Abstract
Wound healing is a very highly organized process where numerous cell types are tightly regulated to restore injured tissue. Myofibroblasts are cells that produce new extracellular matrix and contract wound edges. We previously reported that the human myofibroblasts isolated from normal wound (WMyos) produced microvesicles (MVs) in the presence of the serum. In this study, MVs were further characterized using a proteomic strategy and potential functions of the MVs were determined. MV proteins isolated from six WMyo populations were separated using two-dimensional differential gel electrophoresis. Highly conserved spots were selected and analyzed using mass spectrometry resulting in the identification of 381 different human proteins. Using the DAVID database, clusters of proteins involved in cell motion, apoptosis and adhesion, but also in extracellular matrix production (21 proteins, enrichment score: 3.32) and in blood vessel development/angiogenesis (19 proteins, enrichment score: 2.66) were identified. Another analysis using the functional enrichment analysis tool FunRich was consistent with these results. While the action of the myofibroblasts on extracellular matrix formation is well known, their angiogenic potential is less studied. To further characterize the angiogenic activity of the MVs, they were added to cultured microvascular endothelial cells to evaluate their influence on cell growth and migration using scratch test and capillary-like structure formation in Matrigel®. The addition of a MV-enriched preparation significantly increased endothelial cell growth, migration and capillary formation compared with controls. The release of microvesicles by the wound myofibroblasts brings new perspectives to the field of communication between cells during the normal healing process.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Virtually all cell types release extracellular vesicles (EVs), a common term for both microvesicles (MVs; diameter 100–1000 nm) and exosomes (diameter 30–100 nm). Exosomes are derived from endosomes, whereas MVs are shed from the cell membrane [1]. EVs are considered an important intercellular communication mechanism participating in cell-to-cell communication transferring proteins, lipids and nucleic acids from one cell to another [2].
EVs play significant role in normal and pathological processes such as vascular dysfunction [3], inflammation [4], cancer [5] and fibrosis [6]. Their increased detection in blood in numerous pathologies strengthens the idea that EVs can play a role in disease development. For instance, tumor cells shed EVs that stimulate cancer cell invasion and support angiogenesis to enhance cell exchanges and growth [5, 7] and portal myofibroblasts EVs promote vascular remodeling in cirrhosis [8]. EVs from stem cells but also from blood cells in inflammatory response may act on target cells and modify the response to injury [9].
Even if EVs can be detected in healthy subjects, their role in physiological mechanisms such as wound healing remains poorly studied [1, 10] although an increasing body of evidence indicates that they are capable of mediating tissue repair in models of acute kidney and liver injury [9, 11].
Wound healing is a complex physiological process that involves numerous cell types. Its mechanism is strictly organized via cell interactions [12]. During the formation of new tissue, fibroblasts and myofibroblasts play a crucial role, secreting and remodeling the extracellular matrix [13] while endothelial cells support tissue repair via angiogenesis. Myofibroblasts, which are associated with wound healing, secrete a large quantity of extracellular matrix and have the capability of contracting the wound edges [13]. We have previously demonstrated that myofibroblasts, when cultured with serum or plasma, produce MVs that can stimulate mesenchymal growth [14], a key step during healing to enhance the number of cells producing new matrix. In the absence of serum or plasma, MV production by these cells is very modest in contrast to other cells that need stress such as serum starvation to produce MVs [8, 15, 16]. As MVs have been implicated in many mechanisms as immune modulation, metastasis, cell–stroma interactions and angiogenesis [2, 9, 17, 18], we have further studied the possible action of MVs during wound healing. Our hypothesis was that myofibroblasts can play a central role during healing and that the orchestration of these effects uses MVs. In the present study, we used a proteomic approach to identify the proteins of the MVs produced by the wound healing myofibroblasts and we study their effect on wound healing-associated mechanisms. We show that myofibroblastic MV-enriched preparation promotes angiogenesis by stimulating endothelial cell growth, migration and tube formation, which are crucial steps for tissue repair.
Materials and methods
Cell culture
All procedures involving patients were reviewed and approved by the Research Ethical Committee of the Centre Hospitalier Universitaire de Québec and followed the Declaration of Helsinki protocols. The cells were grown at 37 °C in a humidified incubator with 8% CO2.
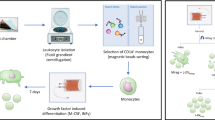
Populations of primary cells were isolated from normal skin and wounds of volunteer donors aged 20–40 years. Normal wound myofibroblasts called WMyos were isolated from experimental granulation tissues made with implants (polyvinyl alcohol sponge inserted into perforated silicone tubes) inserted subcutaneously into the arms of volunteers [19, 20]. Numbers given after each cell population refer to the individuals from which the cells were isolated. The cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 20% fetal bovine serum (FBS; Hyclone, Flow Lab, Mississauga, ON, Canada), 100U/ml penicillin G and 25 mg/ml gentamicin (Schering Inc., Pointe Claire, Canada). Confluent cell cultures between passages 3 and 6 were used in this study.
Human microvascular endothelial cells (HMECs) were isolated from skin biopsies obtained during myofibroblast implant surgery [14]. HMECS between passages 3 and 6 were cultured with EGM™-2MV (EBM-2 with additives and 5% FBS) (Lonza Clonetics, San Diego, CA, USA).
