Abstract
The phage shock protein (Psp) stress-response system protects bacteria from envelope stress through a cascade of interactions with other proteins and membrane lipids to stabilize the cell membrane. A key component of this multi-gene system is PspA, an effector protein that is found in diverse bacterial phyla, archaea, cyanobacteria, and chloroplasts. Other members of the Psp system include the cognate partners of PspA that are part of known operons: pspF||pspABC in Proteobacteria, liaIHGFSR in Firmicutes, and clgRpspAMN in Actinobacteria. Despite the functional significance of the Psp system, the conservation of PspA and other Psp functions, as well as the various genomic contexts of PspA, remain poorly characterized in Actinobacteria. Here we utilize a computational evolutionary approach to systematically identify the variations of the Psp system in ~450 completed actinobacterial genomes. We first determined the homologs of PspA and its cognate partners (as reported in Escherichia coli, Bacillus subtilis, and Mycobacterium tuberculosis) across Actinobacteria. This survey revealed that PspA and most of its functional partners are prevalent in Actinobacteria. We then found that PspA occurs in four predominant genomic contexts within Actinobacteria, the primary context being the clgRpspAM system previously identified in Mycobacteria. We also constructed a phylogenetic tree of PspA homologs (including paralogs) to trace the conservation and evolution of PspA across Actinobacteria. The genomic context revealed that PspA shows changes in its gene-neighborhood. The presence of multiple PspA contexts or of other known Psp members in genomic neighborhoods that do not carry pspA suggests yet undiscovered functional implications in envelope stress response mechanisms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The phage-shock-protein (Psp) system is an envelope stress-response system (Joly et al. 2010; Darwin 2005) that was first identified in Gammaproteobacteria in response to filamentous phage infection (Brissette et al. 1990). PspA, the main component of the system, is conserved in most lineages across the three kingdoms of life (Huvet et al. 2011; Joly et al. 2010; Vothknecht et al. 2012). The phyletic spread of other cognate partners of PspA, which are typically transcriptional regulators and membrane proteins, remains poorly understood.
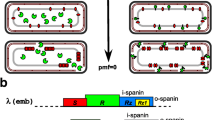
The minimal Psp system in Proteobacteria comprises the transcriptional regulator, PspF, and the three-gene operon pspABC (Fig. 1a) (Huvet et al. 2011; Darwin 2005; Joly et al. 2010). Since the first report of the Psp system in the early 1990s, other variants of the system have been discovered in Firmicutes (Mascher et al. 2004) and Actinobacteria (Datta et al. 2015; Manganelli and Gennaro 2016). In Firmicutes, PspA is part of the lantibiotic stress-response system, the Lia operon (liaIHGFSR, Fig. 1a), which contains a three-component system. In this system, the PspA homolog is called LiaH (Mascher et al. 2004). We recently characterized a functionally similar, but contextually different, Psp system in Mycobacterium tuberculosis and other genera (Datta et al. 2015; Manganelli and Gennaro 2016). In addition to PspA, the system comprises a transcriptional regulator, ClgR, that contains an cHTH domain (Aravind et al. 2005), an integral membrane protein PspM, and PspN, the product of a fourth gene of unknown function (Fig. 1a).
Phyletic spread of known Psp members in Actinobacteria. a The known Psp operons in E. coli, M. tuberculosis and B. subtilis. Key: rectangles, genes; arrows, direction of transcription; block arrows, promoters. b Heatmap of the presence of homologs of known Psp members. The color gradient represents the percentage of genomes within each order that carry a homolog (e.g., the darkest shade, 100%, indicates that all genomes in the order contain a homolog). Columns Known Psp members—PspA, PspM, PspN, PspC, LiaI, LiaG, and LiaF—queried against all sequenced actinobacterial genomes. Rows Actinobacterial orders with sequenced genomes (Actinopolysporales and Jiangellales were not included due to the absence of representative genomes), listed in phyletic order (NCBI taxonomy). The main classes shown are labeled: Actinobacteria, T Thermoleophilia, C Coriobacteriia, R Rubrobacteria, Am Acidimicrobiia
In the present study, we performed a comprehensive search to identify features of the Psp system in Actinobacteria. We started by identifying homologs of all known Psp members and determining whether the encoding genes mapped in close proximity to pspA orthologs. We then explored the conservation and evolution of PspA across all sequenced actinobacterial genomes and the genome contextual changes of PspA.
