Abstract
A novel Gram-stain positive, short rod, forming sub-terminal endospores of ellipsoidal shape, halophilic, alkaliphilic and aerobic bacterium, designated strain KQ-12T, was isolated from a saline–alkaline lake in China, and characterised by a polyphasic taxonomic approach. The isolate grew at 4–40 °C (optimum, 25 °C), at pH 8.0–10.0 (pH 9.0) and in the presence of 0–16% (w/v) NaCl (8%). 16S rRNA gene sequence similarity of KQ-12T to species in the genera Salipaludibacillus ranged from 96.6 to 98.1%. Phylogenetic trees indicated that the strain should be assigned to the genus Salipaludibacillus. The polar lipids of KQ-12T were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, and an unidentified phospholipid and its major cellular fatty acids were anteiso-C15:0, anteiso-C17:0, iso-C15:0, and C16:0. The isoprenoid quinone was MK-7. These key chemotaxonomic properties also confirmed the affiliation of the strain to the genus Salipaludibacillus. However, some physiological, biochemical properties, low average nucleotide identity and low digital DNA–DNA hybridization relatedness values enabled the strain to be differentiated from closely related species of the genus Salipaludibacillus. Thus, KQ-12T can be classified as a novel species in the genus Salipaludibacillus, for which the name Salipaludibacillus keqinensis sp. nov. is proposed. The type strain is KQ-12T ( = ACCC 60430T = KCTC 33935T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Salipaludibacillus was first proposed by Sultanpuram and Mothe (2016) for an isolate from a saline–alkaline lake. At the time of writing, this genus comprises 4 species with validly published names: Salipaludibacillus aurantiacus (Sultanpuram and Mothe 2016), Salipaludibacillus neizhouensis (Sultanpuram and Mothe 2016; Chen et al. 2009), Salipaludibacillus agaradhaerens (Sultanpuram and Mothe 2016; Nielsen et al. 1995), and Salipaludibacillus halalkaliphilus (Amoozegar et al. 2018). Members of the genus Salipaludibacillus are generally characterised to be Gram-stain positive, non-motile, rod shaped, aerobic or facultatively anaerobic, form oval or ellipsoidal endospores at the sub-terminal position, and have anteiso-C15:0, C16:0 and iso-C15:0 as their major fatty acids, MK-7 as their predominant isoprenoid quinone with minor traces of MK-6, and phosphatidylethanolamine, phosphatidylglycerol and diphosphatidylglycerol as the major polar lipids, and show relatively low G + C contents (39.3–42.4 mol%) (Sultanpuram and Mothe 2016).
Saline–alkaline lakes represent a unique ecosystem with extremely high pH and salinity (Sorokin et al. 2011). These haloalkaliphiles under double stress play essential roles and functions in biogeochemical processes and the ecological function (Sorokin et al. 2011). Furthermore, the unique metabolic pathways of haloalkaliphiles can be applied in the biodegradation and (or) biotransformation of a broad range of toxic industrial pollutants, and in the biofuel industry (Zhao et al. 2014). Therefore, it is of great importance to discover novel extremophiles. In the course of surveying the microbial community of the Keqin Lake, Heilongjiang Province, China (46°18′32′′N, 123°25′58′′E), a novel strain, strain KQ-12T, was isolated. As a result of testing using different taxonomic approaches, we consider the strain to represent a novel species of the genus Salipaludibacillus, and here name it Salipaludibacillus keqinensis sp. nov.
