Abstract
Tioman Island is one of many sources for underexplored actinobacterial diversity in Malaysia. Selective isolation, molecular profiling, 16S rRNA gene sequencing and phylogenetic analyses were carried out to highlight the diversity of the marine actinobacterial community in a sediment collected off Tioman Island. A high number of diverse actinobacteria were recovered using skim milk/HEPES pre-treatment on a mannitol-based medium. A total of 123 actinobacterial strains were isolated, including thirty obligate marine actinobacteria putatively identified as Salinispora spp. Molecular fingerprinting profiles obtained with a double digestion approach grouped the remaining non-Salinispora-like strains into 24 different clusters, with Streptomyces and Blastococcus as the major clusters. A total of 17 strains were identified as novel actinobacterial species within the genera Streptomyces (n = 6), Blastococcus (n = 5), Marinactinospora (n = 3), Nocardiopsis (n = 1), Agromyces (n = 1) and Nonomuraea (n = 1) based on 16S rRNA gene sequence analyses. Polyphasic data from three putative Marinactinospora spp. showed that the strains represent a new genus in the Nocardiopsaceae family. Crude extracts from the strains were also found to inhibit the growth of Gram-positive (Staphylococcus aureus, Bacillus subtilis) and Gram-negative (Providencia alcalifaciens) pathogens. Hierarchical clustering of the bioactivities of an active fraction revealed a unique profile, which is closely related that of fosfomycin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Actinobacteria lineage justifies considerable attention for their biotechnologically important natural products since the discovery of actinomycin in the 1940s (Waksman and Woodruff 1941). Members of the class Actinobacteria are Gram-positive bacteria, most of which contain high G+C content within the genome and are characterised by a homologous insertion of 100 nucleotides between helices 54 and 55 of the 23S rRNA gene (Ventura et al. 2007). Terrestrial originated actinobacteria are extraordinarily diverse, but the ocean that covers three-quarters of the earth’s surface has the greatest diversity. The presence of indigenous marine actinobacteria was supported by the discoveries and descriptions of marine species such as Dietzia maris (Nesteranko et al. 1982; Rainey et al. 1995), Rhodococcus marinonascens (Helmke and Weyland 1984), Salinibacterium amurskyense (Han et al. 2003), Williamsia maris (Stach et al. 2004) and the first obligate marine genus Salinispora (Maldonado et al. 2005). Marine actinobacteria are mostly derived from marine sediments (54%), followed by sponges (21%), marine invertebrates and sea water (Abdelmohsen et al. 2014).
While culture independent or metagenomic studies are the leading trends in actinobacterial diversity and natural product discoveries, culture dependent studies are essential approaches for cultivation of natural actinobacterial strains that can be screened for biological activities of interests and studied for expression of enzymes and bioactive molecules (Vester et al. 2015). However, it is well established that less than 1% of bacteria can be readily cultivated in vitro (Amann et al. 1995). Fastidious growth requirements, including the need for specific nutrients and growth factors, are among the main obstacles in attempts to cultivate the unculturable (Köpke et al. 2005). Moreover, the dominant species on isolation plates introduce strong interspecies competition for nutrients, which further discourages successful isolation of rare actinobacterial species, causing them to be unculturable. Rare and novel actinobacteria represent unique sources of novel biologically active compounds. Selective isolation techniques using various pre-treatments and incorporation of unusual carbon sources were found to favour the growth of certain taxonomic groups of actinobacteria and encouraged isolation of rare actinobacteria species (Bredholdt et al. 2007; Sun et al. 2010).
Actinobacteria are among saprophytes that exhibit a wide extent of survival and adaptive strategies to persist in natural environments. In certain genera such as Actinoplanes, Dactylosporangium, Geodermatophilus, Planomonospora and Spirillospora, spores are harboured in sporangia as motile flagellated zoospores (Garrity et al. 1996). Some of the non-spore forming actinobacteria such as Blastococcus spp. form motile, single flagellated cells. Production of motile spores and cells enables marine actinobacteria to exhibit chemotaxis and access more nutrient sources. Various selective methods have been employed to isolate zoosporic actinobacteria. These include baiting techniques, chemotactic methods that use organic and/or inorganic nutrients as chemoattractants, and centrifugation methods to improve the total number of actinobacterial isolates originating from terrestrial soil samples (Garrity et al. 1996; Otoguro et al. 2001; Dennis et al. 2013). A flooding solution containing skim milk was postulated to stimulate the motility of spores, thus facilitating the isolation of zoosporic actinobacteria (Suzuki 2001). Actinobacteria are also known to resist high doses of ultraviolet (UV) and high frequency irradiation through production of spores and pigments. Bredholdt et al. (2007) demonstrated the efficiency of ultraviolet irradiation and high frequency irradiation to selectively isolate various actinobacteria from shallow water sediments of the Trondheim fjord, Norway.
Tioman Island, located in the state of Pahang, Malaysia, surrounded by the South China Sea, was reported to be an untapped source of rare marine actinobacteria (Vikineswary et al. 2008). The authors isolated diverse actinobacteria from marine sponges and putatively identified selected isolates as Actinoplanes spp., Micromonospora spp., Nocardia spp., Polymorphospora spp., Pseudonocardia spp., Rhodococcus spp., Saccharomonospora spp., Salinispora spp., Sprilliplanes spp. and Verrucosispora spp. Marine actinobacteria were reported to be excellent producers of antimicrobial compounds, in which the backbone of these compounds were synthesized by large enzymes, polyketide synthases and non-ribosomal peptide synthetases (Gomez-Escribano et al. 2016). The objectives of this study were to isolate diverse actinobacteria from a marine sediment sample collected from Tioman Island using various selective cultivation techniques and to investigate the potential antibacterial activity in the isolated strains.