Results and discussion
Phyletic spread of known Psp members in Actinobacteria
PspA, which is the main effector protein, is the conserved member of the Psp stress response system (Flores-Kim and Darwin 2016; Manganelli and Gennaro 2016). Therefore, we first explored the presence of PspA in nearly 450 sequenced genomes of actinobacterial species and found that most orders and genera carry one or more copies of PspA (Fig. 1, column PspA). PspA homologs are apparently rare in the Coriobacteria class of Actinobacteria, with no copies in completely sequenced Coriobacteriales genomes (however, incompletely assembled genome sequences reveal the presence of homologs in representatives of this class). In addition, PspA is not found in the order Eggerthellales, with only Slackia carrying PspA in our dataset. Having determined that PspA was ancestrally present in Actinobacteria, we next investigated the phyletic spread of the other known components of the Psp system. We queried all sequenced actinobacterial genomes with each of the partners of PspA identified in Actinobacteria, Firmicutes and Proteobacteria, namely, PspM, PspN, PspB, PspC, LiaI, LiaG, and LiaF. We excluded from the analysis the transcriptional regulators identified in these three systems (ClgR in Actinobacteria, PspF in Proteobacteria, and the two-component system LiaRS in Firmicutes) due to the over-representation of their constituent domains in bacteria (Aravind et al. 2005). The results obtained from similarity searches (see “Methods” section) are summarized based on order membership and the number of paralogs per species (Fig. 1, Table A1).
We found that PspM, the integral membrane protein identified in Actinobacteria (Rv2743c in M. tuberculosis H37Rv), is present only in a few orders within the class Actinobacteria, whereas the constituent domain of PspN, a gene of unknown function (Rv2742c in M. tuberculosis H37Rv), is found in almost all the orders in the class (Fig. 1). Other actinobacterial classes such as Coriobacteriia or Acidimicrobiia carry neither PspM nor PspN. Among the proteobacterial Psp proteins, we used PspB and PspC as representative queries, since these proteins are found in the “minimal” Psp operon in these bacteria (Huvet et al. 2011; Darwin 2005). We found no actinobacterial protein significantly similar to PspB (hence absent in Fig. 1), while we observed that PspC is present in many orders of Actinobacteria (Fig. 1). Indeed, some orders contain multiple paralogs of PspC (Table A1). When we analyzed the PspA partner proteins identified in Firmicutes, we found that the transmembrane and globular proteins within the Lia operon—LiaI, LiaF, and LiaG—are present in most actinobacterial genera (Fig. 1, Table A1).
Predominant genomic contexts of PspA in Actinobacteria
The function of bacterial proteins is governed not only by protein sequence and structure but also by the genomic context in which the corresponding genes map (Huynen et al. 2000; Overmars et al. 2013; Rogozin et al. 2004; Koonin and Wolf 2008; Korbel et al. 2004). We therefore characterized the neighborhoods (± 7 genes) of the PspA homologs in all actinobacterial genomes. Four different contexts were identified (Fig. 2). The predominant context in Actinobacteria (~42% of genomes) is the one previously identified in mycobacteria: ClgR, PspA, and PspM (Manganelli and Gennaro 2016) (Fig. 2, configuration #1). Studies in M. tuberculosis (Datta et al. 2015) indicate that PspA directly interacts with ClgR and PspM. These protein–protein interactions presumably determine the dual function of PspA in this system (regulatory when bound to ClgR and effector [envelope-stabilizing] when bound to PspM), as seen in proteobacteria (Flores-Kim and Darwin 2016). Since a homologous system is found in an identical genomic context in actinobacterial orders such as Corynebacteriales and Pseudonocardiales, it is likely that similar functions are also expressed in these microorganisms.
