Abstract
Two moderately halophilic strains, designated SL013A34A2T and SL013A24A, were isolated from oil-contaminated saline soil from Shengli Oilfield, eastern China. Cells were found to be Gram-staining negative, aerobic, rod-shaped with a single polar flagellum. The isolates were found to grow at 10–40 °C (optimum 35 °C), pH 6.0–9.0 (optimum pH 8.0), and NaCl concentrations of 0.5–18.0 % (w/v) (optimum 3.0–6.0 NaCl). The 16S rRNA gene sequence analysis indicated that the isolates belong to the genus Marinobacter. Strain SL013A34A2T shares the highest 16S rRNA gene sequence similarities with strain SL013A24A (99.3 %), followed by M. hydrocarbonoclasticus CGMCC 1.7683T (97.8 %), M. vinifirmus CGMCC 1.7265T (97.8 %), and M. excellens KMM 3809T (97.4 %), respectively, but low similarities (93.8–96.4 %) with type strains of the other numbers of genus Marinobacter. DNA–DNA relatedness values of strain SL013A34A2T with strains SL013A24A, M. hydrocarbonoclasticus CGMCC 1.7683T, M. vinifirmus CGMCC 1.7265T and M. excellens KMM 3809T were 88.7, 29.2, 33.4 and 29.4 %, respectively. The major fatty acids of strain SL013A34A2T were identified as C18:1 ω9c, C16:0, C12:03-OH, C12:0, C16:1 ω9c and 10-methyl C18:0. The major respiratory quinone of strain SL013A34A2T was found to be ubiquinone-9, and its predominant polar lipids were identified as diphosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol and unidentified glycolipid. The genomic DNA G + C content was found to be 56.1 mol %. Based on the phenotypic, genetic and chemotaxonomic characteristics, these two isolates are representatives of a novel species of the genus Marinobacter, for which the name Marinobacter shengliensis sp. nov. is proposed. The type strain is SL013A34A2T(=LMG 27740T = CGMCC 1.12758T).
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The genus Marinobacter, belonging to the family Alteromonadaceae, in Proteobacteria, was first proposed by Gauthier et al. (1992). The type species is Marinobacter hydrocarbonoclasticus, which is Gram-staining negative, aerobic, flagellated, halophilic, hydrocarbon-degrading and rod-shaped bacterium (Gauthier et al. 1992). At the time of writing, 34 species have been validly named (http://www.bacterio.net/marinobacter.html), which were isolated from diverse environments, including seawater (Gauthier et al. 1992; Gorshkova et al. 2003; Yoon et al. 2003; 2004; Romanenko et al. 2005; Shivaji et al. 2005; Antunes et al. 2007; Roh et al. 2008; Huo et al. 2008; Zhang et al. 2008; Xu et al. 2008; Zhuang et al. 2009; Kharroub et al. 2011; Qu et al. 2011; Lee et al. 2012), marine sediment(Gorshkova et al. 2003; Romanenko et al. 2005; Kim et al. 2006; Guo et al. 2007; Montes et al. 2008; Liu et al. 2012; Gao et al. 2013), saline lake (Aguilera et al. 2009; Bagheri et al. 2013), saltern (Yoon et al. 2007; Wang et al. 2009), saline soil (Martín et al. 2003; Gu et al. 2007), hot spring (Shieh et al. 2003), hydrothermal sediment(Handley et al. 2009), wastewater (Liebgott et al. 2006) and others organisms (Romanenko et al. 2005; Green et al. 2006; Kaeppel et al. 2012). The DNA G+C content of this genus ranges from 52.7 to 59.6 mol %.The predominant fatty acids are C18:1 ω9c, C16:0,C16:1 ω9c and C12:03-OH. The major respiratory quinone is Q-9.
During investigation into microbial communities in petroleum related environments by both culture- dependent and independent analyses, we found numerous novel bacterial lineages (Gu et al. 2007; Wang et al. 2007a; Wang et al. 2007b; Wu et al. 2009; Wang et al. 2010; Cai et al. 2011a; Cai et al. 2011b; Lv et al. 2014; Pan et al. 2014) and several isolates closely related to Marinobacter (Tang et al. 2012; Sun et al. 2014). In this study, we describe two novel Marinobacter strains isolated from an oil polluted saline soil in Shengli Oilfield in eastern China. The physiological, biochemical, and phylogenetic analyses revealed that the isolates represent a novel species of the genus Marinobacter.