Materials and methods
Sampling and pre-treatment of marine sediment sample
A marine sediment sample was collected on 13 March 2013, from a depth of 7 m by scuba diving, from Pirate Reef, Tioman Island, Pahang, Malaysia (N: 02°49′27.1″, E: 104°09′25.0″). Temperature, salinity and pH of the sampling site were recorded. The sediment sample was homogenized by vigorous vortexing and pre-treated separately using three different methods: (a) UV irradiation (Bredholdt et al. 2007), (b) skim milk/HEPES (0.1% skim milk in 10 mmol/l of HEPES) treatment (modified from Xin et al. 2009) and (c) skim milk/HEPES treatment followed by enrichment in humic acid vitamin broth (HVB) (modified from Xin et al. 2009). Briefly, 200 µl of skim milk/HEPES treated suspension from (b) was transferred into 1.8 ml HVB and incubated at 28 °C with shaking at 200 rpm for 24 h prior to serial dilution.
Isolation of actinobacteria from marine sediment
Ten-fold serial dilutions and isolation media were prepared using 3% artificial sea water (Instant Ocean®, Aquarium Systems, Sarrebourg, France). The sediment suspension (100 µl) was spread on the surface of three different isolation media: peptone-asparagine agar (M3) (Zhang et al. 2008), mannitol-arginine agar [modified from medium M2 (Zhang et al. 2008)], and humic acid vitamin agar (HVA) (Xin et al. 2009). The mannitol-arginine medium was prepared using 0.5% mannitol, 0.1% l-arginine, 0.1% K2HPO4 and 0.05% MgSO4. All isolation media were supplemented with nalidixic acid (15 µg/ml) and nystatin (25 µg/ml) and pH of the media were adjusted to 7.5. All the isolation plates were incubated at 28 °C for up to 8 weeks. Colonies were examined under an inverted microscope. Purified actinobacterial strains were maintained on either modified yeast extract malt extract agar (ISP2, Shirling and Gottlieb 1966) or modified Bennett’s agar (MBA) slopes (Tan et al. 2006) and preserved in 20% glycerol suspension at −20 and −80 °C.
Molecular characterisation
Isolated actinobacterial strains were first grouped into Salinispora-like and non-Salinispora-like strains based on colony colour and presence of aerial mycelium on both ISP2 and MBA plates. Restriction fragment length polymorphism (RFLP) fingerprinting of 16S rRNA gene and its adjacent 16S-23S internal transcribed spacer (ITS) region was used to dereplicate these two groups of marine actinobacterial strains. Four to five days old cultures were used for genomic DNA extraction which was performed with NucleoSpin® Tissue genomic DNA extraction kit (Macherey–Nagel, Germany), according to the manufacturer’s instruction. Amplification of the 16S rRNA gene and the adjacent 16S-23S ITS region using the primer pair pA and BL235R (Lanoot et al. 2005) was carried out on all non-Salinispora-like strains. Amplicons were digested using HaeIII (10 U) at 37 °C for 5 min and subsequently with BstU1 (10 U) at 60 °C for 5 min, following the online protocol recommended by New England BioLabs (NEB, Massachusetts, USA; https://nebcloner.neb.com/#!/redigest) and the fragments were resolved on 2% agarose gel electrophoresis in 1 × TAE buffer. The RFLP banding profiles of non-Salinispora-like actinobacterial strains were analysed with the BioNumerics software (version 7.1, Applied Maths, Belgium). Bands from 100 to 1000 bp were included for analysis for 16S ITS RFLP fingerprinting using band-based similarity Dice coefficient. Band tolerance positions were set at 0.11%. UPGMA dendogram was derived from the resultant similarity matrixes. The 16S rRNA ITS region of the Salinispora-like strains was amplified according to Jensen et al. (1993). The resulted PCR product was subjected to restriction enzyme digestion with 5U of BanI (NEB) and incubated at 37 °C for 15 min, followed by gel electrophoresis on a 2.5% agarose gel (Freel et al. 2012). Amplification of the 16S rRNA gene was carried out as described by Vidgen et al. (2012) using MyTaq™ Red DNA Polymerase (Bioline, UK). Sequencing of the 16S rRNA gene was performed by First Base Laboratories Sdn Bhd (Malaysia) using the BigDye® Terminator v3.1 cycle sequencing kit chemistry. Closest matches were identified using EzBioCloud database (Yoon et al. 2017) and corresponding 16S rRNA gene sequences were retrieved from the database for phylogenetic analysis using MEGA version 6.0 (Tamura et al. 2013). Cut-offs for classification of potential novel taxa were based on 16S rRNA gene sequence similarity values of 98.7% (Sangal et al. 2016). Phylogenetic trees were constructed using neighbour-joining algorithm (Saitou and Nei 1987). Evolutionary distance matrices were generated as described by Tamura (1992) and Tamura and Nei (1993). Bootstrap values were calculated based on 1000 re-samplings (Felsenstein 1985).
Phenotypic characterisation
Colony morphology of the isolated marine actinobacterial strains, including colour of aerial and substrate mycelium and production of diffusible pigmentation, were examined on ISP media number 2 and MBA on day 7 and day 14 of incubation at 28 °C (Shirling and Gottlieb 1966) using the ISCC-NBS colour charts (Kelly 1958). Novel marine actinobacterial strains were tested for their ability to tolerate various growth temperature, pH and sodium chloride. The ISP2 medium served as the basal medium for growth. Growth of purified actinobacterial strains was assessed at ten different temperatures (°C): 4, 10, 15, 20, 25, 32, 37, 45, 50 and 55. The pH range for growth was examined at pH 5.0 to 13.0 in increments of 1 pH unit (Xu et al. 2005). Tolerance to sodium chloride (NaCl) was tested at concentrations up to 10% in increment of 1% NaCl (w/v). Growth of all actinobacterial strains were observed and recorded after 14 days of incubation at 28 °C.