Predominant genomic contexts of PspA in Actinobacteria. Representation of the predominant contexts identified by neighborhood searches (± 7 genes flanking each PspA homolog). Contexts were numbered in descending order of occurrences. Protein names are as in Fig. 1a, NYN (ribonuclease), Trx (thioredoxin). DUF (Domain of Unknown Function), MP (membrane protein). Key: Direction of arrow indicates the direction of transcription based on predicted ORFs (a bidirectional arrow means that the indicated gene could be found in both orientations); ‘X’ designates absence of indicated genes in some species; the triangle with an arrow indicates an optional insertion of one gene (or genes when the triangle contains an ellipsis). The numbers in the rightmost column indicate the number of occurrences of the context (numerator) relative to total number of queried actinobacterial genomes (denominator)
The second most frequent genomic configuration of PspA is seen in Streptomycetales (~12% of genomes). In this order, the most frequently occurring proteins in the neighborhood are homologs of the two-component system LiaRS (Fig. 2, configuration #2) found in Firmicutes (see also Hutchings et al. 2004)). This two-component system might contribute to the transcriptional regulation of pspA in this actinobacterial system, presumably even when the liaRS homologs are expressed in the opposite orientation.
The third configuration, which is often seen with the second copy of PspA in Corynebacteriales, contains NYN/Trx (ribonuclease, thioredoxin; Fig. 2, configuration #3; ~13% occurrence). Given its restricted phyletic spread and the opposite orientation of the NYN-coding gene relative to the rest of the neighborhood, this genomic context is likely to have limited functional relevance. An even rarer configuration (Fig. 2, configuration #4; 6% occurrence) contains PspA near transmembrane proteins other than PspM; one example is Bifidobacterium. We also found genomic contexts in which PspA is located near proteins carrying transmembrane domains or HTH domains, suggesting that integral membrane partners or transcriptional regulators different from those previously reported may exist (data not shown).
Conservation and evolution of PspA in Actinobacteria
We next correlated the genomic neighborhood with the evolution of PspA. To do so, we performed a multiple sequence alignment of PspA homologs from ~450 actinobacterial genomes and used it to build a phylogenetic tree (Fig. 3). Each leaf corresponds to one PspA homolog per genome; thus, paralogs feature as separate leaves. In the tree, the mycobacterial and corynebacterial PspAs previously identified (Manganelli and Gennaro 2016; Datta et al. 2015) are part of the largest cluster (Fig. 3a, top, Corynebacteriales). We also observed that the extent of PspA sequence similarity correlated with the proximity of orders in the phylogenetic tree. For example, PspA sequences in closely related orders such as Corynebacteriales and Pseudonocardiales were similar (top cluster of the tree in Fig. 3). In contrast, the PspA sequences found in Coriobacteriia and Streptomycetales, which are located distantly from Corynebacteriales on the tree, were the most divergent from the mycobacterial PspA.
Phylogenetic tree of PspA homologs in Actinobacteria. a PspA homologs were determined using iterative BLAST searches and multiple starting points (see “Methods” section). The phylogenetic tree was constructed based on a multiple sequence alignment performed using PspA homologs across the phylum. As with Fig. 1, the key actinobacterial orders were labeled next to distinct clusters of similar PspA proteins. b Genomic contexts of PspA overlaid on the PspA phylogenetic tree. The tree was constructed as described in panel A. The genomic contexts identified in Fig. 3 (numbers retained) were overlaid atop the phyletic positions of the corresponding PspA proteins on the tree. The colors blue, orange, green and purple correspond to context configurations 1–4 described in Fig. 2. In both panels, the tree leaves are labeled by species and genomic context
Another notable characteristic associated with PspA in Actinobacteria is the presence of paralogs in several orders, including Corynebacteriales, Frankiales, Micrococcales and Streptomycetales [Fig. 3, Table A1 (Datta et al. 2015; Vrancken et al. 2008; Manganelli and Gennaro 2016)]. The position of the paralogs on the tree have notable implications for their origin and functions. For example, the paralogs in Corynebacterium and Frankia are significantly dissimilar and occupy distant positions on the tree (Fig. 3a, C1/C2, F1/F2). In contrast, the PspA paralogs in Arthrobacter and Streptomyces are similar, as demonstrated by their membership to the same cluster in the tree (Fig. 3a, A1/A2, S1/S2). The closely clustering paralogs might have arisen from recent lineage-specific gene duplication whereas dissimilar paralogs may have resulted from lateral gene transfer events.
Next, we investigated the relationship between genomic context and evolution of PspA by overlaying genomic configurations on the PspA phylogenetic tree. The predominant PspA configuration (#1 in Fig. 2) is present in the largest cluster of PspA homologs (Fig. 3b, blue cluster corresponding to configuration #1 from Fig. 2). The contexts bearing NYN/Trx, the two-component system, or alternative membrane proteins/transcriptional regulators form smaller and less distinct groups (Fig. 3b, green, orange, and purple leaves, respectively). These analyses may provide insight about the functional evolution of PspA. For example, when two copies of PspA are present, as in Corynebacterium, they are part of two different genomic neighborhoods. One of them likely represents the envelope stress-response system. These results suggest that PspA paralogs embedded in different genomic neighborhood may be functionally different, even in the same organism.
