Abstract
Vasculogenic mimicry (VM) formed by aggressive tumor cells to mimic vasculogenic networks plays an important role in the tumor malignancy of HCC. However, the pathogenesis underlying VM is complex and has not been fully defined. m6A is a common mRNA modification and has many biological effects. However, the relationship between m6A and VM remains unclear. In this research, we found that m6A methyltransferase METTL3 in HCC tissues was positively correlated with VM. The m6A level of mRNA significantly increased in 3D cultured cells treated with VEGFa and was related to VM formation. Transcriptome sequencing analysis of 3D cultured cells with knockdown Mettl3 showed that the Hippo pathway was involved in m6A-mediated VM formation. Further mechanism research indicated that the m6A modification of YAP1 mRNA affected the translation of YAP1 mRNA. In conclusion, m6A methylation plays a key role in VM formation in HCC. METTL3 and YAP1 could be potential therapeutic targets via impairing VM formation in anti-metastatic strategies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Liver cancers are the fourth most common cause of cancer-related deaths and rank sixth in terms of incident cases worldwide. Hepatocellular carcinoma (HCC) accounts for the majority of primary liver cancers [1]. Although much progress has been made in the treatment of HCC in recent years, the prognosis of patients with HCC remains very poor. Angiogenesis is essential to promote HCC growth and metastasis. Previous studies showed that blood vessels in tumors are only formed by endothelial cells. Since the 1990s, many pharmaceutical companies have focused on developing compounds that curtail angiogenesis to starve tumor cells. Several drugs, such as Avastin and Nexavar [2, 3], have been approved in the U.S. for cancer treatment. However, these drugs only temporarily slow tumor growth and do not completely kill tumor cells. Moreover, many types of tumors treated with these drugs often develop resistance or metastasis [4,5,6].
Vasculogenic mimicry (VM) was discovered by Maniotis [7]. He believes that functional blood supply channels in tumors may not be involved in endothelial cells but are formed by aggressive tumor cells. This finding provides a new perspective on the blood supply of tumors. To date, VM has been found in a variety of cancer types, such as melanoma [8,9,10], ovarian cancer [11, 12], colorectal cancer [9, 13], laryngeal squamous cell carcinoma [14, 15], and HCC, and has been associated with poor prognosis in patients.
The development of drugs based on VM theory is predicted to change the cancer treatment for patients, but the mechanism of VM formation remains unclear. N6-Methyladenosine (m6A) modifications of RNA were discovered in the 1970s [16]. Of the many RNA modifications of eukaryotes, m6A has the most abundant and has unique distribution characteristics on transcripts [17, 18]. Jia et al. identified the first RNA demethylase, fat mass and obesity-associated (FTO) gene, indicating that RNA modification is reversible [19]. Since then, many studies on m6A dynamic modification have been conducted. Methyltransferase-like 3 (METTL3), METTL14, and Wilm’s tumor-1-associated protein (WTAP) have been proved to act as m6A methyltransferases (“writers”) in mammalian cells [20, 21]. The m6A-binding proteins with the YTH domain, including cytoplasmic protein YTHDF1, YTHDF2, YTHDF3, and nuclear protein YTHDC1, have been identified to be the “readers” of m6A and modulate mRNA stability and translation to mediate downstream effects [22, 23]. In recent years, a number of studies have focused on biological function of m6A-modified mRNA. The m6A modification was found to be involved in a variety of biological processes and played an important role in cancer progression. However, the relationship between m6A and VM formation remains unclear.
In this study, we found that the expression of m6A writer METTL3 is positively correlated with VM in HCC tissues. The m6A mRNA level increased in 3D cultured cells treated with VEGFa and was related to VM formation and VM-related makers. RNA sequencing and functional studies indicated the involvement of the Hippo pathway in m6A-mediated VM formation. The m6A modification can promote the translation of YAP1 mRNA and open the Hippo pathway. Moreover, m6A-dependent YAP1 expression can promote VM formation, migration, invasion in vitro, and in vivo. Our findings indicate the critical role of m6A modification in VM formation in HCC. METTL3 and YAP1 could be potential therapeutic targets via impairing VM formation in anti-metastatic strategies.
Material and methods
Patient samples and ethics
HCC tissue microarrays from 75 and 96 cases were purchased from US Biomax for IHC. Each single tissue spot on every array slide was individually examined by certified pathologists according to the WHO published standards for diagnosis, classification, and pathological grading.
Cell culture, treatments, and transfection
Human cancer HepG2 and MHCC-97H cells were purchased from KeyGen Biotech (Nanjing, China). SK-HEP-1-Luc (luciferase labeled) cells were obtained from OBIO (Shanghai, China). The cells were maintained in Dulbecco’s modified Eagle medium (DMEM, Bioind, ISR) or Minimum Essential Medium α (MEM α, Gibco, USA) with 10% fetal bovine serum (FBS, Bioind, ISR) and 1% penicillin/ streptomycin (Hyclone, USA). All cells were tested negative for mycoplasma contamination before use. SK-HEP-1-Luc cells were transfected with psi-LVRU6GP-shMETTL3 or vector control via lipofectamine 2000 to establish stable METTL3 knockdown cell lines. The reference sequence as follows: CTG CAA GTA TGT TCA CTA TGA [20]. The transfected cells were selected with puromycin (2 µg/mL, Gibco, USA) for 2 weeks. The surviving cells were collected and seeded into a 96-well plate for formation of cell clones and further expansion. The expanded monoclonal cell populations were named as SK-HEP-1-Luc-Vector and SK-HEP-1-Luc-shMETTL3 cells. For overexpression of YAP1, the cells were transfected with pcDNA3.1-YAP1 by lipofectamine 2000. For METTL3 knockdown, the cells were transfected with siMETTL3 or nontargeting scrambled siRNAs (Santa Cruz Biotechnology, sc-92172 and sc-37007, respectively) by using RNAiMax (Thermo Fisher, USA).