Materials and methods
Strain and culture conditions
KQ-12T was isolated from mixed water and sediment samples collected from Keqin Lake (28 mM Na+, 0.47 mM Mg2+, pH 8.7) in Heilongjiang Province, China (46°18′32′′N, 123°25′58′′E). Collected samples were transferred immediately to sterile serum bottles, tightly sealed with blue butyl-rubber stoppers, kept at room temperature during transportation and subsequently stored at 4–8 °C for up to 2 weeks until ready for use. While studying the cultivable bacterial diversity of saline ecosystems of Keqin Lake, KQ-12T was isolated using serial dilutions up to 10−5 from the mixed water and sediment sample on solid medium. The isolation medium contained (l−1): NaCl (100 g), NH4Cl (1.0 g), KCl (K+, 13.4 mM) (1.0 g), KH2PO4 (K+, 2.2 mM) (0.3 g), MgSO4·7H2O (0.1 g), Na2CO3 (0.1283 M Na+) (6.8 g), NaHCO3 (0.0452 M Na+) (3.8 g), Yeast extract (Difco) (4 g), Casamino acids (Difco) (0.5 g). The medium was adjusted to pH 9.2 with NaHCO3/Na2CO3 buffer (100 mM in deionized water; pH 9.2) at room temperature and 2% agar was added. After autoclaving at 121 °C for 45 min, 0.2% (w/v) filter-sterilized glucose was added to the medium before pouring plates. KQ-12T was maintained on slant tubes at 4–6 °C and preserved as 15% (w/v) glycerol suspensions at − 80 °C. Unless otherwise stated, cells for physiological and biochemistry analyses were obtained through cultivation in shake flasks at 150 rpm with the aforementioned liquid culture medium at 25 °C for 48 h.
Phenotypic characteristics
General cell morphology was examined by light microscopy (BH-2, Olympus Co., Japan) and transmission electron microscopy (Hitachi H-600, Japan) using cells from exponentially growing cultures. Gram-staining test was examined according to the methods described by Smibert and Krieg (1994), in parallel with the KOH lysis method (Gregersen 1978). Motility was observed by stab-culture in semi-solid medium according to the procedure of Gerhardt et al. (1981). The ISCC-NBS colour charts (Kelly 1964) were used to assess the colony colour. Growth at different temperatures (4–55 °C) and NaCl tolerance (0–30% (w/v)) were tested using LB as the basal medium. The pH range (pH 5.5–11.5, with intervals of 0.5, with MES buffer for pH 5.5–6.5, HEPES buffer for pH 7.0–8.0, TAPS buffer for pH 8.0–9.0, CHES buffer for pH 9.0–10.0 and CAPS buffer for pH 10.0–11.5). Anaerobic growth test was performed according to previously described method (Zhang et al. 2016). Hydrolysis of aesculin, casein, cellulose, gelatin, starch, tweens 20 and 80, citrate utilization, methyl-red reaction, production of indole and H2S, and observation of endospores were tested as described by Dong and Cai (2001). The Voges–Proskauer reaction, reduction of nitrate, and urease activity were determined according to the methods described by Pettersson et al. (1996). Catalase activity was assessed by a bubble production in 3.0% (v/v) H2O2 (Ohta and Hattori 1983). Oxidase activity was determined with 1% (w/v) tetramethyl-p-phenylenediamine (Cappuccino and Sherman 2002). DNase test was conducted with DNase test agar (Difco). Other enzyme activities and substrate oxidation patterns were assayed using the API ZYM kits (bioMérieux) and GP2 MicroPlates (Biolog), respectively, according to the manufacturer’s instructions with 8% (w/v) NaCl and pH 9.0.
Phylogenetic analysis
Extraction of genomic DNA and amplification of the 16S rRNA gene were carried out as previously reported by Wang et al. (2018). Amplification products were cloned into the vector pMD 19-T (TaKaRa) and then sequenced. The 16S rRNA gene sequence was compared with those of Salipaludibacillus and Bacillus species available in the EzBioCloud server (www.ezbiocloud.net/) (Yoon et al. 2017). Multiple alignments with closely related sequences were performed using the clustal_w program integrated in the mega 7.0 software (Kumar et al. 2016). Phylogenetic trees were reconstructed by the neighbour-joining (Saitou and Nei 1987), maximum-likelihood (Felsenstein 1981) and minimum-evolution (Rzhetsky and Nei 1992) methods with bootstrap values based on 1000 replications. Evolutionary distances among the related taxa were calculated according to Kimura’s two-parameter model (Kimura 1980).