In conclusion, several Psp proteins are prevalent in the phylum Actinobacteria, in addition to PspA. These include PspM and PspN (originally discovered in Mycobacteria), PspC (reported initially in Gammaproteobacteria) and the Lia proteins (studied in Firmicutes). The analysis of genomic neighborhoods shows that PspA occurs in four main contexts in Actinobacteria, with clgRpspAM being the predominant configuration. Moreover, our results indicate that PspA may have been adapted to alternative genomic contexts or derived via lateral transfer from other lineages. The analysis of PspA paralogs suggests that the second pspA copy may have been acquired by gene duplication or lateral transfer. In addition, since we find pspC orthologs in actinobacterial genomic contexts that do not contain pspA orthologs, it is possible that the PspC membrane domain may sense and respond to stress independently of PspA, as previously suggested (Kleine et al. 2017; Flores-Kim and Darwin 2015). Furthermore, the conservation of the Psp system across bacterial phyla—regardless of differences in envelope composition—may be explained by the consideration that PspA is an inner membrane protein, and biochemical differences in outer layers may not be relevant to the Psp function.
The results of the present study give rise to multiple questions. For example, how do different neighborhoods influence PspA function and/or determine its redundancy? What can be learned from the genomic contexts of the distinct variants of all other Psp members? How can evolutionary studies point to mechanisms by which Psp protein homologs express similar stress response functions? Are there functions of the Psp proteins that are independent from stress responses?
Methods
Query and subject selection
All known Psp members—PspA (from Escherichia coli, M. tuberculosis, two copies from Bacillus subtilis); PspM (Rv2743c) and PspN (Rv2742c) (from M. tuberculosis); PspB and PspC (from E. coli); LiaI, LiaG, and LiaF (from B. subtilis)—were queried against all sequenced actinobacterial genomes (~450 of a total of ~6500 completed bacterial genomes; NCBI NR database; Homologs listed in Table A1). This set of genomes contained representative sequences from all actinobacterial classes and orders, except Actinopolysporales and Jiangellales. The phyletic order (sequence) was obtained from NCBI taxonomy and PATRIC (Wattam et al. 2014).
Identification and characterization of protein homologs
To ensure identification of a comprehensive set of homologs (close and remote) for each queried protein, we performed iterative searches using PSIBLAST (Altschul et al. 1997) and sequences of both full-length proteins and corresponding constituent domains. For each protein, searches were conducted using homologous copies from multiple species as starting points. Search results were aggregated and the numbers of homologs per species and of genomes carrying each of the query proteins were recorded (Table A1). These proteins were clustered into orthologous families using the similarity-based clustering program BLASTCLUST (ftp://ncbi.nih.gov/blast/documents/blastclust.html). HHPred, SignalP, TMHMM, Phobius, JPred, Pfam and custom profile databases (Soding et al. 2005; Cole et al. 2008; Sonnhammer et al. 1997; Mistry and Finn 2007; Nielsen 2017; Finn et al. 2011; Kall et al. 2004; Sonnhammer et al. 1998) were used to identify signal peptides, transmembrane regions, known domains and the secondary protein structures in every genome.
Neighborhood search
Bacterial gene neighborhoods (± 7 genes flanking each protein homolog) were retrieved from GenBank (Benson et al. 2013). Gene orientation, domains and secondary structures of the neighboring proteins were characterized using the same methods applied to query homologs.
Phylogenetic analysis
Multiple sequence alignment of the identified homologs was performed using Kalign (Lassmann et al. 2009) and MUSCLE (Edgar 2004). The phylogenetic tree was constructed using FastTree 2.1 with default parameters (Price et al. 2010); this tree was used to overlay the genomic context. Data analyses and visualizations were carried out using R (https://www.r-project.org).
