacterium, designated as A25T, was isolated from a mangrove sediment sample collected in Shenzhen, China. Cells of strain A25T were found to be Gram-staining negative, rod-shaped, flagella-harboring, and oxidase- and catalase-positive. The isolate was able to grow at 4–40 °C (optimum 28 °C) and pH 5.0–9.0 (optimum pH 6.0), and in 0–10% NaCl concentration (w/v) (optimum 1%). Strain A25T was capable of reducing Fe(III) citrate under anaerobic conditions. The major fatty acids of this strain was C16:1ω7c/C16:1ω6c (summed feature 3), C17:1ω8c and iso-C15:0. Results of phylogenetic analyses based on 16S rRNA gene sequences indicated that strain A25T is affiliated with the genus Shewanella, showing the highest similarity to Shewanella seohaensis S7-3T (98.4% similarity). The average nucleotide identity and digital DNA-DNA hybridization values between the genomes of strain A25T and its closely related strains were ≤ 79.0% and ≤ 22.8%, respectively. Based on its phenotypic, phylogenetic properties and physiological and biochemical characteristics, strain A25T (= JCM 34900T = GDMCC 1.2731T) was designated as the type strain of a novel species of the genus Shewanella, for which the name Shewanella shenzhenensis sp. nov. was proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Shewanella, belonging to the family Shewanellaceae within the order Alteromonadales of the class Gammaproteobacteria, was first described by MacDonell and Colwell (MacDonell and Colwell 1985). At the time of writing, this genus contains 76 species with validly published names (https://lpsn.dsmz.de/genus/shewanella). Members of the genus Shewanella have been isolated from diverse environments such as mangrove sediment (Zhang et al. 2021), seawater (Bae et al. 2020), brown algae (Kim et al. 2016), fish (Satomi et al. 2006), activated sludge (Xu et al. 2005), tidal flat sediment (Yoon et al. 2012) and Antarctic coastal areas (Bozal et al. 2002). The genus Shewanella compose a group of facultative anaerobic bacteria that are physiologically diverse, and the hallmark is the ability to utilize a diverse electron acceptors (e.g. Fe(III) and nitrate) in the absence of oxygen, which allows them to survive in diverse habitats (Wang et al. 2010). In light of their fascinating physiology, the genus Shewanella have several biotechnological uses, from bioremediation of chlorinated compounds and other environmental pollutants to energy-generating biocatalysis (Hau and Gralnick 2007).
Mangroves are highly productive ecosystems of tropical and subtropical coastlines which are of great interest for their ecological, sociological and economical roles (Holguin et al. 2001). In mangrove ecosystems, iron performs vital roles as it is an essential nutrient for pants and marine plankton, and the iron bioavailability is directly affected by microbial Fe(III) reduction in mangrove sediments (Queiroz et al. 2022). Up to date, several Shewanella strains have been isolated from the mangrove sediments, and with these isolates as type strains, a total of 5 Shewanella species have been established (Liu et al. 2015, 2021; Zhang et al. 2021). However, none of these species are reported to have the Fe(III) reduction capability.
In the present study, a bacterium of the genus Shewanella, designated A25T, was isolated from a mangrove sediment sample, and was identified using polyphasic taxonomic approaches. As a result, a novel species Shewanella shenzhenensis sp. nov. is proposed using strain A25T as the type strain. Since strain A25T was capable of reducing Fe(III), the study of strain A25T would have important implication for understanding the biogeochemical process of iron in mangrove ecosystem.
Materials and methods
Isolation and culture conditions
Strain A25T was isolated from a mangrove sediment sample in Futian district Shenzhen city (22° 30′–22° 32′ N, 113° 56′–114° 3′ E). In a sterile environment, mangrove sediment sample was diluted with physiological saline, and spread on LB agar containing (/L) 10.0 g peptone, 5.0 g NaCl, 1.0 g glucose, and 5.0 g yeast paste powder (pH 7.0 ± 0.2). After aerobic incubation at 30 °C for one week, the colonies were picked for bacterial purification by plate streaking on LB agar repeatedly. The resulting pure strain was maintained as aqueous glycerol suspensioin (50%; v/v) at − 80 °C and was then deposited in the Guangdong Microbial Culture Collection Center (GDMCC) and Japan Collection of Microorganisms (JCM).