Assessment of antibacterial activity
Antibacterial activity of marine actinobacterial strains were assayed by agar plug diffusion. The strains were cultured on five different production media supplemented with artificial sea salt and 1.5% agar: PM3 (Bredholt et al. 2008), soybean meal glucose (Zheng et al. 2000), MMS (Ismet et al. 2004), Waksman’s glucose agar (ATCC medium 241) and SYP (Bose et al. 2015). Agar plates were incubated at 28 °C for up to 21 days. Four test pathogens (Bacillus subtilis ATCC 23857, Staphylococcus aureus ATCC 29213, Escherichia coli ATCC 47076, Pseudomonas aeruginosa ATCC 27853) were cultured at 37 °C for 18 h and the turbidity adjusted to 0.5 McFarland standard prior to use. Lawn cultures of pathogens with the agar plugs were incubated at 37 °C for 18 h. Antibacterial activity was recorded as positive when diameter of inhibition zone was ≥ 10 mm. Selected actinobacterial strains which showed potent antibacterial activity against tested pathogens were cultured in soybean meal glucose broth supplemented with 20 g per litre of Amberlite XAD-16 resins. The cultures were incubated at 28 °C for 21 days in an orbital shaker and crude extract was obtained by solvent extraction using dichloromethane: methanol (50:50, v/v). Dried crude extracts were subjected to solid phase extraction (SPE) using step gradient elution with methanol and water mixture from 20:80 to 100:0 and a final column flush with 100% ethyl acetate to yield six fractions. SPE fractions were dried and re-constituted in dimethyl sulfoxide (DMSO) and screened for antibacterial activity against a panel of fifteen pathogens which include Gram-positive pathogens: B. subtilis ATCC 23857, Listeria ivanovii ATCC BAA-139, Enterococcus faecium ATCC 6569, Staphylococcus epidermidis ATCC 14990T, S. aureus ATCC 29213 and S. aureus ATCC BAA-44 (MRSA) and Gram-negative pathogens: E. coli ATCC 47076, Providencia alcalifaciens ATCC 9886T, Ochrobactrum anthropi ATCC 49687, Enterobacter aerogenes ATCC 35029, Acinetobacter baumannii ATCC 19606T, P. aeruginosa ATCC 27853, Salmonella enterica subsp. enterica serovar Typhimurium ATCC 700720, Vibrio cholerae O1 (biotype EI Tor A1552) and Yersinia pseudotuberculosis (IP2666 pIBI). Overnight cultures of pathogens were diluted 1:1000 and seeded in a volume of 30 µl/well in sterile clear propylene 384-wells assay plates. Each well was fed with 300 nl of DMSO fractions using a high-throughput pinning robot (Perkin Elmer Janus MDT). Growth curves of pathogens were measured at OD600 at hourly intervals over 24 h in an automated plate reader/shaker (EnVision, Perkin Elmer). DMSO fractions that inhibited Gram-positive and Gram-negative pathogens were selected for parallel screening of twofold dilution series to determine the MIC values. Data were normalized and a BioMap profile was created according to Wong et al. (2012). Normalized MIC values were indicated as 0 or 1. The BioMap profile of the fraction was compared to profiles of training set of antibiotics by hierarchical clustering using Cluster 3.0 software. The cluster plot was displayed as red–black color scheme with a gradient from inactive (black) to most active (red) using TreeView (v1.1.6).
Results
Selective isolation of marine actinobacteria
The marine sediment sample collected from Tioman Island, Pahang, Malaysia had a pH of 7.5 and salinity of 34 parts-per-thousand. Viable colony counts of actinobacteria in the wet sediment sample were in the range of 1.3 × 103 to 3.0 × 104 cfu/g, depending on the type of pre-treatment and isolation media. A total of 123 putative actinobacterial strains was isolated from the marine sediment sample and the strains were initially separated into two groups based on colony morphology. The first group comprised of 76 non-Salinispora-like strains, whereas the second group consisted of 47 Salinispora-like strains. The non-Salinispora-like strains were colonies with black, blue, brown, gray, red, white, yellow, and green coloured aerial mycelia. The Salinispora-like strains were bright orange colonies lacking aerial mycelium, similar to morphological features of the members of the genus Salinispora.
A total 114 putative marine actinobacterial strains was isolated using the pre-treatment method with skim milk/HEPES solution (Table 1). This method coupled with the use of a mannitol-based isolation medium and HVA recovered high numbers of actinobacterial strains. This pre-treatment also supported the isolation of high numbers of Salinispora-like and non-Salinispora-like strains on mannitol-arginine agar (n = 37) and HVA plates (n = 41), respectively (Table 1). The remaining nine strains were recovered from the pre-treatment with UV irradiation (n = 2) and HVB enrichment of skim milk/HEPES treated sample (n = 7) with only four and three strains from mannitol-arginine agar and HVA plates, respectively (data not shown).