LC–MS/MS quantification of m6A in poly(A)-mRNA
The cells were coincubated with VEGFa (5 ng/mL). After incubation, mRNA was extracted from total RNA by using a PolyATtract® mRNA Isolation Systems (Promega, USA). mRNA concentration was measured by ultraviolet absorbance at 260 nm. mRNA (200 ng) was first denatured by heating at 95 °C for 5 min, chilled on ice for 2 min, and added with 1/10 volume of S1 nuclease buffer (30 mM CH3COONa, pH 4.6; 280 mM NaCl; 1 mM ZnSO4) and 150 units of S1 nuclease (Thermo Fisher Scientific, USA). The mixture (20 μL) was incubated at 37 °C for 5 h. The solution was subsequently added to 1/10 volume of alkaline phosphatase buffer (50 mM Tris–HCl, 10 mM MgCl2, pH 9.0) and 15 units of alkaline phosphatase (Takara, Dalian, China). The cells were continuously incubated at 37 °C for an additional 2 h, followed by extraction with an equal volume of chloroform twice. The resulting aqueous layer was collected, lyophilized to dryness, and reconstituted in 100 μL of water. The sample was diluted to 1 mL and then filtered (0.22 μm pore size, 4 mm diameter, Millipore). The solution (10 μL) was injected into the LC–MS/MS device. The nucleosides were separated by reverse-phase ultra-performance liquid chromatography on a C18 column with online mass spectrometry detection using Waters XEVO TQ-STM (Waters, USA) triple quadrupole mass spectrometer in positive electrospray ionization mode. All nucleosides were quantified using retention time and ion mass transitions of 268.0 to 136.0 (A) and 282.1 to 150.0 (m6A). Quantification was conducted using standard curves ran in the same batch. Ratio of m6A to A was calculated based on the calibration curves.
In vivo orthotopic implantation model
Animal experiments were conducted in accordance with the National Institutes of Health Animal Use Guidelines. All experimental protocols were approved by the Institutional Animal Care and Use Committee at Tianjin International Joint Academy of Biomedicine. Male BALB/c nude mice aged 4–6 weeks were used in the experiments. According to institutional guidelines, all animals were maintained in a specific pathogen-free animal care facility. An orthotopic liver tumor model in nude mice was established to assess metastasis. In brief, 1 × 107 SK-HEP-1-Luc-shControl or SK-HEP-1-Luc-shMETTL3 stable cells(as Fig. 6a clone#1) were suspended in 300 μL of PBS and injected orthotopically into the left liver lobe of nude mice. The mice were killed, and the lungs and liver were preserved in 10% formalin solution for further study. Tumor growth and lung metastases were detected in vivo after injection of 150 mg/kg D-Luciferin and potassium salt substrate (Yeasen, CN) in PBS into anesthetized mice with NightOWLIILB983 (Berthold Technologies).
Diethylnitrosamine-induced Mettl3 +/− mouse model of HCC
Animal experiments were conducted in accordance with the National Institutes of Health Animal Use Guidelines. All experimental protocols were approved by the Institutional Animal Care and Use Committee at Tianjin International Joint Academy of Biomedicine. Mettl3+/− mice with C57BL6/J background were generated using CRISPR–Cas9 system. Exon2-Exon3 of Mettl3 is the knockout region. sgRNA was transcribed in vitro. Cas9 and sgRNA were microinjected into the fertilized eggs of C57BL/6 J. Mice fertilized eggs were transplanted to obtain positive F0 mice which were confirmed by PCR(PCR primers: F1:TCC CTG GGA AAC ATA ATC ACA TCC,F2: CTC CCT AAA AGA GTG TAA CTT CCC C, WT: 1975 bp, Target: 520 bp). Fifteen-day-old mice were injected with diethylnitrosamine (DEN; 20 mg/kg body weight; Sigma, USA) to initiate tumor generation. Beginning at 29-day-old, the mice were injected with TCPOBOP (3 mg/kg body weight, intraperitoneally, Sigma-Aldrich, St. Louis, MO) biweekly for 3–15 weeks for a total of seven times to promote tumor growth. Once HCC was established, Mettl3+/− mice were divided into three groups assigned to receive the transposon system. The DNA mixtures of transposons (H6552 or H6552-YAP1 plasmids) and transposase-encoding vector (H6281, molar ratio of transposon plasmids to transposase-encoding vector of 2:1) were suspended in Lactated Ringer’s solution and injected into the lateral tail veins of mice (0.1 mL/g body weight) in less than 7 s. After 5 weeks, the mice were euthanized. Liver and lung tissue samples were collected and processed for histological analysis.
Statistical analysis
Experiments were repeated at least three times independently. Measured data were represented as mean ± SEM. One-way analysis of variance (ANOVA) or two-tail Student t test was applied to compare quantitative data. Overall survival was analyzed with Kaplan–Meier method by using log-rank test to determine difference. p values for each analysis are marked on figures, and the level of statistical significance was set as p < 0.05 (*p < 0.05; **p < 0.01; ***p < 0.001).
The other methods for m6A immunofluorescence, bioinformatics analysis, METTL3 knockdown transcriptome sequencing, MeRIP-qPCR, luciferase reporter assays, polysome profiling analysis, protein stability assays, western blot, RNA extraction and quantitative real-time PCR for gene expression, RNA stability assays, scanning electron microscopy, three-dimensional culture assay, wound healing, invasion assays, hematoxylin–eosin (H&E) and immunohistochemistry are described in detail in the Supplementary Materials and Methods.
Results
m6A writer METTL3 expression in clinical HCC tissues is associated with VM and poor prognosis
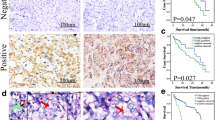
CD34 or CD31 and periodic acid–Schiff (PAS) dual staining was applied to distinguish the matrix-rich morphological pattern of VM. First, a retrospective study of 75 patients with HCC was conducted. After CD31-PAS double staining and IHC staining (Fig. 1a, Table S3), the pathological group was divided into four subgroups based on the number of VM cases and METTL3 staining intensity: VM + / METTL3 + , VM − / METTL3 + , VM + /METTL3 − , and VM − /METTL3 − . Survival analysis indicated that the survival rate of the double-positive group was significantly lower than that of the double-negative group (Fig. 1 b). The staining intensity of METTL3 was also strongly correlated with the number of VM (Fig. 1c). Moreover, coexpression analysis in 96 patients with HCC was conducted. We found that METTL3 was negatively correlated with the expression of epithelial marker E-cadherin and positively correlated with VM markers, namely, VE-cadherin, VEGFR1, and VEGFR2; mesenchymal marker Vimentin; and tumor VM microenvironmental markers, namely, MMP2 and MMP9 (Fig. 1d, e, Fig.S1 a-b, Table S4). The TCGA data showed that the expression of METTL3 in liver cancer was significantly higher than those of normal tissues (Fig.S1 c), and was associated with pathological grade and clinical stage (Fig.S1 d-e). Moreover, the expression of METTL3 in 364 patients with liver cancer was positively correlated with VM-related proteins, namely, CDH5, VIM, FLT1, KDR, MMP2, MMP9, FN1, and SERPINE2 (Fig. 1f, Fig.S1 f-g). In addition, the patients with were separated based on the mRNA expression of METTL3 into two groups, namely, METTL3 high expression and METTL3 low expression. The differential genes between the two groups were enriched by GSEA to find m6A writer METTL3-related pathways. The VEGFa signaling pathway was enriched (Fig. 1g). These data showed that METTL3 was involved in the formation of HCC VM and closely related to poor prognosis.