Phylogeny analysis based on 16S rRNA gene sequences
Genomic DNA of strain A25T was extracted using a DNA extraction Kit (Takara, Japan), and was used for sequencing of the 16S rRNA gene and the whole genome. The 16S rRNA gene was amplified by PCR using primers 27F and 1492R (Weisburg et al. 1991). The resulting PCR product was purified and cloned into the pMD19-T vector (Takara) and sequenced using Sanger sequencing. The 16S rRNA gene sequence obtained was subsequently compared with other type strains using the EzBiocloud platform (Yoon et al. 2017a, b). The 16S rRNA gene sequences of closely related species were subsequently aligned with strain A25T by ClustalW algorithm of MEGA X (Kumar et al. 2018). Phylogenetic trees were reconstructed based on the miximum-likelihood and neighbour-joining algorithms using MEGA X with Psychrobium conchae BJ-1T as outgroup. For the neighbour-joining tree, the substitution model maximum composite likelihood method was chosen, and for the maximum-likelihood tree, the Tamura-Nei model was applied. The topology of the trees was evaluated by performing a bootstrap analysis based on 1000 replications.
Genome sequencing and analysis
For genome analyses of strain A25T, the whole genomes were sequenced using Illumina Hiseq 2000 technology and were assembled by SOAPdenovo (Guangzhou Meige Biotechnology Co., Ltd.). The draft genome sequence of strain A25T has been deposited in Genbank database under the accession numbers of JAKOGF000000000. Genome annotation was carried out using Rapid Annotation using Subsystem Technology (RAST) server (Aziz et al. 2008) and NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al. 2016). For functional classification, the predicted protein sequences were searched against the NCBI non-redundant database using BLASTP, and the outputs were imported into BLAST2GO V5.2.5 for GO term mapping (Conesa et al., 2005). The results of BLAST2GO were submitted to the WEGO for GO classification (Ye et al., 2006). The predicted protein sequences were submitted to BlastKOALA for functional annotation based on KEFF orthology (KO), and the KO number were then submitted to KEGG Mapper to reconstruct the pathway (Kanehisa et al. 2016; Kanehisa 2017).
The relatedness between the genomic sequences of strain A25T and type strains of Shewanella species was estimated based on the average nucleotide identity (ANI), and digital DNA–DNA hybridization (dDDH) using the ANI calculator in EzBioCloud platform (Yoon et al. 2017a, b) and Genome-to-Genome Distance Calculator (GGDC) version 2.1 (Meier-Kolthoff et al. 2013), respectively.
For genome phylogenetic analysis, the automated multi-locus species tree (autoMLST) online server was used to automatically identify house-keeping genes (Alanjary et al. 2019). The closely related type species of the new isolates and outgroup Phenylobacterium zucineum HLK1T were selected to construct a high-resolution species tree. The final phylogenetic tree based on 100 core genes was reconstructed by MEGA X software using maximum-likelihood algorithm.
Phenotypic and chemotaxonomic characterization
Cell morphology was observed with a transmission electron microscope after cultivation on LB plate for 24 h. Clone morphology was observed after cultivation on LB agar for 48 h. The Gram reaction was determined by a Gram staining kit HB8278 (Qingdao Hope-Bio Technology Co., Ltd; China). Oxidase activity was determined using an oxidase reagent (BioMérieux), and catalase activity was determined by observing bubble production in 3% (v/v) hydrogen peroxide solution. Hydrolysis of Tween 20, Tween 40, Tween 80, starch and casein were performed on MA with a final concentration of 1% (w/v). The temperature range for growth was determined in the range 0–42 °C (at intervals of 0, 4, 10, 15, 20, 25, 28, 30, 37 and 42 °C). The pH range for growth was determined at pH 4.0–10.0 (at intervals of 1.0 pH unit). The tolerance for NaCl concentrations was determined with 0–12% (w/v) NaCl (with increments of 1.0%). The anaerobic growth was tested in mineral medium containing (L-1) 0.04 g CaCl2·2H2O, 0.10 g MgSO4·7H2O, 1.80 g NaHCO3, 0.43 g Na2CO3, 0.42 g KH2PO4, 0.22 g K2HPO4, 0.20 g NH4Cl, 0.38 g KCl, 10.0 ml vitamin stock solution and 10.0 ml mineral stock solution (Lovley and Phillips 1988) supplementary with lactate (10 mM) as electron donor and Fe(III) citrate (50 mM) or nitrate (20 mM) as electron acceptor; The optical density at 600 nm (OD600) was measured to evaluate cell growth. Other physiological characteristics were characterized with the API 20NE systems (BioMérieux) according to the manufacturer’s instructions. Disc diffusion was used to test the strain's sensitivity to antibiotics. The following antibiotics were tested: erythromycin (15 μg), neomycin (30 μg), penicillin (10 μg), kanamycin (30 μg), gentamicin (10 μg), carbenicillin (100 μg), clindamycin (2 μg), lincomycin (2 μg), polymixinB (300 iu), rifamipcin (5 μg), chloramphenicol (30 μg), ofloxacin (5 μg), norfloxacin (10 μg), and ciprofloxacin (5 μg). Sensitivity is assessed by determining the size of the diameter of the antibacterial ring.
