Abstract
A novel bacterium, strain L3T, was isolated from an activated sludge sample retrieved from a municipal wastewater treatment plant in Huangdao, China. On the basis of 16S rRNA gene sequence similarity studies, strain L3T was affiliated to the genus Sinorhodobacter, being most closely related to Sinorhodobacter ferrireducens (98.0 %). The 16S rRNA gene sequence similarity of strain L3T to other related species, Thioclava atlantica DLFJ1-1T (96.5 %), Rhodobacter capsulatus ATCC 11166T (96.3 %), Paenirhodobacter enshiensis DW2-9T (96.3 %) and Rhodobacter viridis JA737T (96.0 %) is less than 96.5 %. Chemotaxonomic characterization further supported classification of the strain to the genus Sinorhodobacter. The major polar lipid profile consists of diphosphatidyglycerol, phosphatidylglycerol and phosphatidylethanolamine. The major fatty acids are C18:1 ω7c (66.3 %), C16:0 (12.9 %) and C18:0 (8.0 %). The major quinone is Q-10. The G+C content of the genomic DNA of strain L3T is 68.0 mol %. DNA–DNA relatedness value between L3T and the closely related type strain S. ferrireducens SgZ-3T was 35.2 %. Based on these results, a new species Sinorhodobacter huangdaonensis is proposed. The type strain is L3T (= CGMCC 1.12963T = KCTC 42823T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The proposal of new genus Sinorhodobacter was based on the differential phenotypic including the lack of phototrophic growth and the absence of bacteriochlorophyll α and carotenoids, phylogenetic, and chemotaxonomic properties of the in type strain SgZ-3T (Yang et al. 2013). However, the proposed genus and species names are yet to be validated.
During an investigation of the bacterial composition of activated sludge from a municipal wastewater treatment plant, we isolated strain L3T, which showed morphological characteristics typical to the genus Sinorhodobacter. In this paper, we report the taxonomic characterization of strain L3T as a novel species of the genus Sinorhodobacter. An emended description of the genus Sinorhodobacter is also proposed.
Materials and methods
Bacterial strain isolation and growth conditions
Strain L3T was isolated from an activated sludge sample, which was collected from a municipal wastewater treatment plant in Qingdao, China (36°35′ 16.04″N 120°10′ 40.89″E) in October 2014. The sediment sample was well-mixed at room temperature, serially diluted, and plated on yeast extract-malt extract agar [ISP 2, medium 2 of the International Streptomyces Project (Küster 1959)]. After 5 days of incubation at 30 °C, the strain L3T was isolated from a single colony and purified on ISP 2 medium.
Type strain S. ferrireducens SgZ-3T was used for comparative purposes in this study and was kindly provided by Prof. Yuan of Guangdong Institute of Eco-Environmental and Soil Sciences, Guangzhou, China.
Phenotypic and chemotaxonomic analysis
Cell growth under anaerobic and photo-organoheterotrophic conditions was tested according to the method described by Yang et al. (2013). The in vivo absorption spectrum of intact bacteria cells in 60 % sucrose (w/v) and the absorption spectrum of pigments extracted with acetone were recorded using UV-2450 Visible spectrophotometer (Shimadu, Japan). Cultural and morphological characteristics of strain L3T were observed on tryptic soy agar (TSA, Oxoid) or in tryptic soy broth (TSB, Oxoid) following incubation for 2–7 days at 30 °C. Cell morphology was investigated using a transmission electron microscope (JEM-1400, JEOL). The Gram-reaction was determined by the conventional Gram staining method (Smibert and Krieg 1994). Media and procedures used to study physiological and biochemical features (growth conditions, catalase, β-galactosidase, urease, oxidase, esterase, arginine dihydrolase, tryptophan dehydrogenase, gelatin hydrolysis, H2S production, nitrate reduction, acid formation, and carbon assimilation) were as described elsewhere (Dong and Cai 2001). The reduction of Fe3+ was monitored by measuring the change in the total Fe2+ concentration (Wu et al. 2010). Biomass for fatty acid analysis was collected by scraping colonies from TSA medium after strain L3T was grown at 30 °C for 3 days. Biomass for other chemotaxonomic studies was obtained after strain L3T was grown TSB at 30 °C for 3–7 days in a rotary shaker (model, brand). Standard analytical procedures were used to extract and analyze the isomeric forms of quinones (Collins et al. 1985, 1977; Tamaoka 1986) and polar lipids (Collins and Jones 1980; Minnikin et al. 1979). Fatty acids were analyzed using the standard MIDI (Microbial Identification, Sherlock version 6.0) procedure (Sasser, 1990) and an Agilent GC 6890 gas chromatograph. The resulting profiles were identified using the database TSBA6, version 6.0. The G+C content of genomic DNA was determined by using the HPLC method (Jin and Komagata 1984).