m6A writer METTL3 expression in clinical HCC tissues is associated with VM and poor prognosis of patients. a Evidence and morphologic observation of VM and Endothelial Vessel by CD31 (brown) /PAS (pink) /hematoxylin (blue) staining in HCC. b Immunostaining for METTL3 in HCC tissue microarrays (left). Kaplan–Meier curves for the OS of HCC patients (right), VM + / METTL3 + ( n = 21), VM − / METTL3 + ( n = 18), VM + /METTL3 − (n = 19), and VM − /METTL3 − (n = 17). c correlation analysis between VM and METTL3 (n = 75). d Representative images of METTL3, VE-cadherin, E-cadherin, Vimentin, MMP2, and MMP9 expression in VM negative and positive HCC tissue samples. e Correlation analysis between VE-cadherin, E-cadherin, Vimentin, MMP2, and MMP9 expression (n = 96). f Correlation analysis between METTL3 and VM maker by using TCGA dataset (n = 364). g GSEA enrichment analysis of low expression (n = 182) and high expression (n = 182) of METTL3 from TCGA dataset. Black scale bar = 50 μm. All data were represented as mean ± SEM. *p < 0.05, ** p < 0.01, ***p < 0.001
VM in HCC cell line is regulated by m6A mRNA level
Although VM can be induced by various extracellular ligands and microenvironment, VEGFa has been considered as the major inducer of this process in HCC cells [24]. We treated 3D cultured HepG2 and MHCC-97H cells with VEGFa (5 ng/mL) for 24 h. The tube formation ability was significantly increased. We then investigated the variations in the m6A mRNA levels in 3D cultured cells. By using LC–MS/MS, we identified that the m6A mRNA levels isolated from HepG2 and MHCC-97H cell treated with VEGFa were statistically (p < 0.05, t test) more abundant than those in their corresponding control cells. The m6A/A mRNA levels in HepG2 and MHCC-97H cells treated with VEGFa increased by 49% and 59%, respectively (Fig. 2a). These data showed that HCC cell lines undergoing VM increased the m6A levels of mRNA.
VM formation is regulated by m6A level of mRNAs. a MHCC-97H and HepG2 cells were treated with or without 5 ng/ml VEGFa for 24 h, the m6A/ A ratio of mRNA were determined by LC–MS/MS. b m6A IF assay of 3D cultured cells transfected with siNC or siMETTL3 (left) and quantitatively analyzed by Wimasis Image Analysis (right). White scale bar = 200 μm. c The morphologic detect with scanning electron microscopy in 3D culture assay. White scale bar = 100 μm. d, e Protein (d) and mRNA (e) expressions of E-Cad, VE-Cad, and METTL3 in MHCC-97H and HepG2 cells were measured by qRT-PCR and western blot analysis, respectively. f MHCC-97H and HepG2 cells were transfected with siNC or siMETTL3 for 24 h, and then were treated with or without 5 ng/ml VEGFa for another 24 h, protein expression was determined by western blot analysis. g m6A IF assay of 3D cultured cells transfected by siNC or overexpression ALKBH5 (left) and quantitatively analyzed by Wimasis Image Analysis (right). White scale bar = 200 μm. All data were represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001
To characterize the roles of m6A in VM, we reduced the m6A level by using siRNA pools of METTL3 and investigated the tube formation ability in HCC cell lines. The m6A level was detected by immunofluorescent assay. Silenced METTL3 cells had significantly lower levels of m6A and weaker tube formation ability than siRNA-negative control cells (Fig. 2b), which also confirmed the roles of METTL3 as the m6A “writer” in VM. The ultramicrostructure of VM was observed by scanning electron microscope. The silenced METTL3 cells were not connected well and formed a curved shape (Fig. 2c). Then we evaluated VM-related makers, namely, E-cadherin and VE-cadherin, in silenced METTL3 cells. The results of western blot and qPCR analysis showed that the knockdown of METTL3 suppressed the expression of VE-cadherin and promoted the expression of E-cadherin in HepG2 and MHCC-97H cells (Fig. 2d, e). To rule out the possibility of off-target effect, we also chose another shRNA of METTL3 to inspect its effect on VM formation. The results showed that silenced METTL3 cells transfected with this plasmid had significantly lower levels of m6A and weaker tube formation ability than shcontrol cells (Fig. S2. a). The western blot results showed that knocking down METTL3 suppressed the expression of VE-cadherin and promoted the expression of E-cadherin(Fig. S2. b).
Furthermore, VEGFa induced downregulation of E-Cadherin and upregulation of VE-Cadherin were rescued by knocking down METTL3 in HepG2 and MHCC-97H cells (Fig. 2f). Overexpression ALKBH5 (a m6A eraser) cells had significantly lower levels of m6A and weaker tube formation ability than control cells (Fig. 2g). Together, our data indicated that the total m6A levels mRNA may regulate VM formation in HCC in vitro.
Hippo pathway is inhibited by interfering METTL3
To investigate the m6A-related pathway undergoing VM process of HCC. RNA-seq was used for transcriptome mapping in 3D cultured HepG2 cells treated by knocking down METTL3 and their corresponding control cells (Fig. 3a). A total of 149 differentially expressed genes were identified. Overlapping these differential genes and m6A-modified genes from GSE110320 [25], we found that the mRNA of most of the differential genes (75%) were modified by m6A (Fig. 3b). The overlapping genes were enriched by Gene Ontology (GO) analysis (Fig. 3c). The VEGFa–VEGFR2 pathway were enriched, which was reported to be associated with VM formation. Moreover, as an important pathway, Hippo signaling was enriched and played an important role in the malignant progression of cancer [26]. Then the involvement of the Hippo pathway in m6A-mediated VM formation was investigated. The analysis of the MeRIP data indicated the high abundance of m6A modification in the YAP1 mRNA, particularly in the CDS region (Fig. 3d). MeRIP-qPCR experiments were conducted to enrich m6A-modified mRNA. The m6A modification level of YAP1 mRNA was significantly reduced in knockdown METTL3 cells (Fig. 3e). In summary, the Hippo pathway is involved in the m6A-mediated VM formation and that the mRNA of YAP1 is modified by m6A.