For fatty acid profile analysis, cells of strain A25T and its reference strains grown in LB medium to exponential growth phase were collected by centrifugation at 12,000 rpm at 4 °C, and freeze-dried using the vacuum freeze drying apparatus (Scientz-10 N, Ningbo, China). The fatty acids in whole cells were saponified, methylated and extracted according to the standard protocol of MIDI (Sherlock Microbial Identification System, version 6.0B). The fatty acids were analyzed with GC (Agilent Technologies 6850) and identified using the TSBA6.0 database of the Microbial Identification System.
Results and discussion
Phylogenetic analysis based on 16S rRNA gene and core genomes
The nearly complete 16S rRNA gene sequence of strain A25T (1526 bp) was obtained. Sequence comparison showed that strain A25T shared the highest 16S rRNA gene sequence similarity with Shewanella seohaensis S7-3T (98.4% similarity) followed by Shewanella decolorationis S12T (98.0% similarity). These values were below the threshold (98.7%) for species delineation (Chun et al., 2018). In the maximum-likelihood and neighbour-joining phylogenetic trees based on 16S rRNA gene sequences (Supplementary Fig. S1 and S2), strain A25T fell within the clade of the genus Shewanella and formed an internally independent branch with S. seohaensis S7-3T and S. decolorationis S12T. Above results suggested that strain A25T represents a different species from known species of the genus Shewanella.
The genome sequence of strain A25T was 4,821,510 bp with G+C content of 47.3 mol%. The results of dDDH and ANI analysis indicated that strain A25T showed the highest dDDH and ANI values with S. decolorationis S12T (22.8% for dDDH and 79.0% for ANI), which were lower than the standard cutoff value for species delineation (70% for dDDH and 95% for ANI) (Chun et al. 2018). As shown in phylogenetic tree based on core genomes (Fig. 1), strain A25T was located in a cluster with S. decolorationis S12T and was far from other type strains of the genus Shewanella. These results supported the proposal that this strain represented a novel species of the genus Shewanella (Fig. S1 and S2).
Phylogenetic tree based on the core genomes showing the position of strain A25T and related type species of family Shewanellaceae. Phenylobacterium zucineum HLK1T was selected as outgroup. Numbers at branch refer to bootstrap values based on 1000 replicates and only values above 50% were given at nodes. Bar, 0.10 substitutions per nucleotide position
Phenotypic characteristics
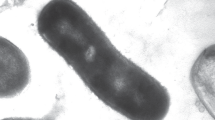
Colonies of strain A25T were 1–2 mm in diameter, pink, circular and smooth. Cells were observed to be Gram-staining-negative and facultative anaerobic. Under electron microscope (Fig. 2), the cells were rod-shaped, 1.7–2.1 μm in length, and 0.5–0.6 μm in diameter and a polar flagellum was detected.
Strain A25T was able to grow under the temperature range of at 4–40 °C (optimum 28 °C) and pH 5.0–9.0 (optimum pH 6.0), and in 0–10% NaCl (w/v). The optimal pH for growth of strain A25T was lower than that of its reference strains S. seohaensis S7-3T (optimal pH 7–8) and S. decolorationis S12T (optimal pH 7) (Table 1), but such low optimal pH was common for some Shewanella species such as Shewanella mangrovi (optimal pH 6) and Shewanella yunxiaonensis (optimal pH 5) which were sourced from mangrove sediments (Liu et al. 2015, 2021). Strain A25T could hydrolyze Tween 20, Tween 40, Tween 80 and gelatin but not casein and starch. Activities of oxidase and catalase were found to be positive. Nitrate cannot be reduced under anaerobic conditions. In API 20NE test, activities of β-glucosidase and urease, reduction of nitrate, and utilization of d-glucose, l-arabinose, N-acetyl-glucosamine, maltose, gluconate, adipic acid and malic acid were positive, but activities of arginine dihydrolase and β-galactosidase, production of indole, production of acid from glucose, and utilization of d-mannose, mannitol, capric acid, citrate and phenylacetic acid were negative. Strain A25T was susceptible to neomycin, penicillin, kanamycin, gentamicin, clindamycin, lincomycin, rifamipcin, erythromycin, chloramphenicol, polymixinB, ofloxacin, norfloxacin, and ciprofloxacin, but resistant to carbenicillin. The physiological properties differentiated strain A25T from its closest neighbours were listed in Table 1.