Diversity of marine actinobacterial strains
Analyses of the RFLP profiles from 68 non-Salinispora-like strains generated 24 clusters (Fig. 1). The 16S rRNA-ITS region amplified by primer pair pA/BL235R has an amplicon size of approximately 1.8 kb and up to ten fragments between 100 bp to 1 kbp were generated from the double RE digestion. The remaining eight non-Salinispora-like actinobacterial strains did not produce any amplicons using the primer pair pA/BL235R. Subsequent 16S rRNA gene sequence analyses revealed that the strains were closely related to Marinactinospora spp. (n = 3), Rhodococcus spp. (n = 2), Nocardiopsis spp. (n = 1), Agromyces spp. (n = 1) and Saccharomonospora spp. (n = 1) (Table 2).
Thirty of the 47 Salinispora-like strains profiled using BanI digestion of the 16S-23S ITS amplicons showed the same banding pattern as Salinispora arenicola CNH-643T. The 16S rRNA gene sequences of four randomly selected strains were found to share 100% similarity with the corresponding sequence of S. arenicola CNH-643T. The remaining 17 strains which had different restriction banding profiles from S. arenicola CNH-643T were found to be closely related to Micromonospora spp. (n = 8), Nocardia spp. (n = 2), Mycobacterium spp. (n = 2), Jishengella spp. (n = 1), Plantactinospora spp. (n = 1), Pseudonocardia spp. (n = 1), Nonomuraea spp. (n = 1) and Rhodococcus spp. (n = 1) based on 16S rRNA gene sequence analyses (data not shown).
Characterisation of novel marine actinobacterial strains
A total of 17 actinobacterial strains were found to represent novel taxa (Table 3, Figs. 2, 3). Fourteen of the novel species were isolated using skim milk/HEPES pre-treated sample, one from HVB enrichment of skim milk/HEPES pre-treated sample and two from sample pre-treated with UV irradiation.
Neighbour-joining tree based on almost full length 16S rRNA gene sequences of strains TPS3, TPS4, TPS114, TPS137, TPS143, TPS183 and closely related Streptomyces spp. Evolutionary distances were computed using Tamura 3-parameter method (Tamura 1992). Bootstrap values are denoted at nodes on branches based on 1000 re-sampling. Only values higher or equal to 50% are indicated. Bar represents 2% sequence divergence
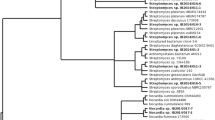
Neighbour-joining tree based on almost full length 16S rRNA gene sequences of eleven novel strains and closely related members of the families Geodermatophilaceae, Microbacteriaceae, Nocardiopsaceae and Streptosporangiaceae. Evolutionary distances were calculated using the algorithm of Tamura-Nei model. Bootstrap values are denoted at nodes on branches based on 1000 re-sampling. Only values higher or equal to 50% are indicated. Bar represents 2% sequence divergence
Physiological and morphological characteristics of the novel actinobacterial strains are summarised in Table 3. All strains grew well at pH range from 6 to 12, except for strains TPS3, TPS4, TPS114 and TPS358a which only tolerated up to pH 11. Strains TPS2, TPS3, TPS16, TPS81 and TPS83 were able to grow at pH 5. All strains were able to grow without the presence of NaCl. Only one strain, TPS2, could tolerate up to 10% NaCl (w/v). In terms of temperature, all strains showed good growth at 25, 28, 32 and 37 °C. Streptomyces sp. TPS143, TPS137, TPS114 and TPS183 were able to grow at 15 °C or lower. Two strains, TPS183 (Streptomyces sp.) and TPS92 (Agromyces sp.), were able to grow at 4 °C. In contrast, Blastococcus sp. TPS166 and TPS418 as well as strains TPS16, TPS81 and TPS83 were able to grow at 50 °C. One Blastococcus strain TPS418 also showed good growth at 55 °C.
Three marine actinobacteria, strains TPS16, TPS81 and TPS83, producing blue aerial mycelia and diffusible pigmentation, were identified to be novel species within the family Nocardiopseaceae (Table 3) based on 16S rRNA gene sequence analyses. These strains share 100% similarity of 16S rRNA gene sequence (1399–1433 bp) between each other and below 98.7% similarities to the closest species, Marinactinospora thermotolerans SCSIO00652T. All three strains formed a distinct and tight cluster within members of the Nocardiopsaceae family in the neighbour-joining phylogenetic tree (Fig. 3).
Assessment of antibacterial activity
A total of 13 strains from the Streptomyces cluster excluding the novel species were observed to exhibit antibacterial activity against at least one pathogen (Table 4). Strains TPS6, TPS10, TPS12 and TPS17 inhibited the growth of Gram-negative as well as Gram-positive pathogens. Strains TPS10, TPS12 and TPS17 growing on Waksman’s glucose agar and SYP media inhibited the growth of E. coli ATCC 47076 and S. aureus ATCC 29213. Another Streptomyces strain TPS6 inhibited the growth of P. aeruginosa ATCC 27853, S. aureus ATCC 29213 and B. subtilis ATCC 23857. Strain TPS37 which was closely related to Saccharopolyspora hirsuta subsp. hirsuta ATCC 27875T was able to inhibit S. aureus ATCC 29213 and B. subtilis ATCC 23857 when cultured on PM3.
The orange coloured strains TPS111 and TPS121 were among the Salinispora-like species that exhibited antibacterial activity. Both strains were identified to be Micromonospora spp. that inhibited the growth of Gram-positive S. aureus ATCC 29213 and B. subtilis ATCC 23857. Strain TPS111 produced activity on four of the five production media, whereas strain TPS121 produced activity only on SYP. All the 30 strains which were highly similar to S. arenicola CNH-643T exhibited antibacterial activity against Gram-positives S. aureus ATCC 29213 and B. subtilis ATCC 23857. A total of 24 strains produced antibacterial activity on all five production media. Interestingly, strain TPS178 showed positive inhibitory activity against Gram negative P. aeruginosa ATCC 27853. Another strain TPS104 also inhibited the growth of P. aeruginosa ATCC 27853 along with the Gram-positive B. subtilis ATCC 23857 and S. aureus ATCC 29213 when it was cultured on Waksman’s glucose medium and SYP; however, it inhibited only S. aureus ATCC 29213 when cultured on PM3 and soybean meal glucose medium (Table 4).