Hippo pathway is involved in m6A-dependent VM formation. a Schematic illustration of the experimental procedure. b Venn diagram showing the shared gene between Differential genes of siMETTL3 and genes modified by m6A (GSE110320). A total of 113 shared gene were observed. c Metascape enrichment analysis of the above shared genes. d m6A peaks were enriched in CDS of YAP1 genes from m6A RIP-seq data (GSE110320). e m6A RIP-qPCR analysis of YAP1 mRNA in MHCC-97H and HepG2 cells by transfected with siNC and siMETTL3. All data were represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001
YAP1 promotes VM formation and malignant progression via a m6A-dependent way in vitro
YAP1 plays an important role in cancer progression. The involvement of YAP1 in VM formation and its relation to m6A were further analyzed. First, we detected YAP1 expression by using western blot after changing the m6A level in MHCC-97H and HepG2 cell lines. YAP1 was downregulated after the decline of m6A by knocking down METTL3 and upregulated after the rise of m6A by overexpressing ALKBH5 (an important m6A demethylase) and inducing by VEGFa (Fig. 4a–c). In addition, the overexpression of YAP1 significantly promoted the expression of VM marker VE-cadherin and reduced the expression of epithelial marker E-cadherin. Knocking down METTL3 significantly rescued the increase in the VE-cadherin level caused by the overexpression of YAP1 (Fig. 4a). In view of the possible role of YAP phosphorylation in signaling during VM formation. We detected phosphorylated YAP, named pYAP(Ser127), level using western blot during VM formation. The results showed that pYAP and YAP have the same expression pattern in cells treated by VEGFa or transfected with siMETTL3(Fig.S3 a-b). These data indicated that m6A could not influence the phosphorylation in signaling during VM formation. The cell function of YAP1 was then evaluated. YAP1 significantly increased VM formation and enhanced cell migration and invasion (Fig. 4d–i). This process was rescued by knocking down METTL3. In summary, YAP1 promotes VM formation and cancer progression via a m6A-dependent way in vitro.
YAP1 promote VM and cancer progression via a m6A-dependent way in vitro. a Expression of YAP1, METTL3, VE-cad and E-Cad in MHCC-97H and HepG2 cells transfected with or without pcDNA-3.1–3 × Flag/YAP1 and siNC/siMETTL3 for 48 h. b Expression of YAP1 and ALKBH5 in MHCC-97H and HepG2 cells transfected with or without pcDNA3.1-AKLBH5. c Expression of YAP1 in MHCC-97H and HepG2 cells treated with VEGFa (24 h) or not. d, e The tube formation of MHCC-97H and HepG2 cells transfected with or without pcDNA-3.1-YAP1 and siNC/siMETTL3 for 48 h were recorded (left) and quantitatively analyzed (right). f, g wound healing of MHCC-97H and HepG2 cells. h, i Transwell of MHCC-97H and HepG2 cells. All data were represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.by Student’s t test
m6A enhances the translation efficiency of YAP1 mRNA
The mechanisms regulating YAP1 expression through m6A methylation were further investigated. The pre-mRNA and mRNA expression levels of YAP1 after interfering with METTL3 were detected. Surprisingly, the transcription level of YAP1 increased after interfering with METTL3 (Fig. 5a). Then we treated siNC and siMETTL3 cells with actinomycin-D to block transcription. The results showed that the half-life of pre-mRNA and mRNA was significantly prolonged after interfering with METTL3 (Fig. 5b, c). The remaining mRNA and pre-mRNA were significantly higher in siMETTL3 group than its corresponding control after transcription inhibition for 6 h(Fig.S4. a-b), which suggested that m6A modification may trigger the splicing of pre-mRNA and degradation of mature mRNA of YAP1 in HCC. The dyssynchrony between the mRNA and protein levels of YAP1 leaded us to suspect that m6A modification may be related to the translation or stability of the YAP1 protein. METTL3 knockdown cells and corresponding control cells were treated with cycloheximide, a protein translation inhibitor. The Western blot assay indicated no significant change in the half-life of the two experimental proteins (Fig. 5d, e). The translation efficiency of the YAP1 protein was measured using dual-luciferase assay. The CDS sequence of YAP1 was constructed into the F-luciferase protein downstream of pGL6-miR for fusion expression. The translation efficiency of the YAP1 protein was significantly reduced after knocking down METTL3 (Fig. 5f, g). It indicated that m6A-induced YAP1 expression is related to regulation of protein translation.
m6A triggers translation of YAP1 mRNA. a Precursor and mature mRNA level of YAP1 in HepG2 cells by transfected with or without siNC/siMETTL3 for 48 h. b, c After transfected siNC or siMETTL3 for 24 h, HepG2 cells were treated with Act-D for 90 min, then precursor (b) or mature (c) YAP1 mRNA were detected at indicated times using qPCR. TI, Transcription Inhibition. The RNA lifetime (t1/2) can be calculated by nonlinear regression analysis. d, e After transfection with siNC or siMETTL3 for 24 h, HepG2 cells were treated with CHX for the indicated times, and protein expression of YAP1 was analyzed by western blot. f, g HepG2 cells were transfected with pGL6-miR-YAP1-CDS reporter and siMETTL3 for 48 h, and the translation efficiency of YAP1 is defined as the quotient of reporter protein production (F-luc/R-luc). h Polysome profiling of HepG2 cells were analyzed after knockdown METTL3. i Analysis of YAP1 mRNA in non-ribosome portion (< 40S), 40S, 60S, 80S, and polysome for transfected siMETTL3 or siNC in HepG2 cells. All data were represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, by Student’s t test
Total RNA of HepG2 was separated by differential centrifugation with sucrose density gradient. We obtained nontranslating fraction (< 40 S), translation initiation fraction (including 40S ribosomes, 60S ribosomes, 80S monosomes, and < 80S), and translation active polysomes (> 80S) by ribosome profiling. The experimental results showed the significant decrease in the assembly of the ribosome translation initiation complex (Fig. 5h). The qPCR test showed the significantly reduced mRNA level of YAP1 in the translation-activated polysome state in siMETTL3 cells (Fig. 5i). In summary, m6A primarily changes the expression of the YAP1 protein by affecting translation efficiency.