Chemotaxonomic analysis
The whole-cell fatty acid profile of strain A25T contained the major cellular fatty acids (> 10%) of C16:1ω7c/C16:1ω6c (summed feature 3) (19.3%), C17:1ω8c (17.2%) and iso-C15:0 (14.4%), which was the same pattern found in the two reference strains (S. seohaensis S7-3T and S. decolorationis S12T) (Table S1) and several type strains of other Shewanella species (Cha et al. 2020; Verma et al. 2011).
Capacity of Fe(III) reduction
Under anaerobic conditions, strain A25T was capable of reducing about 50 mM of Fe(III) citrate with lactate as the sole electron donor within 5 days, and the Fe(III) reduction could support the cell growth of strain A25T (Supplementary Fig. S3). It has been reported that members of the genus Shewanella such as S. decolorationis S12T (relative of strain A25T) and Shewanella oneidensis MR-1 (well-studied model of Fe(III) reduction of the genus Shewanella) have the ability of iron reduction which play important roles in the global iron and carbon cycle (Fu et al. 2016; Li et al. 2012). Although there are more than 20 Shewanella species which are capable of reducing Fe(III), none of them were obtained from mangrove sediments. Therefore, strain A25T might represent the first mangrove-sourced Shewanella species capable of reducing Fe(III).
Genomic features
A total of 4885 genes were predicted in the genomes of strains A25T including 4723 protein-coding genes, 108 RNA genes (24 rRNA genes, 79 tRNA genes and 5 ncRNA genes) and 54 pseudogenes. The GO analysis indicated that these genes were assigned to a wide range of functional categories with three main categories: biological process, molecular function and cellular components (Supplementary Fig. S4). The majority of GO terms were assigned to the biological process in which the top five subgroups were metabolic process, cellular process, single-organism process, localization and biological regulation.
Mangrove habitats are subjected to stressors such as high salinity, hypoxia and strong tidal flows (Soldan et al. 2019), and the bacteria in mangrove sediments have evolved several stress response mechanisms to adapt to such conditions (Liu et al. 2021). In the genome of strain A25T, there are 12 genes associated with antioxidant activity (Supplementary Fig. S4) which confer the stress tolerance ability to bacteria, 4 associated with potassium ion homeostasis which are involved in osmoregulation and pH homeostasis (MacGilvary et al. 2019). These genes might be essential for survival of strain A25T in mangrove sediment, because they are crucial for detecting and responding to perturbations caused by environmental stress. The homologous genes of such stress-associated genes had been ever been reported in other Shewanella species including Shewanella avicenniae, Shewanella sedimentimangrovi and Shewanella yunxiaonensis which were sourced from mangrove sediments (Liu et al. 2021).
Strain A25T was detected to be positive for nitrate reduction under aerobic conditions, and thereby the genes responsible for nitrate reduction were analyzed using KEGG. Results showed that there is a complete pathway of dissimilatory nitrate reduction in the A25T genome, which could mediate the reduction of nitrate to ammonia. In detail, the genes associated with dissimilatory nitrate reduction include napAB (L9G16_05415, L9G16_05420, L9G16_13485 and L9G16_13490), nirBD (L9G16_06820 and L9G16_06825) and nrfAH (L9G16_04630, L9G16_07330).
As reported, cytochromes are key components for Fe(III) reduction of Shewanella members (Shi et al. 2009). To explore the Fe(III) reduction mechanism of strain A25T, the cytochrome genes in A25T genome were analyzed. Results showed that the A25T genome contains 60 cytochrome encoding genes among which most have homologous genes in the genomes of strain MR-1 and strain S12T and 3 (MCH1933042, MCH1933053 and MCH1932204) are unique in the genome of strain A25T (Table S2). In addition, homologous genes (MCH1928961, MCH1928962 and MCH1928963) of the mtrABC gene cluster which is known in strain MR-1 are also detected in the genome of strain A25T. In a word, the abundant cytochrome genes might be responsible for Fe(III) reduction ability of strain A25T.