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM (2005) The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29:231–262
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2013) GenBank. Nucleic Acids Res 41:D36–D42
Brissette JL, Russel M, Weiner L, Model P (1990) Phage shock protein, a stress protein of Escherichia coli. Proc Natl Acad Sci USA 87:862–866
Cole C, Barber JD, Barton GJ (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36:W197–W201
Darwin AJ (2005) The phage-shock-protein response. Mol Microbiol 57:621–628
Datta P, Ravi J, Guerrini V, Chauhan R, Neiditch MB, Shell SS, Fortune SM, Hancioglu B, Igoshin OA, Gennaro ML (2015) The Psp system of Mycobacterium tuberculosis integrates envelope stress-sensing and envelope-preserving functions. Mol Microbiol 97:408–422
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37
Flores-Kim J, Darwin AJ (2015) Activity of a bacterial cell envelope stress response is controlled by the interaction of a protein binding domain with different partners. J Biol Chem 290:11417–11430
Flores-Kim J, Darwin AJ (2016) The phage shock protein response. Annu Rev Microbiol. https://doi.org/10.1146/annurev-micro-102215-095359
Hutchings MI, Hoskisson PA, Chandra G, Buttner MJ (2004) Sensing and responding to diverse extracellular signals? Analysis of the sensor kinases and response regulators of Streptomyces coelicolor A3(2). Microbiology 150:2795–2806
Huvet M, Toni T, Sheng X, Thorne T, Jovanovic G, Engl C, Buck M, Pinney JW, Stumpf MP (2011) The evolution of the phage shock protein response system: interplay between protein function, genomic organization, and system function. Mol Biol Evol 28:1141–1155
Huynen M, Snel B, Lathe W, Bork P (2000) Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res 10:1204–1210
Joly N, Engl C, Jovanovic G, Huvet M, Toni T, Sheng X, Stumpf MP, Buck M (2010) Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol Rev 34:797–827
Kall L, Krogh A, Sonnhammer EL (2004) A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036
Kleine B, Chattopadhyay A, Polen T, Pinto D, Mascher T, Bott M, Brocker M, Freudl R (2017) The three-component system EsrISR regulates a cell envelope stress response in Corynebacterium glutamicum. Mol Microbiol 106(5):719–741
Koonin EV, Wolf YI (2008) Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 36:6688–6719
Korbel JO, Jensen LJ, von Mering C, Bork P (2004) Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat Biotechnol 22:911–917
Lassmann T, Frings O, Sonnhammer EL (2009) Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res 37:858–865
Manganelli R, Gennaro ML (2016) Protecting from envelope stress: variations on the phage-shock-protein theme. Trends Microbiol. https://doi.org/10.1016/j.tim.2016.11.010
Mascher T, Zimmer SL, Smith TA, Helmann JD (2004) Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob Agents Chemother 48:2888–2896
Mistry J, Finn R (2007) Pfam: a domain-centric method for analyzing proteins and proteomes. Methods Mol Biol 396:43–58
Nielsen H (2017) Predicting secretory proteins with SignalP. Methods Mol Biol 1611:59–73
Overmars L, Kerkhoven R, Siezen RJ, Francke C (2013) MGcV: the microbial genomic context viewer for comparative genome analysis. BMC Genom 14:209
Price MN, Dehal PS, Arkin AP (2010) FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490
Rogozin IB, Makarova KS, Wolf YI, Koonin EV (2004) Computational approaches for the analysis of gene neighbourhoods in prokaryotic genomes. Brief Bioinform 5:131–149
Soding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248
Sonnhammer EL, Eddy SR, Durbin R (1997) Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405–420
Sonnhammer EL, Eddy SR, Birney E, Bateman A, Durbin R (1998) Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res 26:320–322
Vothknecht UC, Otters S, Hennig R, Schneider D (2012) Vipp1: a very important protein in plastids?! J Exp Bot 63:1699–1712
Vrancken K, van Mellaert L, Anne J (2008) Characterization of the Streptomyces lividans PspA response. J Bacteriol 190:3475–3481
Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJ, Yoo HS, Zhang C, Zhang Y, Sobral BW (2014) PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 42:D581–D591
Acknowledgements
This work was supported in part by a grant from the National Institutes of Health (R01AI104615) to MLG. We thank members of the Gennaro and Aravind laboratories for valuable discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ravi, J., Anantharaman, V., Aravind, L. et al. Variations on a theme: evolution of the phage-shock-protein system in Actinobacteria. Antonie van Leeuwenhoek 111, 753–760 (2018). https://doi.org/10.1007/s10482-018-1053-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-018-1053-5