Phylogenetic analyses
Extraction of genomic DNA was performed as described by Chun and Goodfellow (1995). PCR amplification of the 16S rRNA gene was performed using a bacterial universal primer set (27F and 1492R) (Lane 1991). The PCR product was purified using the TIANgel Midi Purification Kit DP209 (Beijing, China), ligated into the PMD19-T vector and transformed into Escherichia coli DH5α using a PMD19-T cloning kit (Takara Bio). The 16S rRNA gene sequence was determined by Sangon Biotech (Shanghai, China). The isolate was identified using the EzTaxon server on the basis of 16S rRNA sequence data (Kim et al. 2012). Phylogenetic trees were constructed using MEGA version 6.0 (Tamura et al. 2013) based on approximately 1350 nucleotides from each related type strain (retrieved from the GenBank/EMBL/DDBJ database) mentioned in this study. Rhodospirillum rubrum ATCC 11170T was used as an out-group. The neighbour-joining (NJ) method (Saitou and Nei 1987), maximum-parsimony (MP) (Fitch 1971) and maximum-likelihood (ML) (Felsenstein 1981) were used for phylogenetic analysis. The topologies of the phylogenetic trees were determined using bootstrap analyses (Felsenstein 1985) based on 1000 replications.
DNA–DNA relatedness
The DNA G+C content was determined using the thermal denaturation method (Marmur and Doty 1962) using Escherichia coli K12 as calibration standard. DNA–DNA hybridizations between isolate L3T and S. ferrireducens SgZ-3T were performed using the liquid renaturation method (De Ley et al. 1970) and according to modification described by Huss et al. (1983). DNA–DNA hybridizations were carried out in 2× SSC at 81 °C. Both experiments were performed at 260 nm with a model Lambda 35 UV/VIS spectrometer equipped with a Peltier System (PTP 1+1) (Perkin–Elmer). The hybridization value was calculated from triplicate experiments.
Results and discussion
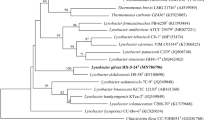
Analysis of the almost-full length 16S rRNA gene sequence (1435 bp) confirmed that strain L3T belongs to the genus Sinorhodobacter. It formed a coherent cluster (bootstrap value of 100 %) with type strain Sinorhodobacter ferrireducens (98.0 %), and grouped together with Thioclava atlantica DLFJ1-1T (96.5 %), Rhodobacter capsulatus ATCC 11166T (96.3 %), Paenirhodobacter enshiensis DW2-9T (96.3 %) and Rhodobacter viridis JA737T (96.0 %) as shown on the phylogenetic trees (NJ tree (Fig. 1), MP tree (Supplementary Fig. S1) and ML tree (Supplementary Fig. S2)). The 16S rRNA gene sequence similarity between this strain and other phylogenetically related species is <96.0 %.
Neighbour-joining tree based on 16S rRNA gene sequences showing the phylogenetic position of strain L3T and representatives of other related taxa. Bootstrap values (Interior Branch Test of Phylogeny, bootstrap = 1000) 50 % are shown at the branch points. Bar, 0.02 substitutions per nucleotide position
The morphological and physiological properties of the new bacterium are given in species description and summarized in Table 1. The G+C content of the genomic DNA of strain L3T is found to be 68.0 mol %. The DNA–DNA hybridization experiments revealed that strain L3T shared 35.2 ± 5.1 % DNA relatedness with S. ferrireducens SgZ-3T. This value is notably lower than the 70 % cut-off value recommended for species differentiation (Wayne et al. 1987); this result confirmed that L3T can be considered as a distinct species of the genus Sinorhodobacter.
The chemotaxonomic characteristics include the presence of the principal quinone Q-10, which is typical quinone reported for the genera Sinorhodobacter (Yang et al. 2013), Rhodobacter (Imhoff et al. 1984) and Thioclava (Sorokin et al. 2005); the major fatty acids such as C18:1 ω7c (66.3 %, 66.7 %), C16:0 (12.9 %, 11.7 %), C18:0 (8.0 %, 6.8 %), C10:0 3-OH (4.3 %, 5.7 %), C18:0 3-OH (4.3 %, 3.7 %) and C12:0 3-OH (1.2 %, 1.5 %), which are detected in both strain L3T and S. ferrireducens SgZ-3T.