The draft genome of KQ-12T and S. neizhouensis KCTC 13187T were sequenced using the Hiseq 4000 sequencing platform with paired-end read length of 2 × 150 bp and de novo assembled using MicrobeTrakr plus v. 0.9.1 (http://www.microbetrakr.com). The obtained genomes were submitted to the GenBank database, and the DNA G + C content was gained directly from the genome sequence. The level of pairwise genome-based similarity was evaluated using average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values, which were achieved by using the Orthologous Average Nucleotide Identity Tool (www.ezbiocloud.net/tools/orthoani) and Genome-to-Genome Distance Calculator software version 2.1 (http://ggdc.dsmz.de/) with Formula 2, respectively.
Chemotaxonomy
For cellular fatty acid analysis, KQ-12T and the three related reference strains were cultured on LB medium at pH 9.0, 25 °C and 8% (w/v) NaCl for 48 h. Fatty acids were purified, identified and quantified by GC using the Sherlock Microbial Identification System (MIDI) (Kämpfer and Kroppenstedt 1996). MIDI Sherlock version 6.0 and the TSBA6 database were employed for this analysis. Isoprenoid quinones were extracted from lyophilized cells, purified by thin-layer chromatography (TLC) and investigated by HPLC (Collins 1985) using the menaquinones of the reference type strains as standards. Preparation of cell walls and determination of peptidoglycan structure were analysed as described by Hasegawa et al. (1983). Polar lipids were extracted following Minnikin et al. (1984), separated by two-dimensional TLC and detected by spraying individual plates with: molybdophosphoric acid, molybdenum blue, ninhydrin, p-anisaldehyde.
Results and discussion
Cells of KQ-12T were observed to be Gram-stain positive, aerobic, motile, producing endospores which are ellipsoidal and located sub-terminally, rod-shaped and 0.7–0.9 × 1.5–2.4 µm in size (Fig. S1). Other phenotypic and physiological characteristics are presented in the species description. Differential characteristics between KQ-12T and the closely related species in the genus Salipaludibacillus are given in Table 1.
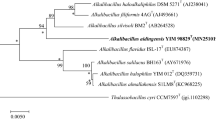
The almost-complete 16S rRNA gene sequence (1491 bp) of KQ-12T has been deposited as MH939198 in the GenBank/EMBL/DDBJ databases. Identification using the EzTaxon server revealed that KQ-12T is closely related to S. aurantiacus S9T (98.1%, with 16S rRNA gene sequence similarity), followed by S. neizhouensis JSM 071004T (97.7%), S. agaradhaerens DSM 8721T (97.6%), S. halalkaliphilus GASy1T (96.6%). These values are at the level suggested to allocate this strain to a new species (Kim et al. 2014). The neighbour-joining tree demonstrated that KQ-12T formed a separate branch with S. neizhouensis JSM 071004T and S. halalkaliphilus GASy1T, and is closely related to other members of the genus Salipaludibacillus (Fig. 1). The same cluster was recovered when the trees were reconstructed using minimum-evolution (Fig. S2) and maximum-likelihood (Fig. S3) algorithms. Phylogenetic analysis showed KQ-12T is a member of the genus Salipaludibacillus.