Taxonomic conclusion
In sum, phylogenetic analysis based on the 16S rRNA genes and the genomes, the phenotypic characteristics, and the fatty acid profile supported the classification of strain A25T as a member of the genus Shewanella, and suggested that the new isolate represents a novel species of the genus Shewanella, for which the name Shewanella shenzhenensis sp. nov. is proposed.
Description of Shewanella shenzhenensis sp. nov.
Shewanella shenzhenensis (shen.zhen.en'sis. N.L. fem. adj. shenzhenensis, referring to Shenzhen, the city where the type strain was isolated).
Cells are Gram-strain-negative, facultative aerobic and rod-shaped, with 1.7–2.1 μm long and 0.5–0.6 μm wide. Colonies are 1–2 mm in diameter, pink, circular and smooth after incubation on LB agar for 2 days. Bacteria growth occurs in the range of 4–40 °C (optimum 28 °C), pH 5.0–9.0 (optimum pH 6.0) and 0–10% (w/v) NaCl (optimum 1%). Nitrate can be reduced under aerobic conditions but not under anaerobic conditions. Anaerobic growth occurs using Fe(III) citrate as electron acceptor and lactate as electron donor. Cells are positive for oxidase, catalase and hydrolysis of Tween 20, Tween 40, Tween 80 and gelatin, but negative for production of indole and hydrolysis of starch and casein. The predominant cellular fatty acids (> 5%) contain summed feature 3, C17:1ω8c, iso-C15:0, and C16:0. The G + C content of genomic DNA of the type strain is 47.3 mol%.
The type strain A25T (= JCM 34900T = GDMCC 1.2731T) is isolated from a mangrove sediment in Futian district, Shenzhen, China. The 16S rRNA gene sequence and the whole genome sequence of the type strain are available from GenBank with accession number MZ477861 and JAKOGF000000000, respectively.
Data availability
All data generated or analyzed during this study are included in this published article, its supplementary information file and GenBank/EMBL/DDBJ. The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene and the genome sequences of strain A25T are MZ477861 and JAKOGF000000000, respectively. Supplementary file including Neighbour-joining phylogenetic tree based on 16S rRNA gene sequences, transmission electron microscopy image strain A25T, and reduction of Fe(III) citrate is available with the online version of this paper.
References
Alanjary M, Steinke K, Ziemert N (2019) AutoMLST: an automated web server for generating multi-locus species trees highlighting natural product potential. Nucleic Acids Res 47:W276–W282
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al (2008) The RAST server: rapid annotations using subsystems technology. BMC Genom 9:1–15
Bae SS, Jung Y, Baek K (2020) Shewanella maritima sp. nov., a facultative anaerobic marine bacterium isolated from seawater, and emended description of Shewanella intestini. Int J Syst Evol Microbiol 70:1288–1293
Bozal N, Montes MJ, Tudela E, Jiménez F, Guinea J (2002) Shewanella frigidimarina and Shewanella livingstonensis sp. nov. isolated from Antarctic coastal areas. Int J Syst Evol Microbiol 52:195–205
Cha Q, Ren X, Sun Y, He X, Su H, Chen X et al (2020) Shewanella polaris sp. nov., a psychrotolerant bacterium isolated from Arxtic brown algae. Int J Syst Evol Microbiol 70:2096–2102
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, Costa MS et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Conesa A, Götz S, Garcíagómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Fu L, Li S, Ding Z, Ding J, Lu Y, Zeng RJ (2016) Iron reduction in the DAMO/Shewanella oneidensis MR-1 coculture system and the fate of Fe(II). Water Res 88:808–815
Hau HH, Gralnick JA (2007) Ecology and biotechnology of the genus Shewanella. Annu Rev Microbiol 61:237–258
Holguin G, Vazquez P, Bashan Y (2001) The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecocyctems: an overview. Biol Fertil Soils 33:265–278
Kanehisa M, Sato Y, Morishima K (2016) BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731
Kanehisa M (2017) Enzyme annotation and metabolic reconstruction using KEGG. In: Protein function prediction. Humana Press, New York, pp 135–145
Kim J, Yoo H, Lee D, Park S, Kim Y, Oh D (2016) Shewanella algicola sp. nov., a marine bacterium isolated from brown algae. Int J Syst Evol Microbiol 66:2218–2224
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetic analysis across computing platforms. Mol Biol Evol 35:1547–1549