Among the 17 novel strains, the Streptomyces spp. TPS114 and TPS137 were active against B. subtilis ATCC 23857 and S. aureus ATCC 29213, whereas strain TPS143 was only active against B. subtilis ATCC 23857 (Table 4). All three strains produced antibacterial activity on production medium MMS. Three strains belonging to the family Nocardiopsaceae, TPS16, TPS81, and TPS83, grown on soybean meal glucose medium, were shown to exhibit antibacterial activity against E. coli ATCC 47076, B. subtilis ATCC 23857 and S. aureus ATCC 29213. The strain TPS83 was thus selected for further study based on novelty and spectrum of activities. Examination of the crude SPE fractions of TPS83 revealed the antibacterial potential of this novel strain. The fraction TPS83_D eluted with 80% methanol–water appeared to be the most potent among the six fractions as it inhibited the growth of Gram-positive pathogens and Gram-negative P. alcalifaciens ATCC 9886T. MIC values of crude SPE fraction TPS83_D were determined as 100 µM for B. subtilis ATCC 23857, E. faecium ATCC 6569, S. aureus ATCC 29213 and S. aureus ATCC BAA-44, 25 µM for S. epidermidis ATCC 14990T and 6.25 µM for P. alcalifaciens ATCC 14990T by parallel screening of the twofold dilution series of the fraction. The BioMap profile of fraction TPS83_D formed a distinct cluster from the training set of antibiotics suggesting a different antibiotic profile (Fig. 4).
Fraction TPS83_D was found to be the most active among the six fractions, showing inhibitory activity against six out of 15 tested pathogens. Hierarchical clustering of active fractions with reference antibiotics based on normalized MIC values revealed a single and distinct cluster of all TPS83 crude fractions, which was closely related to fosfomycin based on the activity profile. Potency of fractions was represented by a red–black colour scheme: inactive (black) and most active (red). (Color figure online)
Discussion
Selective isolation of diverse actinobacteria
In this study, skim milk/HEPES pre-treatment with centrifugation at 1000×g was shown to improve isolation of diverse actinobacteria from the sediment sample, yielding members of 18 actinobacterial genera. Shannon–Wiener index (H′) for the pre-treated sample indicated that mannitol-arginine agar and HVA recovered greater diversity of actinobacteria than peptone-asparagine agar (Fig. 5). By coupling skim milk treatment with centrifugation at 1000–1500×g, numbers of non-motile actinobacteria and Streptomyces spp. colonies decreased, thereby facilitating isolation of rare actinobacteria (Suzuki et al. 1999; Hayakawa 2008). However, the HVB enrichment of skim milk/HEPES treated sample encouraged the growth of higher populations of fast growing non-actinobacterial strains. Thus, the recovery rate of actinobacteria was much lower compared to the skim milk/HEPES pre-treatment without enrichment in HVB. Humic acid vitamin agar was found to support growth of non-Salinispora-like strains. Humic acid is a sole carbon and nitrogen source that encourages growth of spore-forming actinobacteria including various rare actinobacteria, while reducing growth of non-filamentous bacterial colonies (Hayakawa 2008).
Diversity of actinobacteria recovered from Tioman marine sediment following skim milk/HEPES treatment on peptone-asparagine, mannitol-arginine and humic acid vitamin agars. Shannon–Wiener index (H′) and total number of actinobacterial strain (n) isolated from each isolation medium are also indicated
In addition, most of the Salinispora-like strains were isolated using a mannitol-based medium modified for this study. Isolation medium with low concentrations of mannitol had been reported to yield marine strains including Salinispora spp. from marine sediments (Jensen et al. 2005). Highly UV-resistant actinobacteria of the genera Arthrobacter, Curtobacterium and Geodermatophilus were isolated from desert rock samples treated with UV irradiation (Kuhlman et al. 2005). These actinobacteria were found to appear in clusters or aggregates. In this study, the only strains recovered from the UV irradiation pre-treatment method were Blastococcus spp. which were observed as irregularly shaped coccoid cell aggregates.
Diversity and characterisation of isolated actinobacteria
In general, analyses of the 16S rRNA gene sequences of marine actinobacterial strains indicated close relationships to members of 18 genera: Actinomadura, Agromyces, Blastococcus, Jishengella, Marinactinospora, Micromonospora, Mycobacterium, Nocardia, Nocardiopsis, Nonomuraea, Plantactinospora, Pseudonocardia, Rhodococcus, Saccharomonospora, Saccharopolyspora, Salinispora, Streptomyces, and Streptosporangium. Almost half and a quarter of the total actinobacteria isolated were Streptomyces spp. (47.97%) and Salinispora spp. (23.58%), respectively.
The non-Salinispora-like strains were grouped into two major clusters: the Streptomyces cluster (Clusters 1–5, Cluster 7, Clusters 9–21, Cluster 23) and the Blastococcus cluster (Cluster 24) (Fig. 1). The Streptomyces cluster was found to share less than 10 RFLP banding patterns suggesting high diversity between the isolated strains of this genus. Analyses of 16S rRNA gene sequences indicated that Streptomyces spp. were isolated in a high number on mannitol-arginine and HVA media. Strains of Salinispora spp. and Blastococcus spp. were recovered exclusively from mannitol-arginine agar regardless of pre-treatments. Genera that were also recovered from mannitol-arginine agar included Nonomuraea, Saccharomonospora, Nocardiopsis, Plantactinospora and Pseudonocardia. Although the Plantactinospora spp. isolated from this study shared 99.4% similarity of 16S rRNA gene sequence to Plantactinospora endophytica YIM 68255T, this is the first report on isolation of Plantactinospora sp. from a marine sediment.