Neighbor-joining tree showing the phylogenetic position of the novel species based on 16S rRNA gene sequences. Bootstrap values more than 50% based on 1000 replications are shown at branching points. Marinococcus halophilus DSM 20408T was used as an outgroup. Bar, 0.01 substitutions per nucleotide position
The draft genome size of KQ-12T is 4,150,426 bp with a G + C content of 39.6 mol%. The draft genome size of S. neizhouensis KCTC 13187T is 5,397,042 bp with a G + C content of 37.2 mol%. The genomic G + C content (39.6 mol%) of KQ-12T is within the range of the genus salipaludibacillus (39.3–42.4 mol%) (1). The ANI values between strain KQ-12T (GeneBank: PDOD00000000) and its related species S. aurantiacus S9T (FOGT00000000), S. neizhouensis KCTC 13187T (PDOE00000000) and S. agaradhaerens DSM 8721T (MTIU00000000) were 72.5, 71.6 and 72.0%, respectively, which are much lower than the accepted ANI species cut-off value of 94–96% (Richter and Rosselló-Móra 2009). Furthermore, the dDDH values of KQ-12T with the selected reference strains S. aurantiacus S9T, S. neizhouensis KCTC 13187T and S. agaradhaerens DSM 8721T were 19.6, 20.5 and 22.3%, respectively, well below the threshold of 70% (Wayne et al. 1987), indicating that KQ-12T does not belong to any of these related species.
Chemotaxonomic characteristics of KQ-12T also supported its classification as a member of the genus salipaludibacillus. The peptidoglycan cell wall of KQ-12T contained meso-diaminopimelic acid (m-DAP) as the diagnostic diamino acid, which is consistent with the results reported for members of the genus Salipaludibacillus (Sultanpuram and Mothe 2016; Amoozegar et al. 2018). The major cellular fatty acids (content ≥ 5%) of KQ-12T were anteiso-C15:0 (47.1%), anteiso-C17:0 (12.1%), iso-C15:0 (6.7%), and C16:0 (5.5%). The fatty acid profile of KQ-12T was similar to those of the three selected reference strains in genus Salipaludibacillus (Table 2). However, some minor differences were observed between KQ-12T and the reference strains, which included the presence of Summed feature 4 (anteiso-C 17:1ω7c and/or iso I), low percentage of Summed feature 3 (C 16:1ω6c and/or C 16:1ω7c;) and a high percentage of anteiso-C 17:0 compared to S. aurantiacus KCTC 33633T (Table 2). As can be seen, KQ-12T showed a similar fatty acid profile to other species of the genus Salipaludibacillus. (Table 2).The isoprenoid quinone profile of KQ-12T was characterised by the predominance of MK-7 (approx. 100%), which was similar to that of S. halalkaliphilus GASy1T (Amoozegar et al. 2018). The polar lipids of KQ-12T were identified as diphosphatidylglycerol, phosphatidylglycerol and phosphatidylethanolamine followed by an unidentified phospholipid (Fig. S4); similar profiles were also reported in the descriptions of the genus Salipaludibacillus (Sultanpuram and Mothe 2016; Amoozegar et al. 2018).
To summarize, KQ-12T shared high 16S rRNA gene sequence similarities with respect to the type strains of the genus Salipaludibacillus, phylogenetic analysis exhibited that the isolate grouped with Salipaludibacillus species, and it should be assigned to this genus. Furthermore, the chemotaxonomic data (the major fatty acids, the predominant menaquinone, the polar lipids and the diagnostic diamino acid) support the affiliation of KQ-12T to the genus Salipaludibacillus. Also, the new isolate can be clearly distinguished from the other recognized species of the genus Salipaludibacillus based on genomic relatedness (ANI and dDDH), and morphological and physiological properties (Table 1). Accordingly, it is evident that KQ-12T should be considered to represent a novel species of the genus Salipaludibacillus, for which the name S. keqinensis sp. nov., is proposed. The Digital Protologue database (Rosselló-Móra et al. 2017) TaxoNumber for strain KQ-12T is TA00789.
Description of Salipaludibacillus keqinensis sp. nov.
Salipaludibacillus keqinensis (ke.qin.en’sis. N.L. masc. adj. keqinensis pertaining to salt lake Keqin in Heilongjiang Province, China, where the type strain was isolated).