Materials and methods
Isolation and cultivation
Strains SL013A34A2T and SL013A24A were isolated from oil-contaminated saline soil in Gudao product (118°50′E; 37°53′N), Shengli Oilfield, China. The two strains were isolated by 10-fold dilution plating technique on the oil production water agar (OPWA, 15 g agar in 985 ml oil production mixture)plates, which were incubated at 30 °C for 7 days (Wang et al. 2007a). Single colonies were streaked on artificial seawater (ASW) agar plate containing (l−1): 5 g peptone; 1 g yeast extract; 24 g NaCl; 4 g Na2SO4; 0.68 g KCl; 0.1 g KBr; 0.025 g H3BO3; 5.4 g MgCl2·H2O; 1.5 g CaCl2·2H2O; 0.024 g SrCl2·6H2O; 0.2 g NaHCO3; 0.04 g Na2HPO4; 0.5 g NH4Cl and 0.002 g NaF, 15 g agar; pH 8.0) (Eguchi et al. 1996) to obtain pure cultures. The reference strains Marinobacter vinifirmus CGMCC 1.7265T and Marinobacter hydrocarbonoclasticus CGMCC 1.7683T were obtained from China General Microbiological Culture Collection Center (CGMCC). Marinobacter excellens KMM 3809T was kindly presented by Prof. E. P. Ivanova, Swinburne University of Technology.
Morphological, physiological, and chemotaxonomic tests
After growth on Luria–Bertani (LB) agar (l−1: 10 g peptone; 5 g yeast extract; 10 g NaCl and 15 g agar; pH 8.0) for 2 days at 30 °C, cell morphology and flagellum were examined by using transmission electron microscopy (TEM, JEM-1230; JEOL).The optimal temperature was assessed in LB (pH 8.0) broth by testing bacterial growth at 4,10,15,20,25,30,35,40,45,50 and 55 °C while NaCl concentration was kept at 3 %. Requirement of NaCl for growth was determined using LB medium supplemented with different concentrations of NaCl (0, 0.5, 1.0, 2.0, 3.0, 4.0,6.0, 8.0, 10.0, 12.0, 14.0, 16.0,18.0 and 20.0 %, w/v),while pH value and temperature were kept at 8.0 and 35 °C, respectively. Requirement of pH for growth was tested with the pH ranged between 4.0 and 10.0 (at 1 pH unit interval) while NaCl concentration and temperature were kept at 3.0 % and 35 °C, respectively. For maintaining the pH of the medium, the sodium phosphate/citric acid buffer (10 mM, for pH 3-8) and glycine/NaOH buffer (10 mM, for pH 8.6–10) were used. All these growth tests were made in triplicates, and growth was determined with optical density (OD600).
Oxidase reagent (bioMérieux, Kovacs 1956) was used for testing oxidase activity, and catalase activity was determined via bubble production after addition of 3 % (v/v) hydrogen peroxide solution (Muurholm et al. 2007). H2S production, and hydrolysis of starch, Tween 80 and gelatin were tested according to the methods described by Williams et al. (1983) and Lányí (1988). Tests of some enzyme activities and other physiological and biochemical properties were carried out by using the API ZYM and API-20NE systems according to manufacturer’s instructions, respectively. The Biolog GEN III MicroPlate System (Biolog Inc., Hayward, CA)was used to perform the 94 phenotypic tests according to the manufacturer’s instruction (http://www.biolog.com), including the assays for utilization of 71 carbon sources and sensitivities to 23 chemicals. Simultaneously, the two reference type strains were also tested, and acid production was determined as described elsewhere (Shivaji et al. 2005).Antibiotic resistance tests were performed on LB (pH 8.0) using strip with the antibiotics according to the method described by Andrews (2008).