All five strains in the Blastococcus cluster (strains TPS166, TPS357, TPS418, TPS448 and TPS459) were putatively identified as novel species based on 16S rRNA sequence analyses. There are only five Blastococcus species validly named to date: Blastococcus aggregatus, Blastococcus capsensis, Blastococcus endophyticus, Blastococcus jejuensis and Blastococcus saxobsidens, which were isolated from brackish water (Ahrens and Moll 1970), archaeological Roman pool (Hezbri et al. 2016), medicinal plant leaves (Zhu et al. 2013), beach sediment (Lee 2006) and monument stones (Urzì et al. 2004), respectively. This study demonstrated the first isolation of members of the genus Blastococcus from a marine sediment sample. The strains were able to tolerate up to 8% NaCl; strains TPS166 and TPS418 showed growth up to 50 and 55 °C, respectively. In contrast, known Blastococcus species are only capable of tolerating up to 3% NaCl and 45 °C. Moreover, all five novel marine Blastococcus spp. derived from the Tioman marine sediment sample were able to grow from pH 6 to 12, whereas B. saxobsidens and B. jejuensis were reported to only tolerate up to pH 8 and pH 10, respectively.
Members within the family of Nocardiopsaceae are known to be halophiles or halotolerant species that tolerated 10% NaCl or above, as showed by members of the genera Haloactinospora and Salinactinospora (Tang et al. 2008; Chang et al. 2012). The type genus Nocardiopsis also contains alkaliphilic members such as Nocardiopsis valliformis and Nocardiopsis dassonvillei subsp. prasina that tolerate up to pH 13 (Miyashita et al. 1984; Yang et al. 2008). Members of the family Nocardiopsaceae are commonly present in terrestrial soil, however, the genus Spinactinospora was only discovered from marine sediments and the type species Spinactinospora alkalitolerans is known to be alkaliphilic (Chang et al. 2012). In this study, four new strains belonging to the Nocardiopsaceae family were isolated from the Tioman marine sediment: strain TPS2 is closely related to Nocardiopsis alba and strains TPS16, TPS81 and TPS83 were closely related to M. thermotolerans. These strains were characterised by their ability to tolerate up to pH 12 and 8-10% NaCl.
Overall, 17 novel species of actinobacteria were isolated from the sediment sample, constituting 21.5% of the total number of non-Salinispora-like strains and 13.5% of the total actinobacterial strains, indicating that marine sediments of Tioman Island is indeed a potential resource of novel and diverse actinobacteria.
Antibacterial activity of novel members of the family Nocardiopsaceae
Novel actinobacteria of the Nocardiopsaceae family, represented by the strains TPS16, TPS81 and TPS83, were recovered from HVA following skim milk/HEPES pre-treatment of the marine sediment sample. The novel strains cultured on soybean meal glucose medium were shown to be able to inhibit the growth of Gram-negative E. coli ATCC 47076 as well as Gram-positives B. subtilis and S. aureus ATCC 29213. Although all three strains shared 100% similarity of their 16S rRNA gene sequences, significantly different antibacterial activity profiles were obtained when cultured different production media. Strains TPS16 and TPS83 were able to inhibit both B. subtilis ATCC 23857 and S. aureus ATCC 29213 while strain TPS81 did not show any antibacterial activity when grown on PM3. In contrast, strain TPS81 was able to inhibit the same pathogens when grown on Waksman’s glucose agar and SYP but strains TPS16 and TPS83 could not. These results represent the possibility of producing strain-specific antibacterial compounds.
Hierarchical clustering of the active fractions obtained from the crude extract of strain TPS83 with BioMap profiles revealed that all the fractions formed a separate and distinct cluster although closely related to fosfomycin. The antibiotic fosfomycin is only one of the few antibiotics that still remained active against broad spectrum targets including the multi-drug resistant and extensively-drug resistant pathogens (Falagas et al. 2016). It is a bactericidal compound that interferes with the formation of UDP N-acetylmuramic acid, the peptidoglycan precursor, which is involved in the first cytoplasmic step of bacterial cell wall synthesis (Borisova et al. 2014). Fraction TPS83_D was shown to inhibit the largest number of pathogens, including Gram-negative P. alcalifaciens ATCC 14990T, as compared to other fractions. These results suggest potential novel antibacterial activity of the fractions and further studies are needed to isolate the bioactive compounds.
Conclusion
Culture dependent techniques are important in recovering bioactive actinobacteria from marine environmental samples. Our study demonstrated successful selective isolation of high numbers and diverse marine actinobacteria from a marine sediment including 17 novel actinobacterial strains based on skim milk/HEPES pre-treatment using mannitol-based and humic acid vitamin media. Future studies are on-going to describe the novel species and to identify their potentially novel antibacterial metabolites.