The polar lipid profile of strain L3T was found to contain diphosphatidyglycerol, phosphatidylglycerol, phosphatidylethanolamine, unidentified aminophospholipid and aminolipid, and lacking diphosphatidyglycerol. This phospholipids composition is characteristic for bacteria of the genus Sinorhodobacter (Fig. S3). The lack of diphosphatidylglycerol is a distinct feature of strain L3T and S. ferrireducens SgZ-3T and could be used to distinguish bacteria of this genus from other phylogenetically related bacteria (Dan et al. 2013; Lai et al. 2014; Raj et al. 2012; Sorokin et al. 2005). Consequently, strain L3T and SgZ-3T demonstrate genus level polar lipid production characteristics; thus, these results support the assignment of strain L3T to the genus Sinorhodobacter.
Based on the results of the phenotypic, phylogenetic, chemotaxonomic analysis and DNA–DNA relatedness, it can be concluded that strain L3T represents a novel species within the genus Sinorhodobacter, for which we propose the name Sinorhodobacter huangdaonensis sp. nov.
Emended description of the genus Sinorhodobacter
Iron may or may not be reduced from Fe3+ to Fe2+ by members of this genus. The major polar lipid profile contains the compounds diphosphatidyglycerol, phosphatidylglycerol and phosphatidylethanolamine.
Description of Sinorhodobacter huangdaonensis sp. nov.
Sinorhodobacter huangdaonensis (huang.dao.nen´sis. N.L. masc. adj. Huangdaonensis of Huangdao, a district of Qingdao city in Shandong province, PR China, where the type strain was first isolated).
Cells are motile, Gram-negative, and ovoid to short rod-shaped, about 0.3–0.6 µm wide and 0.8–1.5 µm long (Fig. 2). Aerobic. Chemoorganotroph. On TSA medium forms smooth grey–white colonies with regular edges and are 2–3 mm in diameter after 72 h of incubation at 30 °C. Growth does not occur via anoxygenic photosynthesis. The growth occurs in media with 0–5 % (w/v) NaCl (optimum 0–1 %), at 15–35 °C (optimum 30 °C) and at pH 5.0–7.0 (optimum pH 6.0). Both bacteriochlorophyll α and carotenoids are absent (Supplementary Figs. S4, S5). Positive for catalase, β-galactosidase, urease, arginine dihydrolase, tryptophan dehydrogenase, hydrolysis of gelatin, H2S production and nitrate reduction and they are able to ferment d-glucose. Negative for Fe3+ reduction, indole production, acetoin production, oxidase and esterase. Assimilate l-serine, lysine, ornithine, d-maltose, d-saccharose and citric acid. Principal fatty acids are C18:1 ω7c, C16:0 and C18:0 and the major quinone is Q-10. The major polar lipid profile contains diphosphatidyglycerol, phosphatidylglycerol and phosphatidylethanolamine. The G+C content of genomic DNA of the type strain is 68.0 mol %.
The type strain L3T (=CGMCC 1.12963T = KCTC 42823T), isolated from an activated sludge sample collected from a municipal wastewater treatment plant. The GenBank accession number for the 16S rRNA gene sequence of S. huangdaonensis L3T is KU042973.