For respiratory quinone analysis, strain SL013A34A2T were harvested from LB (pH 8.0) broth cultivated at 30 °C for 48 h. Cells were washed, lyophilized and extracted with chloroform/methanol (2:1, v/v). Respiratory quinones were then analyzed by a high performance liquid chromatography(HPLC) with a reversed-phase column as described by Komagata and Suzuki (1987).Total lipids were extracted by a chloroform/methanol system and analyzed by two-dimensional thin layer chromatography (TLC), as described previously (Kates 1986). The TLC plate (silica gel 60 F254, Merck) dotted with sample was subjected to two-dimensional development, with the first solvent of chloroform/methanol/water (65:25:4, v/v) followed by second solvent of chloroform–methanol-acetic acid–water (85:12:15:4, v/v). For fatty acid analysis, the cells of SL013A34A2T, SL013A24A, M. hydrocarbonoclasticus CGMCC 1.7683T and M. vinifirmus CGMCC 1.7265T, were firstly cultured on MA (2216E) medium (BD, U S A.; l−1 distilled water: Bacto peptone 5.00 g, Bacto yeast extract 1.00 g, Fe(III) citrate 0.10 g, NaCl 19.45 g, MgCl2 (anhydrous) 5.90 g, Na2SO4 3.24 g, CaCl2 1.80 g, KCl 0.55 g, NaHCO3 0.16 g, KBr 0.08 g, SrCl2 34.00 mg, H3BO3 22.00 mg, Na2SiO3 4.00 mg, NaF 2.40 mg, NH4NO3 1.60 mg, Na2HPO4 8.00 mg, 18 g agar for solid medium, pH 8.0) (Power and Johnson 2009) at 30 °C for 2 days. Fatty acids were then extracted, methylated, detected by a gas chromatography (6890; Hewlett Packard). Peaks were automatically computed using the standard MIDI procedure (Microbial Identification, Sherlock version 6.0).
Phylogenetic analysis
Chromosomal DNA was extracted and purified according to standard methods (Marmur 1961). The 16S rRNA gene was PCR amplified with universal bacterial primers 8F (5′-AGA GTT TGA TCC TGG CTC AG) and 1492R (5′-GGT TAC CTT GTT ACG ACT T) (Embley 1991) and sequenced. Multiple sequence alignments, and phylogenetic analysis of 16S rRNA gene sequence from strains SL013A34A2T and SL013A24A with 34 validly published species of the genus Marinobacter, were performed by MEGA version 5.0 (Tamura et al. 2011). Phylogenetic trees were reconstructed using neighbor-joining method. The neighbor-joining (NJ) algorithm used a matrix of pairwise distances estimated under the Tamura and Nei (1993) model for nucleotide sequences. Maximum-parsimony (Tamura et al. 2011) and maximum likelihood (Felsenstein 1981) algorithms were also used to evaluate the stability of the tree topology. The genome DNA G + C content was determined from melting point (T m) curves (Mandel and Marmur 1968) obtained by using a Lambda35 UV/Vis spectrophotometer (Perkin Elmer) equipped with a temperature program controller (PTP-1 Peltier System). Escherichia coli strain K-12 DNA was used as a control (Marmur and Doty 1962).Homoduplex and heteroduplex DNA–DNA hybridizations were performed in triplicate as described by De Ley et al. (1970) and Huß et al. (1983). The optimum renaturation temperatures (T or ) of the heteroduplex genomic DNA solution were performed at 74.0 °C.
Results and Discussion
The two isolates exhibited similar phenotypic features. Cells were found to be Gram-staining negative, rod-shaped, motile with a single polar flagellum and multiplied by binary fission (Fig. 1). The cell size was found to be 1.5–2.0 μm in length and 0.5 to 0.8 μm in width. Colonies of both of the isolates are smooth, circular, convex and creamy on LB agar (pH 8.0; 3.0 % NaCl). Growth of strain SL013A34A2T occurs at 0.5–18.0 % (w/v) NaCl (optimum 3.0–6.0 % NaCl), pH 6.0–9.0 (optimum pH 8.0) and 10–45 °C (optimum 35 °C),while strain SL013A24A grows at 0.5–16.0 % (w/v) NaCl (optimum 1.0–3.0 % NaCl), pH 6.0–9.0 (optimum pH 8.0) and 15–40 °C (optimum 30 °C). The detailed phenotypic and physiological differences are presented in Table 1, which clearly differentiate the two isolates from the close phylogenetic neighbours. Both strains were found to be resistant to vancomycin, fusidic acid, rifamycin SV and aztreonam. Sensitive to amikacin, amoxicillin, ampicillin, carbenicillin, cefalexin, cefotaxime, ceftazidime, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, kanamycin, lincomycin, minocycline, nalidixic acid, norfloxacin, ofloxacin, roxithromycin, penicillin G, polymyxin B, streptomycin, sulfamethoxazole, tetracycline and tobramycin.