References
Abdelmohsen UR, Bayer K, Hentschel U (2014) Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat Prod Rep 31:381–399
Ahrens R, Moll G (1970) A new budding bacterium from the Baltic Sea. Arch Mikrobiol 70:243–265
Amann RI, Ludwig W, Schleifer K (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Borisova M, Gisin J, Mayer C (2014) Blocking peptidoglycan recycling in Pseudomonas aeruginosa attenuates intrinsic resistance to fosfomycin. Microb Drug Resist 20:231–237
Bose U, Hewavitharana AK, Ng YK, Shaw PN, Fuerst JA, Hodson MP (2015) LC-MS-Based metabolomics study of marine bacterial secondary metabolite and antibiotic production in Salinispora arenicola. Mar Drugs 13:249–266
Bredholdt H, Galatenko OA, Engelhardt K, Fjærvik E, Terekhova LP, Zotchev SB (2007) Rare actinomycete bacteria from the shallow water sediments of the Trondheim fjord, Norway: isolation, diversity and biological activity. Environ Microbiol 9:2756–2764
Bredholt H, Fjærvik E, Johnsen G, Zotchev SB (2008) Actinomycetes from sediments in the Trondheim Fjord, Norway: diversity and biological activity. Mar Drugs 6:12–24
Chang X, Liu W, Zhang XH (2012) Salinactinospora qingdaonensis gen. nov., sp. nov., a halophilic actinomycete isolated from a salt pond. Int J Syst Evol Microbiol 62:954–959
Dennis PG, Seymour J, Kumbun K, Tyson G (2013) Diverse populations of lake water bacteria exhibit chemotaxis towards inorganic nutrients. ISME J 7:1661–1664
Falagas ME, Vouloumanou EK, Samonis G, Vardakas KZ (2016) Fosfomycin. Clin Microbiol Rev 29:321–347
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Freel KC, Edlund A, Jensen PR (2012) Microdiversity and evidence for high dispersal rates in the marine actinomycete “Salinispora pacifica”. Environ Microbiol 14:480–493
Garrity GM, Heimbuch BK, Gagliardi M (1996) Isolation of zoosporogenous actinomycetes from desert soil. J Ind Microbiol 17:260–267
Gomez-Escribano JP, Alt S, Bibb MJ (2016) Next generation sequencing of actinobacteria for the discovery of novel natural products. Mar Drugs 14:78
Han SK, Nedashkovskaya OI, Mikhailov VV, Kim SB, Bae KS (2003) Salinibacterium amurskyense gen. nov., sp. nov., a novel genus of the family Microbacteriaceae from the marine environment. Int J Syst Evol Microbiol 53:2061–2066
Hayakawa M (2008) Studies on the isolation and distribution of rare actinomycetes in soil. Actinomycetologica 22:12–19
Helmke E, Weyland H (1984) Rhodococcus marinonascens sp. nov., an actinomycete from the sea. Int J Syst Bacteriol 34:127–138
Hezbri K, Louati M, Nouioui I, Gtari M, Rohde M, Spröer C, Schumann P, Klenk H, Ghodhbane-Gtari F, Montero-Calasanz MD (2016) Blastococcus capsensis sp. nov., isolated from an archaeological Roman pool and emended description of the genus Blastococcus, B. aggregatus, B. saxobsidens, B. jejuensis and B. endophyticus. Int J Syst Evol Microbiol 66:4864–4872
Ismet A, Vikineswary S, Paramaswari S, Wong WH, Ward A, Seki T, Fiedler HP, Goodfellow M (2004) Production and chemical characterization of antifungal metabolites from Micromonospora sp. M39 isolated from mangrove rhizosphere soil. World J Microbiol Biotechnol 20:523–528
Jensen MA, Webster JA, Straus N (1993) Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol 59:945–952
Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W (2005) Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ Microbiol 7:1039–1048
Kelly LK (1958) Central notations for the revised ISCC-NBS color-name blocks. J Res Natl Bur Stand 61:427–431
Köpke B, Wilms R, Engelen B, Cypionka H, Sass H (2005) Microbial diversity in coastal subsurface sediments: a cultivation approach using various electron acceptors and substrate gradients. Appl Environ Microbiol 71:7819–7830
Kuhlman KR, Allenbach LB, Ball CL, Fusco WG, La Duc MT, Kuhlman GM, Anderson RC, Stuecker T, Erickson IK, Benardini J, Crawford RL (2005) Enumeration, isolation and characterization of ultraviolet (UV-C) resistant bacteria from rock varnish in the Whipple Mountains, California. Icarus 174:585–595
Lanoot B, Vancanneyt M, Hoste B, Vandemeulebroecke K, Cnockaert MC, Dawyndt P, Liu Z, Huang Y, Swings J (2005) Grouping of streptomycetes using 16S-ITS RFLP fingerprinting. Res Microbiol 156:755–762
Lee SD (2006) Blastococcus jejuensis sp. nov., an actinomycete from beach sediment, and emended description of the genus Blastococcus Ahrens and Moll 1970. Int J Syst Evol Microbiol 56:2391–2396
Maldonado LA, Fenical W, Jensen PR, Kauffman CA, Mincer TJ, Ward AC, Bull AT, Goodfellow M (2005) Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int J Syst Evol Microbiol 55:1759–1766
Miyashita K, Mikami Y, Arai T (1984) Alkalophilic actinomycete, Nocardiopsis dassonvillei subsp. prasina subsp. nov., isolated from soil. Int J Syst Evol Microbiol 34:405–409
Nesteranko OA, Nogina TM, Kasumova SA, Kvasnikov EI, Batrakov SG (1982) Rhodococcus luteus nom. nov. and Rhodococcus maris nom. nov. Int J Syst Bacteriol 32:1–14
Otoguro M, Hayakawa M, Yamazaki T, Iimura Y (2001) An integrated method for the enrichment and selective isolation of Actinokineospora spp. in soil and plant litter. J Appl Microbiol 91:118–130
Rainey FA, Klatte S, Kroppenstedt RM, Stackebrandt E (1995) Dietzia, a new genus including Dietzia maris comb. nov., formerly Rhodoccous maris. Int J Syst Bacteriol 45:32–36