References
Chun J, Goodfellow M (1995) A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int J Syst Bacteriol 45:240–245
Collins MD, Jones D (1980) Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2, 4-diaminobutyric acid. J Appl Microbiol 48:459–470
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Collins MD, Goodfellow M, Minnikin DE, Alderson G (1985) Menaquinone composition of mycolic acid-containing actinomycetes and some sporoactinomycetes. J Appl Bacteriol 58:77–86
Dan W, Hongliang L, Shixue Z, Gejiao W (2013) Paenirhodobacter enshiensis gen. nov., sp. nov., a non-photosynthetic bacterium isolated from soil, and emended descriptions of the genera Rhodobacter and Haematobacter. Int J Syst Evol Microbiol 64:551–558
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Dong X, Cai M (2001) Manual of systematic and determinative bacteriology. Academic Press, Beijing
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Huss VAR, Festl H, Schleifer K-H (1983) Studies on the spectrometric determination of DNA hybridisation from renaturation rates. Syst Appl Microbiol 4:184–192
Imhoff JF, Truper HG, Pfennig N (1984) Rearrangement of the species and genera of the phototrophic “Purple Nonsulfur Bacteria”. Int J Syst Evol Microbiol 34:340–343
Jin T, Komagata K (1984) Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett 25:125–128
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Küster E (1959) Outline of a comparative study of criteria used in characterization of the Actinomycetes. Int Bull Bacteriol Nomencl Taxon 9:97–104
Lai Q, Li S, Xu H, Jiang L, Zhang R, Shao Z (2014) Thioclava atlantica sp. nov., isolated from deep sea sediment of the Atlantic Ocean. Antonie Van Leeuwenhoek 106:919–925
Lane DJ (1991) 16S/23S rRNAsequencing. In: Stackebrandt ER, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty Acid and Polar Lipid Composition in the Classification of Cellulomonas, Oerskovia and Related Taxa. J Appl Microbiol 47:87–95
Raj PS, Ramaprasad EVV, Vaseef S, Sasikala C, Ramana CV, Raj PS, Vaseef S (2012) Rhodobacter viridis sp. nov., a phototrophic bacterium isolated from Western Ghats of India. Int J Syst Evol Microbiol 63:181–186
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, vol 101. MIDI technical noteMIDI, Newark
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for General and Molecular Bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Sorokin D, Tourova T, Spiridonova EFA, Muyzer G (2005) Thioclava pacifica gen. nov., sp. nov., a novel facultativelyautotrophic, marine, sulfur-oxidizing bacterium from a near-shoresulfidic hydrothermal area. Int J Syst Evol Microbiol 55:1069–1075
Tamaoka J (1986) Analysis of bacterial menaquinone mixtures by reverse-phase high-performance liquid chromatography. Method Enzymol 123:251–256
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Wayne LG, Brenner DJ, Colwell RR et al (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Wu CY, Zhuang L, Zhou SG, Li FB, Li XM (2010) Fe(III)-enhanced anaerobic transformation of 2,4-dichlorophenoxyacetic acid by an iron-reducing bacterium Comamonas koreensis CY01. FEMS Microbiol Ecol 71:106–113
Yang G, Chen M, Zhou S, Liu Z, Yuan Y (2013) Sinorhodobacter ferrireducens gen. nov., sp. nov., a non-phototrophic iron-reducing bacterium closely related to phototrophic Rhodobacter species. Antonie Van Leeuwenhoek 104:715–724
Acknowledgments
We would like to thank Prof. Yong Yuan (Guangdong Institute of Eco-Environmental and Soil Sciences, Guangzhou, China) for providing type strain S. ferrireducens SgZ-3T. And this research is funded by the National Natural Science Foundation of China (Grant No. 31400005), the Natural Science Foundation of Shandong Province (Grant No. ZR2013CL028), and the Fundamental Research Funds for the Central Universities of China (Grant No. 15CX02014A and 16CX02044A).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (PDF 2952 kb)
Supplementary Fig. S1 Maximum-Parsimony tree based on 16S rRNA gene sequences showing the phylogenetic position of strain L3T and representatives of other related taxa. Bootstrap values (bootstrap=1000) 50 % are shown at the branch points
Supplementary Fig. S2 Maximum-Likelihood tree based on 16S rRNA gene sequences showing the phylogenetic position of strain L3T and representatives of other related taxa. Bootstrap values (bootstrap=1000) 50 % are shown at the branch points. Bar, 0.01 substitutions per nucleotide position
Supplementary Fig. S3 Polar lipid profiles of strains L3T and ‘S. ferrireducens’ SgZ-3T, separated by two-dimensional thin layer chromatography and detected with molybdatophosphoric acid. Abbreviations: DPG, diphosphatidylglycerol; PE, phosphatidylethanolamine; PG, phosphatidylglycero; APL, aminophospholipid; AL, aminolipid
Supplementary Fig. S4 Whole-cell absorption spectrum of strain L3T
Supplementary Fig. S5 Acetone spectrum of extracted pigments of strain L3T
Rights and permissions
About this article
Cite this article
Xi, L., Qiao, N., Zhang, Z. et al. Sinorhodobacter hungdaonensis sp. nov. isolated from activated sludge collected from a municipal wastewater treatment plant. Antonie van Leeuwenhoek 110, 27–32 (2017). https://doi.org/10.1007/s10482-016-0770-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-016-0770-x