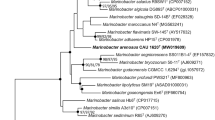
The 16S rRNA gene sequences (1,463 bp) of strains SL013A34A2T and SL013A24A show 99.3 % similarity to each other and strain SL013A34A2T was found to have the highest 16S rRNA gene sequence similarities to M. hydrocarbonoclasticus CGMCC 1.7683T (97.8 %), M. vinifirmus CGMCC 1.7265T (97.8 %) and M. excellens KMM 3809T (97.4 %), while those to the type strains of the other recognized species of the genus Marinobacter are between 93.8 and 96.4 %. Phylogenetic analysis showed that the two strains form a separate cluster within a subgroup containing M. vinifirmus CGMCC 1.7265T, M. excellens KMM 3809T, M. litoralis SW-45T, M. daepoensis SW-156T and M. hydrocarbonoclasticus CGMCC 1.7683T (Fig. 2 and Supplementary Fig. S1 & S2).The bootstrap resampling analysis showed that the association was relatively stable.
Neighbor-joining phylogenetic tree based on the 16S rRNA gene sequences, showing the positions of strains SL013A34A2T and SL013A24A, and all the type strains of genus Marinobacter. Oceanospirillum linum ATCC 11336T was used as an out group. Bootstrap values >50 % are shown at nodes. Bar, 0.01 substitutions per nucleotide position
DNA–DNA relatedness values of strain SL013A34A2T with strains SL013A24A, M. hydrocarbonoclasticus CGMCC 1.7683T, M. vinifirmus CGMCC 1.7265T and M. excellens KMM 3809T were 88.7 ± 1.3, 29.2 ± 2.9, 33.4 ± 0.3 and 29.4 ± 3.0 %, respectively. These values are below the DNA–DNA hybridization threshold value of 70 % which is used for separate species delineation by Wayne et al. (1987). Strains SL013A34A2T and SL013A24A showed 88.7 ± 1.3 % DNA–DNA relatedness with each other, which further confirmed that they belong to the same species.
The genomic DNA G + C contents of the strains SL013A34A2T and SL013A24A were found to be 56.1 and 55.7 mol % (Table 1), respectively, compared to 53.8–59.9 mol % for the closest type species.
The major cellular fatty acids of the two isolates were identified as C18:1 ω9c (37.6–37.7 %), C16:0 (27.3–28.9 %), C12:03-OH(7.5–8.2 %), C12:0(5.6–6.0 %) and C16:1 ω9c (4.3–5.4 %), which are in line with those of strains M. hydrocarbonoclasticus CGMCC 1.7683T and M. vinifirmus CGMCC 1.7265T. The 10-methyl C18:0 (5.0–5.3 %) fatty acid was found to be present only in the two isolates, while C17:1 ω8c fatty acid, which is found in other reference strains, was found to be absent in the two isolates (Table 2). The predominant respiratory quinone of strains SL013A34A2T and SL013A24A were determined to be Q-9, which is typical for Marinobacter species except for M. lutaoensis, which contains Q-8 (Shieh et al. 2003).The polar lipid profiles of strains SL013A34A2T and SL013A24A was found to consist of diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), phosphatidylglycerol (PG) and unidentified glycolipid(GL); strain SL013A34A2T also has phosphatidylcholine (PC), while strain SL013A24A does not. Strains SL013A34A2T and SL013A24A are different from M. hydrocarbonoclasticus CGMCC 1.7683T with regatd to an unidentified aminolipid, which was absent from the two isolates. The polar lipid profiles for both new isolates was found to be similar to that of M. vinifirmus CGMCC 1.7265T (Table 1 and Supplementary Fig. S3).