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sangal V, Goodfellow M, Jones AL, Schwalbe EC, Blom J, Hoskisson PA, Sutcliffe IC (2016) Next-generation systematic: An innovative approach to resolve the structure of complex prokaryotic taxa. Sci Rep 6:1–12
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Stach JE, Maldonado LA, Ward AC, Bull AT, Goodfellow M (2004) Williamsia maris sp. nov., a novel actinomycete isolated from the sea of Japan. Int J Syst Evol Microbiol 54:191–194
Sun W, Dai S, Jiang S, Wang G, Liu G, Wu H, Li X (2010) Culture-dependent and culture-independent diversity of Actinobacteria associated with the marine sponge Hymeniacidon perleve from the South China Sea. Antonie Van Leeuwenhoek 98:65–75
Suzuki S (2001) Establishment and use of gellan gum media for selective isolation and distribution survey of specific rare actinomycetes. Actinomycetologica 15:55–60
Suzuki S, Okuda T, Komatsubara S (1999) Selective isolation and distribution of Sporichthya strains in soil. Appl Environ Microbiol 65:1930–1935
Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol 9:678–687
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tan GYA, Ward AC, Goodfellow M (2006) Exploration of Amycolatopsis diversity in soil using genus-specific primers and novel selective media. Syst Appl Microbiol 29:557–569
Tang S, Tian X, Zhi X, Cai M, Wu J, Yang L, Xu L, Li W (2008) Haloactinospora alba gen. nov., sp. nov., a halophilic filamentous actinomycete of the family Nocardiopsaceae. Int J Syst Bacteriol 58:2075–2080
Urzì C, Salamone P, Schumann P, Rohde M, Stackebrandt E (2004) Blastococcus saxobsidens sp. nov., and emended descriptions of the genus Blastococcus Ahrens and Moll 1970 and Blastococcus aggregatus Ahrens and Moll 1970. Int J Syst Evol Microbiol 54:253–259
Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, Sinderen D (2007) Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev 71:495–548
Vester JK, Glaring MK, Stougaard P (2015) Improved cultivation and metagenomics as new tools for bioprospecting in cold environments. Extremophiles 19:17–29
Vidgen ME, Hooper JNA, Fuerst JA (2012) Diversity and distribution of the bioactive actinobacterial genus Salinispora from sponges along the Great Barrier Reef. Antonie Van Leeuwenhoek 101:603–618
Vikineswary S, Christabel LJ, Thong KL, Tan GYA, Affendi YA (2008) Sponges of Tioman Island and their actinomycete inhabitants. In: Phang SM, Affendi YM, Ooi JLS, Bin Mydin HAJ (eds) Natural history of the Pulau Tioman group of islands. Institute of Ocean and Earth Science (IOES), University of Malaya, Kuala Lumpur, pp 35–41
Waksman SA, Woodruff BH (1941) Actinomyces antibioticus, a new soil organism antagonistic to pathogenic and non-pathogenic bacteria. J Bacteriol 42:231–249
Wong WR, Oliver AG, Linington RG (2012) Development of antibiotic activity profile screening for the classification and discovery of natural product antibiotics. Chem Biol 19:1483–1495
Xin Y, Wu P, Deng M, Zhang W (2009) Phylogenetic diversity of the culturable rare actinomycetes in marine sponge Hymeniacidon perlevis by improved isolation media. Acta Microbiol Sin 49:859–866
Xu P, Li WJ, Tang SK, Zhang YQ, Chen GZ, Chen HH, Xu LH, Jiang CL (2005) Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int J Syst Evol Microbiol 55:1149–1153
Yang R, Zhang L, Guo L, Shi N, Lu Z, Zhang X (2008) Nocardiopsis valliformis sp. nov., an alkaliphilic actinomycete isolated from alkali lake soil in China. Int J Syst Evol Microbiol 58:1542–1546
Yoon S, Ha S, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zhang H, Zhang W, Jin Y, Jin M, Yu X (2008) A comparative study on the phylogenetic diversity of culturable actinobacteria isolated from five marine sponge species. Antonie Van Leeuwenhoek 93:241–248
Zheng Z, Zeng W, Huang Y, Yang Z, Li J, Cai H, Su W (2000) Detection of antitumor and antimicrobial activities in marine organism associated actinomycetes isolated from the Taiwan Strait, China. FEMS Microbiol Lett 188:87–91
Zhu W, Zhang J, Qin Y, Xiong Z, Zhang D, Klenk H, Zhao L, Xu L, Li W (2013) Blastococcus endophyticus sp. nov., an actinobacterium isolated from Camptotheca acuminata. Int J Syst Evol Microbiol 63:3269–3273
Acknowledgements
The authors would like to acknowledge the Ministry of Natural Resources and Environment for permission to collect marine environmental samples from Tioman Marine Park (permit dated 17 January 2013). This research study was supported by MOSTI-ScienceFund (Project No.: 04-01-03-SF0666) and MOHE Malaysia (HIR-005). We are grateful to scuba divers, Chai Ming Lau and Daicus M. Belabut, for collecting sediment samples and Kavimalar Devaraj for her assistance in sampling and sample processing. We also acknowledge Weng Ruh Wong who provided expertise that greatly assisted in the antibacterial activity screening. Both the authors declare that we have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ng, Z.Y., Tan, G.Y.A. Selective isolation and characterisation of novel members of the family Nocardiopsaceae and other actinobacteria from a marine sediment of Tioman Island. Antonie van Leeuwenhoek 111, 727–742 (2018). https://doi.org/10.1007/s10482-018-1042-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-018-1042-8