Silencing METTL3 suppresses VM formation and cancer progression in an orthotopic transplantation tumor model of HCC
We used an orthotopic transplantation tumor model of HCC to study the potential effects of m6A methylation in HCC during metastasis and VM formation in vivo. First, sh-control and sh-METTL3 SK-Hep1-Luc cells were injected into the liver of nude mice (Fig. 6a). Obviously, the luciferase signaling of the whole body and the lung tissue was inhibited in shMETTL3 nude mouse (Fig. 6b–e). mice. The Kaplan–Meier survival analysis revealed that sh-METTL3 nude mice had favorable prognosis (Fig. 6f). The fixed tissue was stained by HE to eliminate lung metastasis. The counts of tumor foci in sh-METTL3 nude mice were less than those in sh-Control node mouse (Fig. 6g). VM formation in tumor tissues was analyzed by PAS and CD31 dual staining. sh-METTL3 cells exhibited less VM than sh-Control group (Fig. 6h). The IHC staining analysis showed that the expression levels of VM marker VE-cadherin and other makers involved in VM (MMP2, MMP9, and Fibronectin) decreased, while the expression of the epithelial maker E-cadherin increased in the sh-METTL3 group. The expression of YAP1 was also inhibited in the sh-METTL3 group (Fig. 6i). In conclusion, m6A methylation could promote cancer progression and VM formation in vivo.
Silencing of METTL3 suppresses VM formation and cancer progression in a orthotopic transplantation tumor models of HCC. a The expression of METTL3 in stable METTL3 knockdown cell lines by G418 screening. b In vivo Imaging in the BALB/C nude mice. SK-HEP-1-Luc-shMETTL3 (n = 6), and SK-HEP-1-Luc-shControl (n = 6) cell lines were injected into the liver of BALB/C nude mice. c Statistical analysis of the luciferease signal. d Imaging of the luciferase signal of the lungs after dissection. e Statistical analysis of the luciferase signal. f Survival analysis of the BALB/C nude mice. g Formation of HCC metastatic foci in the lung was confirmed by hematoxylin and eosin staining. h Evidence and morphologic observation of VM. Red arrow points to the PAS-positive and CD31-negative vessels lining with tumor cells. Yellow arrow points to the endothelial cell lined vessels(left). Statistical results of VM in shControl and shMETTL3 groups (right). i METTL3, E-cadherin, VE-cadherin, MMP2, MMP9 and YAP1 expression in the tumor tissues of Vector and shMETTL3 groups were analyzed via IHC. All data were represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001
m6A-dependent YAP1 expression promotes VM formation and tumor metastasis in a DEN-induced Mettl3 +/− mouse model of HCC
To further investigate the role of m6A and YAP1 in cancer progression and VM formation in vivo, we generated Mettl3+/− C57BL6/J mice by using CRISPR–Cas9 system. Considering that knock out of METTL3 is embryonically lethal, we obtained mice with just one allele knocked out. However, METTL3 was downregulated significantly in the liver tissues of these mice (Fig. 7a). An HCC model was generated by treating Mettl3+/− mice and wild-type mice with DEN to initiate tumor generation. The mice were then given with TCPOBOP every 2 weeks for a total of seven times to promote tumor growth. At the end of the treatment, half of the Mettl3+/− mice were treated with YAP1 plasmid via hydrodynamic injection into the tail vein [27] (Fig. 7 c), namely Mettl3+/− + YAP1 group, and the other half of the Mettl3+/− mice were treated with the control plasmid, namely Mettl3+/− + Vector group. At the same time, the wild-type mice were also treated with the control plasmid, namely Wt + Vector group (Fig. 7 b). The liver tumor foci were counted after dissection. Mettl3+/− + Vector group mice had less tumor foci than the Wt + Vector group. After the hydrodynamic injection of YAP1, which rescued the decrease of tumor foci due to the knocking down METT3 in Mettl3+/− mice (Fig. 7d). The fixed tissue was stained by HE to eliminate lung metastasis. The counts of metastasis foci were counted. Mettl3+/− + Vector group mice had less foci than the Wt + Vector group. After the hydrodynamic injection of YAP1, which rescued the decrease of foci due to the knocking down METT3 in Mettl3+/− mice (Fig. 7e). VM formation in tumor tissue was analyzed by PAS and endomucin dual staining. The VM counts in Mettl3+/− + Vector group mice decreased significantly compared with those in the Wt + Vector group. Meanwhile, the overexpression of YAP1 due to hydrodynamic injection rescued the decrease resulting from the knocking down of METTL3 (Fig. 7f). The IHC staining analysis showed that the expression of VE-cadherin, MMP2, and MMP9 decreased in Mettl3+/− + Vector mice, while the overexpression of YAP1 rescued the influence of knocking down METTL3. The E-cadherin expression was opposite to the expression pattern of VE-cadherin (Fig. 7g). These data demonstrated that METTL3 knockdown can attenuate the effect on cancer progression and VM formation in a DEN-induced Mettl3+/− mouse model of HCC, and YAP1 could restore the influence of knocking down METTL3.
YAP1 promote VM formation and tumor metastasis in a DEN-induced Mettl3+/− mouse HCC model. a METTL3 expression in wild-type and Mettl3+/− C57BL/6 J mice was detected by Western blot. (#1,# 2, #3 represent three wild-type mice, #4, #5, #6 represent three Mettl3+/− C57BL/6 J mice). b the experimental procedure of DEN-induced Mettl3+/− C57BL/6 J mice HCC model. c Schematic illustration of the experimental procedure. Selected transposons were hydrodynamically delivered to the liver together with plasmids encoding transposase by tail venn injection. d Representative photos of the visible liver nodules and HE staining results.(left). The tumor foci of Wt + Vector (n = 6), Mettl3+/− + Vector(n = 6) and Mettl3+/− + YAP1(n = 6) groups were statistically analyzed (right). e Formation of HCC metastatic foci in the lung was confirmed by hematoxylin and eosin staining(left). The tumor foci were statistically analyzed (right). f Representative photos of PAS and endomucin dual staining(left). Statistical results of VM counts in Wt + Vector, Mettl3+/− + Vector and Mettl3+/− + YAP1 groups. g E-cadherin, VE-cadherin, MMP2, MMP9 and YAP1 expression in the tumor tissues of WT + Vector, Mettl3+/− + Vector and Mettl3+/− + YAP1 groups were analyzed via IHC. All data were represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
RNA m6A methylation plays an important role in cancer generation and progression. In this study, the analysis of clinical case groups showed that m6A write-in protein METTL3 was significantly related to VM formation and expression of VM-related markers. As such, METTL3-mediated m6A modification may be involved in VM formation. Analysis using VEGFa-induced VM formation model indicated that the m6A modification level increased significantly during VM formation. After interfering with METTL3, the VM formation ability decreased significantly. Transcriptome analysis was carried out in 3D cultured tumor cells. The Hippo pathway plays an important role in m6A-dependent VM formation. YAP1 is an important protein in the Hippo pathway, and many m6A modifications are found in the CDS region of YAP1. The cell function data analysis showed that YAP1 can significantly promote VM formation and rescued the decline by knocking down METTL3. In-depth mechanistic study found that m6A can affect the translation efficiency and expression of the YAP1 protein. The analysis of the orthotopic transplantation tumor model and the DEN-induced HCC model in Mettl3+/− mice showed that m6A can significantly affect VM formation and cancer progression. Thus, m6A or m6A writer METTL3 can be used as an indicator prognosis and a target of anti-metastasis drugs in HCC.