The phylogenetically coherent clustering and chemotaxonomic characteristics revealed that the two isolates are members of genus Marinobacter. Based on the low DNA–DNA relatedness to the members of closely-related taxa, their unique branching position in phylogenetic analyses, and differences in physiological characteristics including the NaCl and temperature ranges for growth, ability of nitrate reduction, hydrolysis of certain substrates, the carbon utilization pattern and the enzyme activities, strains SL013A34A2T and SL013A24A represent a novel species within the genus Marinobacter, family Alteromonadaceae, for which the name Marinobacter shengliensis sp. nov. is proposed.
Description of Marinobacter shengliensis sp. nov
Marinobacter shengliensis (sheng.li.en’sis. N.L. masc. adj. shengliensis pertaining to Shengli oilfield, China, from where the type strain was firstly isolated.)
Cells are Gram-staining negative, aerobic, moderately halophilic, motile and rod-shaped (1.5–2.0 × 0.5–0.8 μm) with a single polar flagellum. Colonies grown on LB (pH 8.0) agar for 2 days are creamy, circular and convex with smooth surface (1.5–2.0 mm in diameter). Growth occurs at 0.5–18.0 % (w/v) NaCl, pH 6.0–9.0 and 10–45 °C with the optimum grow that pH 8.0, 35 °C and 3.0–6.0 %(w/v) NaCl. Activities of catalase, oxidase, and hydrolysis of Tween 80 and starch are positive, whereas activities of H2S production, urease, β-galactosidase, glucose fermentation, nitrate reduction, indole production, arginine dihydrolase, and hydrolysis of gelatin are negative. Acid is not produced from fructose, xylose, mannitol, lactose, maltose, sucrose or mannose. Positive for alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, acid phosphatase, phosphohydrolase, α-glucosidase and N-acetyl-β-glucosamindase activities. Negative for urease, arginine dihydrolase, lipase (C14), valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, α-mannosidase and α-fucosidase activities. Can utilise Tween 40, dextrin, glycerol, pectin, d-maltose, d, l-fucose, d-fructose-6-PO4, l-alanine, l-glutamic acid, d-galacturonic acid, l-galactonic acid lactone, d-glucuronic acid, glucuronamide, d-lactic acid methyl ester, l-lactic acid, d, l-malic acid, bromo-succinic acid, α-hydroxy-butryric acid, β-hydroxy-d, l-butyric acid, α-keto-butryric acid, acetoacetic acid, propionic acid and acetic acid as the sole carbon sources.
Q-9 is the predominant respiratory quinone. The major cellular fatty acids are C18:1 ω9c, C16:0, C12:03-OH, C12:0, C16:1 ω9c and 10-methyl C18:0. The major polar lipids are diphosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol and unidentified glycolipid. The DNA G + C content of the type strain is 56.1 mol %.
The type strain, SL013A34A2T (=LMG 27740T = CGMCC 1.12758T), was isolated from oil-contaminated saline soil in Shengli oilfiled, China.
References
Aguilera M, Jiménez-Pranteda ML, Kharroub K, González-Paredes A, Durban JJ, Russell NJ, Ramos CA, Monteoliva-Sánchez M (2009) Marinobacter lacisalsi sp. nov., a moderately halophilic bacterium isolated from the saline-wetland wildfowl reserve Fuente de Piedra in southern Spain. Int J Syst Evol Microbiol 59:1691–1695
Andrews JM, for the BSAC Working Party on Susceptibility Testing (2008) BSAC standardized disc susceptibility testing method (version 7). J Antimicrob Chemother 62: 256–278
Antunes A, França L, Rainey FA, Huber R, Nobre MF, Edwards KJ, Da Costa MS (2007) Marinobacter salsuginis sp. nov., isolated from the brine–seawater interface of the Shaban Deep, Red Sea. Int J Syst Evol Microbiol 57:1035–1040