Cells are Gram-stain positive, aerobic, motile short rods (0.7–0.9 × 1.5–2.4 μm). Colonies are circular, smooth, convex, light yellow in colour and 1.0–2.0 mm in diameter after 48 h of incubation at 25 °C. Growth is observed at 4–40 °C, pH 8.0–10.0 and with up to 16% (w/v) NaCl. Optimal growth occurs at 25 °C, pH 9.0 and in the presence of 8% (w/v) NaCl. Aesculin, casein, cellulose, DNA, gelatin, starch Tween 20 and 80 are not hydrolysed. Positive for catalase activity, nitrate reduction, and Voges-Proskauer test, but negative for oxidase, urease, citrate utilization, methyl red test, indole and H2S production. Enzyme activities are detected for esterase (C4), esterase lipase (C8), α-chymotrypsin, naphthol-AS-BI-phosphohydrolase, β-galactosidase and α-glucosidase; No activity is detected for alkaline phosphatase, lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, acid phosphatase, α-galactosidase, β-glucuronidase, β-glucosidase, N-acetul-β-glucosaminidase, α-mannosidase or α-fucosidase (API ZYM test strips). In Biolog GP2 microplates (48 h incubation), the following substrates yield positive reactions for substrate oxidation: l-arabinose, palatinose, d-psicose, d-ribose and d-xylose; the other substrates are not. Major cellular fatty acids (content ≥ 5%) are anteiso-C15:0, anteiso-C17:0, iso-C15:0 and C16:0. The predominant menaquinone is MK-7. The peptidoglycan cell wall contains meso-diaminopimelic acid. The polar lipids include diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine and one unidentified phospholipid.
The type strain, KQ-12T (= ACCC 60430T = KCTC 33935T), was isolated from Keqin lake in Heilongjiang Province, China. The GenBank/EMBL/DDJB accession number for the 16S rRNA gene sequence and the whole genome sequence of KQ-12T are MH939198 and PDOD00000000, respectively.
References
Amoozegar MA, Shahinpei A, Makzum S, Rafieyan S, Moshtaghi Nikou M et al (2018) Salipaludibacillus halalkaliphilus sp. nov., a moderately haloalkaliphilic bacterium from a coastal-marine wetland. Int J Syst Evol Microbiol 68(7):2214–2219. https://doi.org/10.1099/ijsem.0.002814
Cappuccino JG, Sherman N (2002) Microbiology: a laboratory manual, 6th edn. Pearson Education, Inc. and Benjamin Cummings, San Francisco
Chen YG, Zhang YQ, Wang YX, Liu ZX, Klenk HP et al (2009) Bacillus neizhouensis sp. nov., a halophilic marine bacterium isolated from a sea anemone. Int J Syst Evol Microbiol 59:3035–3039. https://doi.org/10.1099/ijs.0.009522-0
Collins MD (1985) Analysis of isoprenoid quinones. Methods Microbiol 18:329–366. https://doi.org/10.1016/S0580-9517(08)70480-X
Dong XZ, Cai MY (2001) Determination of biochemical properties. In: Dong XZ, Cai MY (eds) Manual for the systematic identification of general bacteria. Science Press, Beijing, pp 370–398 (in Chinese)
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA et al (1981) Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC, pp 25–29
Gregersen T (1978) Rapid method for distinction of gram-negative from gram-positive bacteria. Appl Environ Microbiol 5:123–127. https://doi.org/10.1007/bf00498806
Hasegawa T, Takizawa M, Tanida S (1983) A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 29:319–322. https://doi.org/10.2323/jgam.29.319
Kämpfer P, Kroppenstedt RM (1996) Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can J Micobiol 42:989–1005. https://doi.org/10.1139/m96-128
Kelly KL (1964) Inter-society colour council-national bureau of standards colour-name charts illustrated with centroid colours published in US. US Government Printing Office, Washington, DC
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351. https://doi.org/10.1099/ijs.0.064931-0
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111. https://doi.org/10.1007/BF01731581