The mechanism of cancer metastasis remains unclear. Considerable evidence indicates that m6A plays an important role in cancer metastasis. Epithelial-mesenchymal transition(EMT), an important theory of tumor metastasis established by oncologist Weinberg [28], is related to m6A. METTL3 could promote EMT by METTL3/ZMYM1/E-cadherin signaling. m6A-induced lncRNA RP11 can trigger the dissemination of CRC cells via post-translational upregulation of Zeb1 [29]. m6A can regulate EMT by influencing translation of Snail [30]. m6A demethylase ALKHB5 can inhibit pancreatic cancer motility by demethylating lncRNA KCNK15-AS1 [31]. In addition, m6A was related to many cancer types, including lung cancer [32], leukemia [33, 34], colon cancer [35, 36], gastric cancer [37], glioma [38], bladder cancer [39], nasopharyngeal cancer [40].
The essence of EMT is a transdifferentiation behavior during cancer progression, and VM formation is also a phenotype of tumor cells that transdifferentiate from epithelial cells to endothelial cells. Ten years ago, we proposed that the essence of VM is the process of epithelial–endothelial transition (EET) in tumor cells. Moreover, Twist1, an important transcription factor that drives EMT, can transcriptionally regulate the expression of VM marker VE-cadherin [41]. The present study is the first to describe the role of m6A in the formation of VM and provides new insights for explaining this process and cancer progression.
Various biochemical and physiological characteristics will be changed by RNA m6A modification. Many research works reported that m6A could influence the stability [22, 42], distribution [43], alternative splicing [44] of RNA. The circRNA modified by m6A could be translated as a protein [45]. m6A also could promote R-LOOP formation to inhibit the transcription [46]. In this study, m6A can affect the half-life of YAP1 mRNA, which may be mediated by m6A reader YTHDF2 and be a conservative mechanism for m6A-mediated mRNA stability. Our results showed that the protein expression of YAP1 was more influenced by translation efficiency.
Hippo pathway plays an important role in tumorigenesis and malignant progression. However, the relationship between Hippo pathway and VM has not been reported. Though we find two research works involving Hippo pathway and VM, these results had no focused on the function of Hippo pathway in VM formation. Ou et al. found that Frizzled 2 (FZD2) could induce EMT and VM. RNA sequencing reveals that the Hippo pathway is enriched following stable silencing of Frizzled 2 (FZD2) in vitro [47]. Wei et al. found that Verteporfin suppresses cell survival, angiogenesis and vasculogenic mimicry of pancreatic ductal adenocarcinoma. Mechanism studies found that YAP transcriptionally regulates Ang2 expression to enhance angiogenesis in vitro [48]. Our results firstly showed that YAP1 promotes VM formation and malignant progression via a m6A-dependent way in vitro and in vivo.
The mechanism of m6A in VM formation is complex. Other m6A-related proteins (including METTL14, FTO, ALKBH5 and the recently discovered large number of m6A readers) may be involved in many important pathways and thus affect VM formation. Further research will be conducted to explain the role of m6A-related mechanisms in VM formation to provide important targets for development of VM-based anti-metastasis drugs.
Change history
30 September 2022
A Correction to this paper has been published: https://doi.org/10.1007/s10456-022-09857-2
Abbreviations
- HCC:
-
Hepatocellular carcinoma
- VM:
-
Vasculogenic mimicry
- m6A:
-
N6-methyladenosine
- VEGFa:
-
Vascular endothelial growth factor a
- YAP1:
-
Yes associated protein 1
- DEN:
-
Diethylnitrosamine
- METTL3:
-
Methyltransferase-like 3
- ALKBH5:
-
AlkB homolog 5
- PAS:
-
Periodic acid–Schiff
- IHC:
-
Immunohistochemistry
- CDH5:
-
Cadherin 5
- MMP2:
-
Matrix metallopeptidase 2
- MMP9:
-
Matrix metallopeptidase 9
- FN1:
-
Fibronectin 1
- MeRIP:
-
Methylated RNA immunoprecipitation
References
Villanueva A (2019) Hepatocellular carcinoma. N Engl J Med 380(15):1450–1462. https://doi.org/10.1056/NEJMra1713263
Vogel A, Saborowski A (2020) Current strategies for the treatment of intermediate and advanced hepatocellular carcinoma. Cancer Treat Rev 82:101946. https://doi.org/10.1016/j.ctrv.2019.101946
Yang W, Li Z, Qin R, Wang X, An H, Wang Y, Zhu Y, Liu Y, Cai S, Chen S, Sun T, Meng J, Yang C (2019) YY1 promotes endothelial cell-dependent tumor angiogenesis in hepatocellular carcinoma by transcriptionally activating VEGFA. Front Oncol 9:1187. https://doi.org/10.3389/fonc.2019.01187
Frentzas S, Simoneau E, Bridgeman VL, Vermeulen PB, Foo S, Kostaras E, Nathan M, Wotherspoon A, Gao Z-H, Shi Y, Van den Eynden G, Daley F, Peckitt C, Tan X, Salman A, Lazaris A, Gazinska P, Berg TJ, Eltahir Z, Ritsma L, Van Rheenen J, Khashper A, Brown G, Nystrom H, Sund M, Van Laere S, Loyer E, Dirix L, Cunningham D, Metrakos P, Reynolds AR (2016) Vessel co-option mediates resistance to anti-angiogenic therapy in liver metastases. Nat Med 22(11):1294–1302. https://doi.org/10.1038/nm.4197
Mahase S, Rattenni RN, Wesseling P, Leenders W, Baldotto C, Jain R, Zagzag D (2017) Hypoxia-mediated mechanisms associated with antiangiogenic treatment resistance in glioblastomas. Am J Pathol 187(5):940–953. https://doi.org/10.1016/j.ajpath.2017.01.010
Wang S, Xiao Z, Hong Z, Jiao H, Zhu S, Zhao Y, Bi J, Qiu J, Zhang D, Yan J, Zhang L, Huang C, Li T, Liang L, Liao W, Ye Y, Ding Y (2018) FOXF1 promotes angiogenesis and accelerates bevacizumab resistance in colorectal cancer by transcriptionally activating VEGFA. Cancer Lett 439:78–90. https://doi.org/10.1016/j.canlet.2018.09.026
Maniotis AJ, Folberg R, Hess A, Seftor EA, Gardner LM, Pe'er J, Trent JM, Meltzer PS, Hendrix MJ (1999) Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am J Pathol 155(3):739–752. https://doi.org/10.1016/S0002-9440(10)65173-5