Bagheri M, Amoozegar MA, Didari M, Makhdoumi-Kakhki A, Schumann P, Spröer C, Sánchez-Porro C, Ventosa A (2013) Marinobacter persicus sp. nov., a moderately halophilic bacterium from a saline lake in Iran. Antonie Van Leeuwenhoek 104:1–8
Cai M, Wang L, Cai H, Li Y, Tang YQ, Wu XL (2011a) Rubrimonas shengliensis sp. nov. and Polymorphum gilvum gen. nov., sp. nov., novel members of Alphaproteobacteria from crude oil contaminated saline soil. Syst Appl Microbiol 34:321–327
Cai M, Wang L, Cai H, Li Y, Wang YN, Tang YQ, Wu XL (2011b) Salinarimonas ramus sp. nov. and Tessaracoccus oleiagri sp. nov., isolated from a crude oil-contaminated saline soil. Int J Syst Evol Microbiol 61:1767–1775
Eguchi M, Nishikawa T, Macdonal K, Cavicchioli R, Gottschal JC, Kjelleberg S (1996) Responses to stress and nutrient availability by the marine ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol 62:1287–1294
Embley TM (1991) The linear PCR reaction: a simple and robust method for sequencing amplified rRNA genes. Lett Appl Microbiol 13:171–174
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likeihood approach. J Mol Evol 17:368–376
Gao W, Cui ZS, Li Q, Xu GS, Jia XJ, Zheng L (2013) Marinobacter nanhaiticus sp. nov., polycyclic aromatic hydrocarbon-degrading bacterium isolated from the sediment of the South China Sea. Antonie Van Leeuwenhoek 103:485–491
Gauthier MJ, Lafay B, Christen R, Fernandez L, Acquaviva M, Bonin P, Bertrand JC (1992) Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int J Syst Bacteriol 42:568–576
Gorshkova NM, Ivanova EP, Sergeev AF, Zhukova NV, Alexeeva Y, Wright JP, Nicolau DV, Mikhailov VV, Christen R (2003) Marinobacter excellens sp. nov., isolated from sediments of the Sea of Japan. Int J Syst Evol Microbiol 53:2073–2078
Green DH, Bowman JP, Smith EA, Gutierrez T, Bolch CJS (2006) Marinobacter algicola sp. nov., isolated from laboratory cultures of paralytic shellfish toxin-producing dinoflagellates. Int J Syst Evol Microbiol 56:523–527
Gu J, Cai H, Yu SL, Qu R, Yin B, Guo YF, Zhao JY, Wu XL (2007) Marinobacter gudaonensis sp. nov., isolated from an oil-polluted saline soil in a Chinese oilfield. Int J Syst Evol Microbiol 57:250–254
Guo B, Gu J, Ye YG, Tang YQ, Kida K, Wu XL (2007) Marinobacter segnicrescenssp. nov., a moderate halophile isolated from benthic sediment of the South China Sea. Int J Syst Evol Microbiol 57:1970–1974
Handley KM, Héry M, Lloyd JR (2009) Marinobacter santoriniensissp. nov., an arsenate-respiring and arsenite-oxidizing bacterium isolated from hydrothermal sediment. Int J Syst Evol Microbiol 59:886–892
Huo YY, Wang CS, Yang JY, Wu M, Xu XW (2008) Marinobacter mobilis sp. nov. and Marinobacter zhejiangensis sp. nov., halophilic bacteria isolated from the East China Sea. Int J Syst Evol Microbiol 58:2885–2889
Huß VAR, Festl H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184–192
Kaeppel EC, Gärdes A, Seebah S, Grossart HP, Ullrich MS (2012) Marinobacter adhaerens sp. nov., isolated from marine aggregates formed with the diatom Thalassiosira weissflogii. Int J Syst Evol Microbiol 62:124–128
Kates M (1986) Techniques of lipidology: isolation, analysis, and identification of lipids, 2nd edn. Elsevier, Amsterdam, pp 100–110
Kharroub K, Aguilera M, Jiménez-Pranteda ML, González-Paredes A, Ramos-Cormenzana A, Monteoliva-Sánchez M (2011) Marinobacteroul menensissp. nov., a moderately halophilic bacterium isolated from brine of a salt concentrator. Int J Syst Evol Microbiol 61:2210–2214
Kim BY, Weon HY, Yoo SH, Kim JS, Kwon SW, Stackebrandt E, Go SJ (2006) Marinobacter koreensis sp. nov., isolated from sea sand in Korea. Int J Syst Evol Microbiol 56:2653–2656