Li X, Liu T, Li F, Zhang W, Zhou S, Li Y (2012) Reduction of structural Fe(III) in oxyhydroxides by Shewanella decolorationis S12 and characterization of the surface properties of iron minerals. J Soil Sediment 12:217–227
Liu Y, Shang X, Yi Z, Gu L, Zeng R (2015) Shewanella mangrovi sp. nov., an acetaldehyde-degrading bacterium isolated from mangrove sediment. Int J Syst Evol Microbiol 65:2630–2634
Liu G, Zhang Q, Rao MPN, Yang S, Tang R, Shi H et al (2021) Stress response mechanisms and description of three novel species Shewanella avicenniae sp. nov., Shewanella sedimentimangrovi sp. nov. and Shewanella yunxiaonensis sp. nov., isolated from mangrove ecosystem. Antonie Van Leeuwenhoek 114:2123–2131
Lovley DR, Phillips EJP (1988) Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol 54:1472–1480
MacDonell MT, Colwell RR (1985) Phylogeny of the Vibrionaceae, and recommendation for two new genera, Listonella and Shewanella. Syst Appl Microbiol 6:171–182
MacGilvary NJ, Kevorkian YL, Tan S (2019) Potassium response and homeostasis in Mycobacterium tuberculosis modulates environmental adaptation and is important for host colonization. PLoS Pathog 15(2):e1007591
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60
Queiroz HM, Ferreira TO, Fandiño VA, Bragantini IOBF, Barcellos D, Nóbrega GN et al (2022) Changes in soil iron biogeochemistry in response to mangrove dieback. Biogeochemistry 158:357–372
Satomi M, Vogel BF, Gram L, Venkateswaran K (2006) Shewanella hafniensis sp. nov. and Shewanella morhuae sp. nov., isolated from marine fish of the Baltic Sea. Int J Syst Evol Microbiol 56:243–249
Shi L, Richardson DJ, Wang Z, Kerisit SN, Rosso KM, Zachara JM et al (2009) The roles of outer membrane cytochromes of Shewanella and Geobacter in extracellular electron transfer. Environ Microbiol Rep 1:220–227
Soldan R, Mapelli F, Crotti E, Schnell S, Daffonchio D, Marasco R, Fusi M, Borin S, Cardinale M (2019) Bacterial endophytes of mangrove propagules elicit early establishment of the natural host and promote growth of cereal crops under salt stress. Microbiol Res 223–225:33–43
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L et al (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624
Verma P, Pandey PK, Gupta AK, Kim HJ, Baik KS, Seong CN et al (2011) Shewanella indica sp. nov., isolated from sediment of the Arabian Sea. Int J Syst Evol Microbiol 61:2058–2064
Wang B, Xu M, Sun G (2010) Comparative analysis of membranous proteomics of Shewanella decolorationis S12 grown with azo compound or Fe(III) citrate as sole terminal electron acceptor. Appl Microbiol Biotechnol 86:1513–1523
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Xu M, Guo J, Cen Y, Zhong X, Cao W, Sun G (2005) Shewanella decolorationis sp. nov., a dyedecolorizing bacterium isolated from activated sludge of a waste-water treatment plant. Int J Syst Evol Microbiol 55:363–368
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z et al (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:293–297
Yoon J, Park S, Jung Y, Lee J (2012) Shewanella seohaensis sp. nov., isolated from a tidal flat sediment. Antonie van Leeuwenhoek 102:149–156
Yoon SH, Ha SM, Kown S, Lim J, Kim Y, Seo H, Chun J (2017a) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Yoon SH, Ha SM, Lim JM, Kwon SJ, Chun J (2017b) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Zhang Q, Liu G, Rao MPN, Tang R, Yang S, Ye W et al (2021) Shewanella cyperi sp. Nov., a facultative anaerobic bacterium isolated from mangrove sediment. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.004940
Funding
This work was supported by the National Natural Science Foundation of China (42077211), and the Natural Science Foundation of Guangdong Province (2021A1515012570; 2022A1515011734).
Author information
Authors and Affiliations
Contributions
GQY drafted the manuscript. XYZ and SJY performed isolation, deposition and identifications. XYZ performed the genome analysis. LZ designed all the experiments and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical approval
This study does not describe any experimental work related to human.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Yang, G., Yao, S. et al. Shewanella shenzhenensis sp. nov., a novel Fe(III)-reducing bacterium with abundant possible cytochrome genes, isolated from mangrove sediment. Antonie van Leeuwenhoek 115, 1245–1252 (2022). https://doi.org/10.1007/s10482-022-01763-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-022-01763-3