Delgado-Bellido D, Fernandez-Cortes M, Rodriguez MI, Serrano-Saenz S, Carracedo A, Garcia-Diaz A, Oliver FJ (2019) VE-cadherin promotes vasculogenic mimicry by modulating kaiso-dependent gene expression. Cell Death Differ 26(2):348–361. https://doi.org/10.1038/s41418-018-0125-4
Liang X, Sun R, Zhao X, Zhang Y, Gu Q, Dong X, Zhang D, Sun J, Sun B (2017) Rictor regulates the vasculogenic mimicry of melanoma via the AKT-MMP-2/9 pathway. J Cell Mol Med 21(12):3579–3591. https://doi.org/10.1111/jcmm.13268
Zhang W, Zhou P, Meng A, Zhang R, Zhou Y (2018) Down-regulating Myoferlin inhibits the vasculogenic mimicry of melanoma via decreasing MMP-2 and inducing mesenchymal-to-epithelial transition. J Cell Mol Med 22(3):1743–1754. https://doi.org/10.1111/jcmm.13455
Ayala-Dominguez L, Olmedo-Nieva L, Munoz-Bello JO, Contreras-Paredes A, Manzo-Merino J, Martinez-Ramirez I, Lizano M (2019) Mechanisms of vasculogenic mimicry in ovarian cancer. Front Oncol 9:998. https://doi.org/10.3389/fonc.2019.00998
Wang Y, Tong L, Wang J, Luo J, Tang J, Zhong L, Xiao Q, Niu W, Li J, Zhu J, Chen H, Li X, Wang Y (2019) cRGD-functionalized nanoparticles for combination therapy of anti-endothelium dependent vessels and anti-vasculogenic mimicry to inhibit the proliferation of ovarian cancer. Acta Biomater 94:495–504. https://doi.org/10.1016/j.actbio.2019.06.039
Li W, Zong S, Shi Q, Li H, Xu J, Hou F (2016) Hypoxia-induced vasculogenic mimicry formation in human colorectal cancer cells: involvement of HIF-1a, Claudin-4, and E-cadherin and Vimentin. Sci Rep 6:37534. https://doi.org/10.1038/srep37534
Bedal KB, Grassel S, Spanier G, Reichert TE, Bauer RJ (2015) The NC11 domain of human collagen XVI induces vasculogenic mimicry in oral squamous cell carcinoma cells. Carcinogenesis 36(11):1429–1439. https://doi.org/10.1093/carcin/bgv141
Wang F, Li XK, Xu HY, Shan ZZ, Wang T, Yang ZC, He W, Wang LX, Fan QX (2015) N-cadherin participated in invasion and metastasis of human esophageal squamous cell carcinoma via taking part in the formation of vasculogenic mimicry. Med Oncol 32(2):480. https://doi.org/10.1007/s12032-014-0480-z
Schäfer KP (1982) RNA synthesis and processing reactions in a subcellular system from mouse L cells. Hoppe Seylers Z Physiol Chem 363(1):33–43. https://doi.org/10.1515/bchm2.1982.363.1.33
Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM (2018) MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46(D1):D303–D307. https://doi.org/10.1093/nar/gkx1030
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485(7397):201–206. https://doi.org/10.1038/nature11112
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C (2011) N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7(12):885–887. https://doi.org/10.1038/nchembio.687
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C (2014) A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10(2):93–95. https://doi.org/10.1038/nchembio.1432
Ping X-L, Sun B-F, Wang L, Xiao W, Yang X, Wang W-J, Adhikari S, Shi Y, Lv Y, Chen Y-S, Zhao X, Li A, Yang Y, Dahal U, Lou X-M, Liu X, Huang J, Yuan W-P, Zhu X-F, Cheng T, Zhao Y-L, Wang X, Rendtlew Danielsen JM, Liu F, Yang Y-G (2014) Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 24(2):177–189. https://doi.org/10.1038/cr.2014.3
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C (2014) N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505(7481):117–120. https://doi.org/10.1038/nature12730
Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C (2015) N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161(6):1388–1399. https://doi.org/10.1016/j.cell.2015.05.014
Bai XL, Zhang Q, Ye LY, Liang F, Sun X, Chen Y, Hu QD, Fu QH, Su W, Chen Z, Zhuang ZP, Liang TB (2015) Myocyte enhancer factor 2C regulation of hepatocellular carcinoma via vascular endothelial growth factor and Wnt/β-catenin signaling. Oncogene 34(31):4089–4097. https://doi.org/10.1038/onc.2014.337
Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, Chen Z, Deng X, Xiao G, Auer F, Klemm L, Wu H, Zuo Z, Qin X, Dong Y, Zhou Y, Qin H, Tao S, Du J, Liu J, Lu Z, Yin H, Mesquita A, Yuan CL, Hu YC, Sun W, Su R, Dong L, Shen C, Li C, Qing Y, Jiang X, Wu X, Sun M, Guan JL, Qu L, Wei M, Muschen M, Huang G, He C, Yang J, Chen J (2019) Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 567(7748):414–419. https://doi.org/10.1038/s41586-019-1016-7
Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, Schinzel AC, Sood S, Rosenbluh J, Kim JW, Zwang Y, Roberts TM, Root DE, Jacks T, Hahn WC (2014) KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158(1):171–184. https://doi.org/10.1016/j.cell.2014.06.004
Moon H, Ju HL, Chung SI, Cho KJ, Eun JW, Nam SW, Han KH, Calvisi DF, Ro SW (2017) Transforming growth factor-beta promotes liver tumorigenesis in mice via up-regulation of snail. Gastroenterology 153(5):1378–1391. https://doi.org/10.1053/j.gastro.2017.07.014
Brabletz T, Kalluri R, Nieto MA, Weinberg RA (2018) EMT in cancer. Nat Rev Cancer 18(2):128–134. https://doi.org/10.1038/nrc.2017.118
Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, Zhao G (2019) METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer 18(1):142–142. https://doi.org/10.1186/s12943-019-1065-4
Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, Luo G, Tauler J, Du J, Lin S, He C, Wang H (2019) RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat Commun 10(1):2065–2065. https://doi.org/10.1038/s41467-019-09865-9
He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, Liu D, Tian L, Yin J, Jiang K, Miao Y (2018) ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem 48(2):838–846. https://doi.org/10.1159/000491915
Lin S, Choe J, Du P, Triboulet R, Gregory RI (2016) The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell 62(3):335–345. https://doi.org/10.1016/j.molcel.2016.03.021