Komagata K, Suzuki K (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Kovacs N (1956) Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 178:703
Lányí B (1988) Classical and rapid identification methods for medically important bacteria. Methods Microbiol 19:1–67
Lee OO, Lai PY, Wu HX, Zhou XJ, Miao L, Wang H, Qian PY (2012) Marinobacter xestospongiae sp. nov., isolated from the marine sponge Xestospongia testudinaria collected from the Red Sea. Int J Syst Evol Microbiol 62:1980–1985
Ley JD, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Liebgott PP, Casalot L, Paillard S, Lorquin J, Labat M (2006) Marinobacter vinifirmus sp. nov., a moderately halophilic bacterium isolated from a wine-barrel-decalcification wastewater. Int J Syst Evol Microbiol 56:2511–2516
Liu C, Chen CX, Zhang XY, Yu Y, Liu A, Li GW, Chen XL, Chen B, Zhou BC, Zhang YZ (2012) Marinobacter antarcticus sp. nov., a halotolerant bacterium isolated from Antarctic intertidal sandy sediment. Int J Syst Evol Microbiol 62:1838–1844
Lv XL, Xie BS, Cai M, Geng S, Tang YQ, Wang YN, Cui HL, Liu XY, Ye SY, Wu XL (2014) Glycocaulis albus sp. nov., a moderately halophilic dimorphic prosthecate bacterium isolated from petroleum-contaminated saline soil. Int J Syst Evol Microbiol 64:3181–3187
Mandel M, Marmur J (1968) Use of ultraviolet absorbance temperature profile for determining the guanine plus cytosine content of DNA. Methods Enzymol 12:195–206
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218
Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118
Martín S, Márquez MC, Sánchez Porro C, Mellado E, Arahal DR, Ventosa A (2003) Marinobacter lipolyticus sp. nov., a novel moderate halophile with lipolytic activity. Int J Syst Evol Microbiol 53:1383–1387
Montes MJ, Bozal N, Mercadé E (2008) Marinobacter guineae sp. nov., a novel moderately halophilic bacterium from an Antarctic environment. Int J Syst Evol Microbiol 58:1346–1349
Muurholm S, Cousin S, Päuker O, Brambilla E, Stackebrandt E (2007) Pedobacter duraquae sp. nov., Pedobacter westerhofensis sp. nov., Pedobacter metabolipauper sp. nov., Pedobacter hartonius sp. nov. and Pedobacter steynii sp. nov., isolated from a hard-water rivulet. Int J Syst Evol Microbiol 57:2221–2227
Pan XC, Geng S, Mei R, Wang YN, Cai H, Liu XY, Tang YQ, Nie Y, Ye SY, Wu XL (2014) Nitratireductor shengliensis sp. nov., isolated from an oil-polluted saline soil. Curr Microbiol 69:561–566
Power DA, Johnson JA (2009) Difco™ and BBL™ Manual: manual of microbiological culture media, 2nd edn. Becton Dickinson and Company, Sparks, pp 347–348
Qu LY, Zhu FL, Zhang JX, Gao CL, Sun XQ (2011) Marinobacter daqiaonensis sp. nov., a moderate halophile isolated from a Yellow Sea salt pond. Int J Syst Evol Microbiol 61:3003–3008
Roh SW, Quan ZX, Nam YD, Chang HW, Kim KH, Rhee SK, Oh HM, Jeon CO, Yoon JH, Bae JW (2008) Marinobacter goseongensis sp. nov., from seawater. Int J Syst Evol Microbiol 58:2866–2870
Romanenko LA, Schumann P, Rohde M, Zhukova NV, Mikhailov VV, Stackebrandt E (2005) Marinobacter bryozoorum sp. nov. and Marinobacter sediminum sp. nov., novel bacteria from the marine environment. Int J Syst Evol Microbiol 55:143–148
Shieh WY, Jean WD, Lin YT, Tseng M (2003) Marinobacter lutaoensis sp. nov., a thermotolerant marine bacterium isolated from a coastal hot spring in Lutao, Taiwan. Can J Microbiol 49:244–252
Shivaji S, Gupta P, Chaturvedi P, Suresh K, Delille D (2005) Marinobacter maritimus sp. nov., a psychrotolerant strain isolated from sea water off the subantarctic Kerguelen islands. Int J Syst Evol Microbiol 55:1453–1456