Kuenen JG, Muyzer G (2011) The microbial sulfur cycle at extremely haloalkaline conditions of soda lakes. Front Microbiol 2:44. https://doi.org/10.3389/fmicb.2011.00044
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbbiol Methods 2:233–241. https://doi.org/10.1016/0167-7012(84)90018-6
Nielsen P, Fritze D, Priest FG (1995) Phenetic diversity of alkaliphilic bacillus strains: proposal for nine new species. Microbiology 141:1745–1761. https://doi.org/10.1099/13500872-141-7-1745
Ohta H, Hattori T (1983) Agromonas oligotrophica gen. nov., sp. nov., a nitrogen-fixing oligotrophic bacterium. Antonie Van Leeuwenhoek 49:429–446. https://doi.org/10.1007/BF00399322
Pettersson B, Lembke F, Hammer P, Stackebrandt E, Priest FG (1996) Bacillus sporothermodurans, a new species producing highly heat-resistant endospores. Int J Syst Bacteriol 46:759–764. https://doi.org/10.1099/00207713-46-3-759
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131. https://doi.org/10.1073/pnas.0906412106
Rosselló-Móra R, Trujillo ME, Sutcliffe IC (2017) Introducing a digital protologue: a timely move towards a database-driven systematics of archaea and bacteria. Antonie Van Leeuwenhoek 110:455–456. https://doi.org/10.1007/s10482-017-0841-7
Rzhetsky A, Nei M (1992) A simple method for estimating and testing minimum-evolution trees. Mol Biol Evol 9:945. https://doi.org/10.1093/oxfordjournals.molbev.a040771
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.moldev.a040454
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 607–654
Sultanpuram VR, Mothe T (2016) Salipaludibacillus aurantiacus gen. nov., sp. nov. a novel alkali tolerant bacterium, reclassification of Bacillus agaradhaerens as Salipaludibacillus agaradhaerens comb. nov. and Bacillus neizhouensis as Salipaludibacillus neizhouensis comb. nov. Int J Syst Evol Microbiol 66:2747–2753. https://doi.org/10.1099/ijsem.0.001117
Wang H, Zhang X, Wang S, Zhao B, Lou K et al (2018) Massilia violaceinigra sp. nov., a novel purple-pigmented bacterium isolated from glacier permafrost. Int J Syst Evol Microbiol 68(7):2271–2278. https://doi.org/10.1099/ijsem.0.002826
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O et al (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464. https://doi.org/10.1111/j.1365-2672.1988.tb01872.x
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Zhang S, Li Z, Yan Y, Zhang C, Li J et al (2016) Bacillus urumqiensis sp. nov., a moderately haloalkaliphilic bacterium isolated from a salt lake. Int J Syst Evol Microbiol 66:2305–2312. https://doi.org/10.1099/ijsem.0.001028
Zhao B, Yan Y, Chen S (2014) How could haloalkaliphilic microorganisms contribute to biotechnology? Can J Microbiol 60:717–727. https://doi.org/10.1139/cjm-2014-0233
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 31300101) and Fundamental Research Funds for Central Non-profit Scientific Institution (Grant No. 1610042018005).
Author information
Authors and Affiliations
Contributions
WS, WH and WK wrote the main manuscript text. WH and WK designed the experiments. WS., DL and XS carried out the experiments. WH, ZB and ZX analyzed the data. All authors approved and read the final manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethical statement
No specific ethical or institutional permits were required to conduct sampling and the experimental studies did not involve endangered or protected species.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, S., Dong, L., Zhao, B. et al. Salipaludibacillus keqinensis sp. nov., a moderately halophilic bacterium isolated from a saline–alkaline lake. Antonie van Leeuwenhoek 112, 897–903 (2019). https://doi.org/10.1007/s10482-018-01224-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-018-01224-w