Kwok C-T, Marshall AD, Rasko JEJ, Wong JJL (2017) Genetic alterations of m(6)A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol 10(1):39–39. https://doi.org/10.1186/s13045-017-0410-6
Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, Yu M, Skibbe J, Dai Q, Zou D, Wu T, Yu K, Weng H, Huang H, Ferchen K, Qin X, Zhang B, Qi J, Sasaki AT, Plas DR, Bradner JE, Wei M, Marcucci G, Jiang X, Mulloy JC, Jin J, He C, Chen J (2018) R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell 172(1–2):90–105.e123. https://doi.org/10.1016/j.cell.2017.11.031
Li T, Hu P-S, Zuo Z, Lin J-F, Li X, Wu Q-N, Chen Z-H, Zeng Z-L, Wang F, Zheng J, Chen D, Li B, Kang T-B, Xie D, Lin D, Ju H-Q, Xu R-H (2019) METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer 18(1):112–112. https://doi.org/10.1186/s12943-019-1038-7
Meng Y, Li S, Gu D, Xu K, Du M, Zhu L, Chu H, Zhang Z, Wu Y, Fu Z, Wang M (2019) Genetic variants in m6A modification genes are associated with colorectal cancer risk. Carcinogenesis. https://doi.org/10.1093/carcin/bgz165
Zhu L, Zhu Y, Han S, Chen M, Song P, Dai D, Xu W, Jiang T, Feng L, Shin VY, Wang X, Jin H (2019) Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis 10(6):383–383. https://doi.org/10.1038/s41419-019-1585-2
Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K (2018) Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37(4):522–533. https://doi.org/10.1038/onc.2017.351
Cheng M, Sheng L, Gao Q, Xiong Q, Zhang H, Wu M, Liang Y, Zhu F, Zhang Y, Zhang X, Yuan Q, Li Y (2019) The m(6)A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene 38(19):3667–3680. https://doi.org/10.1038/s41388-019-0683-z
Zhang P, He Q, Lei Y, Li Y, Wen X, Hong M, Zhang J, Ren X, Wang Y, Yang X, He Q, Ma J, Liu N (2018) m(6)A-mediated ZNF750 repression facilitates nasopharyngeal carcinoma progression. Cell Death Dis 9(12):1169–1169. https://doi.org/10.1038/s41419-018-1224-3
Sun T, Zhao N, Zhao XL, Gu Q, Zhang SW, Che N, Wang XH, Du J, Liu YX, Sun BC (2010) Expression and functional significance of Twist1 in hepatocellular carcinoma: its role in vasculogenic mimicry. Hepatology 51(2):545–556. https://doi.org/10.1002/hep.23311
Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, Qin X, Tang L, Wang Y, Hong G-M, Huang H, Wang X, Chen P, Gurbuxani S, Arnovitz S, Li Y, Li S, Strong J, Neilly MB, Larson RA, Jiang X, Zhang P, Jin J, He C, Chen J (2017) FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell 31(1):127–141. https://doi.org/10.1016/j.ccell.2016.11.017
Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, Okamura H (2013) RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155(4):793–806. https://doi.org/10.1016/j.cell.2013.10.026
Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF (2015) HNRNPA2B1 Is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162(6):1299–1308. https://doi.org/10.1016/j.cell.2015.08.011
Tatomer DC, Wilusz JE (2017) An unchartered journey for ribosomes: circumnavigating circular RNAs to produce proteins. Mol Cell 66(1):1–2. https://doi.org/10.1016/j.molcel.2017.03.011
Yang X, Liu Q-L, Xu W, Zhang Y-C, Yang Y, Ju L-F, Chen J, Chen Y-S, Li K, Ren J, Sun Q, Yang Y-G (2019) m6A promotes R-loop formation to facilitate transcription termination. Cell Res. https://doi.org/10.1038/s41422-019-0235-7
Ou H, Chen Z, Xiang L, Fang Y, Xu Y, Liu Q, Hu Z, Li X, Huang Y, Yang D (2019) Frizzled 2-induced epithelial-mesenchymal transition correlates with vasculogenic mimicry, stemness, and Hippo signaling in hepatocellular carcinoma. Cancer Sci 110(4):1169–1182. https://doi.org/10.1111/cas.13949
Wei H, Wang F, Wang Y, Li T, Xiu P, Zhong J, Sun X, Li J (2017) Verteporfin suppresses cell survival, angiogenesis and vasculogenic mimicry of pancreatic ductal adenocarcinoma via disrupting the YAP-TEAD complex. Cancer Sci 108(3):478–487. https://doi.org/10.1111/cas.13138
Acknowledgements
We are grateful to Prof. Chuan He, Prof. Jianhua Yang, and Prof. Jianjun Chen for providing us with the omics data of MeRIP. This study was supported by National Science and Technology Major Project (Grant No. 2018ZX09736005), the National Natural Science Foundation of China (Grant Nos. 81872374, 81972629, 81972746, 81703581), the Tianjin Science and Technology Project (Grant No. 19JCJQJC63200), and the Taishan Scholars Program of Shandong Province (Grant No. tsqn201909193)
Author information
Authors and Affiliations
Contributions
KQ: acquisition, analysis and interpretation of data, drafting the manuscript. YL, HZ: acquisition of data, statistical analysis; ZX, ZC: acquisition of data; HZ, CZ: technical support; XL: acquisition of data; ZL: acquisition of data; CL: acquisition of data; YL: clinicopathological analysis, CY: study concept and design; TS: study concept and design, study supervision, analysis of data, drafting the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There are no competing interests.
Ethical approval
HCC tissue microarrays from 75 and 96 cases were purchased from US Biomax for IHC. Each single tissue spot on every array slide was individually examined by certified pathologists according to the WHO published standards for diagnosis, classification, and pathological grading.
Research involving human and animal rights
Animal experiments were conducted in accordance with the National Institutes of Health Animal Use Guidelines. All experimental protocols were approved by the Institutional Animal Care and Use Committee at Tianjin International Joint Academy of Biomedicine.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Qiao, K., Liu, Y., Xu, Z. et al. RNA m6A methylation promotes the formation of vasculogenic mimicry in hepatocellular carcinoma via Hippo pathway. Angiogenesis 24, 83–96 (2021). https://doi.org/10.1007/s10456-020-09744-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10456-020-09744-8