Sun JQ, Xu L, Zhang Z, Li Y, Tang YQ, Wu XL (2014) Diverse bacteria isolated from microthermoil production water. Antonie Van Leeuwenhoek 105:401–411
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tang YQ, Li Y, Zhao JY, Chi CQ, Huang LX, Dong HP, Wu XL (2012) Microbial communities in long-term, water-flooded petroleum reservoirs with different in situ temperatures in the Huabei Oilfield, China. PLoS ONE 7(3):e33535
Wang YN, Cai H, Chi CQ, Lu AH, Lin XG, Jiang ZF, Wu XL (2007a) Halomonas shengliensis sp. nov., a moderately halophilic, denitrifying, crude-oil-utilizing bacterium. Int J Syst Evol Microbiol 57:1222–1226
Wang YN, Cai H, Yu SL, Wang ZY, Liu J, Wu XL (2007b) Halomonas gudaonensis sp. nov., isolated from a saline soil contaminated by crude oil. Int J Syst Evol Microbiol 57:911–915
Wang CY, Ng CC, Tzeng WS, Shyu YT (2009) Marinobacter szutsaonensis sp. nov., isolated from a solar saltern. Int J Syst Evol Microbiol 59:2605–2609
Wang YN, Chi CQ, Cai M, Lou ZY, Tang YQ, Zhi XY, Li WJ, Wu XL, Du X (2010) Amycolicicoccus subflavus gen. nov., sp. nov., an actinomycete isolated from a saline soil contaminated by crude oil. Int J Syst Evol Microbiol 60:638–643
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Truper HG (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Williams ST, Goodfellow M, Alderson G, Wellington EMH, Sneath PHA, Sackin MJ (1983) Numericalclassification of Streptomyces and related genera. J Gen Microbiol 129:1743–1813
Wu XL, Yu SL, Gu J, Zhao GF, Chi CQ (2009) Filomicrobium insigne sp. nov., isolated from an oil-polluted saline soil. Int J Syst Evol Microbiol 59:300–305
Xu XW, Wu YH, Wang CS, Yang JY, Oren A, Wu M (2008) Marinobacter pelagius sp. nov., a moderately halophilic bacterium. Int J Syst Evol Microbiol 58:637–640
Yoon JH, Shin DY, Kim IG, Kang KH, Park YH (2003) Marinobacter litoralis sp. nov., a moderately halophilic bacterium isolated from sea water from the East Sea in Korea. Int J Syst Evol Microbiol 53:563–568
Yoon JH, Yeo SH, Kim IG, Oh TK (2004) Marinobacter flavimaris sp. nov. and Marinobacter daepoensis sp. nov., slightly halophilic organisms isolated from sea water of the Yellow Sea in Korea. Int J Syst Evol Microbiol 54:1799–1803
Yoon JH, Lee MH, Kang SJ, Oh TK (2007) Marinobacter salicampi sp. nov., isolated from a marine solar saltern in Korea. Int J Syst Evol Microbiol 57:2102–2105
Zhang DC, Li HR, Xin YH, Chi ZM, Zhou PJ, Yu Y (2008) Marinobacter psychrophilus sp. nov., a psychrophilic bacterium isolated from the Arctic. Int J Syst Evol Microbiol 58:1463–1466
Zhuang DC, Chen YG, Zhang YQ, Tang SK, Wu XL, Tan ZC, Li WJ, Cui XL (2009) Marinobacterzhanjiangensis sp. nov., a marine bacterium isolated from sea water of a tidal flat of the South China Sea. Antonie Van Leeuwenhoek 96:295–301
Acknowledgments
This work was financially supported by National Natural Science Foundation of China (31225001) and National High Technology Research and Development Program of China (2012AA02A703)
Author information
Authors and Affiliations
Corresponding author
Additional information
Yi-Jing Luo and Bai-Sheng Xie have contributed equally to this work.
Sequence deposited
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences of strains SL013A34A2T and SL013A24A are KF307780 and KF307779, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luo, YJ., Xie, BS., Lv, XL. et al. Marinobacter shengliensis sp. nov., a moderately halophilic bacterium isolated from oil-contaminated saline soil. Antonie van Leeuwenhoek 107, 1085–1094 (2015). https://doi.org/10.1007/s10482-015-0401-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-015-0401-y