Microvesicle- and exosome-enriched preparations
Confluent WMyos were cultured during 48 h with DMEM + 20% MV-free FBS (achieved by ultracentrifugation at 100,000×g for 18 h at 4 °C). Conditioned media were collected and centrifuged at 300×g for 10 min at 4 °C to remove cells and large debris. The supernatants were centrifuged at 21,000×g for 30 min at 4 °C. Pelleted MVs were washed with phosphate-buffered saline (PBS) and further centrifuged at 21,000×g, 20 min three times. Each supernatant containing exosomes was separately further centrifuged at 100,000×g for 18 h at 4 °C, pelleted and washed with PBS three times. The size distribution of the two subgroups overlapping, differential centrifugation processing only allows obtaining enriched MV and exosome-enriched preparations but not pure preparations. The protein concentration of the MV- and exosome-enriched samples was measured using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Mississauga, ON, Canada) at 280 nm. MVs and exosomes were separately isolated twice from two different populations of WMyos. Each protein evaluation has been performed in triplicate. A similar volume of non-conditioned medium was prepared simultaneously as a control (named con-MV or con-exo).
2D-DIGE experimental design and sample labeling
A total of six independent MVs preparations coming from six different WMyo populations were used for 2D-DIGE analyses. After isolation, one aliquot of each MV preparation was immediately labeled with Cydye to identify extravesicular proteins. Other aliquots were quickly frozen in liquid nitrogen and stored at −80 °C during 10 days before being resuspended in lysis buffer (7 M urea, 2 M thiourea, 30 mM Tris, 4% CHAPS) and processed for total protein labeling. The vesicles were quantified by measuring vesicle-associated proteins using the Bradford method (Bio-Rad, Mississauga, ON, Canada) with bovine serum albumin as the standard.
For each cell population, the extravesicular-labeled proteins and the total labeled proteins from the same MVs were co-separated with the control sample (pool sample) on an individual gel (i.e., six gels in total). Total labeled proteins from the non-conditioned medium sample (con-MV) were co-separated with the pool sample on an individual gel. The protein samples were minimally labeled with fluorescent cyanine dyes for 2D-DIGE analyses, according to the manufacturer’s instructions (Lumiprobe, Hallandale Beach, FL). Briefly, Cy3 and Cy5 (400 pmol each) were used to label 50 µg of the total protein sample and its extravesicular counterpart, respectively. To eliminate any dye-labeling bias, labeling of proteins was randomly performed using Cy3 and Cy5. An internal standard was generated by pooling 50 µg of the total protein extract from the six independent MV preparations (pool sample) and labeling with Cydye 2 (2400 pmol).
2D-DIGE
For isoelectric focusing (IEF), the same quantity (30 µg) of extravesicular-labeled proteins, total labeled proteins and labeled proteins from the pool sample were mixed to obtain 90 µl at 1 µg/µl. The same volume of sample buffer containing 7 M urea, 2 M thiourea, 4% CHAPS, 1%(v/v) IPG buffer pH 3–11 NL, 5 mM 1,4-dithiothreitol (DTT) and 0.001% bromophenol blue was added to each dye-labeled sample mix. After centrifugation, the supernatant volume was completed with 270 µl of rehydration buffer (7 M urea, 2 M thiourea, 4%(w/v) CHAPS, 2.6% DeStreack™ (GE Healthcare Life Science, Mississauga, ON, Canada), 0.5% (v/v) IPG buffer pH 3–11 NL, 0.0005%(w/v) bromophenol blue). IPG strips (24 cm, pH 3–11 NL) (GE Healthcare Life Science) were rehydrated overnight with these diluted dye-labeled samples mixes. Proteins were separated by IEF for a total of 40 000 Vh according to the manufacturer’s specification for these strips using an Ettan IPGphor3 apparatus (GE Healthcare Life Science). Prior to the second-dimension SDS-PAGE, each strip gel was equilibrated in 20 ml equilibration buffer (6 M urea, 75 mM Tris–HCl pH 8.8, 30% glycerol, 2% SDS, 1% (w/v) DTT) for 20 min followed by a second step equilibration in the same buffer except that DTT was replaced by 4.5% (w/v) iodoacetamide. IPG strips were then placed on 10–18% gradient polyacrylamide gels and cast in low-fluorescent glass plates using an Ettan-DALT caster (GE Healthcare Life Science). The second-dimension electrophoresis was performed at 0.5 W/gel for 3h20 followed by 1 W/gel overnight at 21 °C. The migration was stopped once the bromophenol blue dye front reached the bottom of the gel.
After analysis, a new 2D-DIGE gel prepared with MVs isolated from the WMyo12 population (also containing the pool sample) was used to excise protein spots for LC–MS/MS identification. Following electrophoresis and gel imaging, the gel was fixed with 30% methanol/10% acetic acid for 30 min and then washed (three times) for 10 min with deionized H2O prior to protein visualization using blue–silver staining [21].
2D-DIGE analyses
Gel imaging
Gels containing Cydye-labeled proteins were visualized by scanning on a Typhoon Trio + variable mode imager (GE Healthcare Life Science). Characteristic excitation/emission wavelengths of 488/520, 532/580 and 633/670 nm were used to scan for Cy2, Cy3 and Cy5 components, respectively, resulting in the generation of 20 images in total (three per gel for the MV samples and two from the control gel).
Analysis and spot selection criteria
Analyses were only performed on the total labeled protein samples. Spot map image files were analyzed in batch processor mode using Delta 2D software version 4.2 (Decodon GmbH, Greifswald, Germany). The resulting spot pattern was assigned to each of the three individual 2D images (Cy2, Cy3, Cy5) of each gel in the experiment. For further analysis, uniform spot quantification on all gels and internal normalization was conducted. The spot information of the Cy5/Cy2 and Cy3/Cy2 ratios was used to calculate an average volume ratio for corresponding spots from each gel. The results obtained with the control gel (con-MV) were subtracted from those for other gels, eliminating any spots resulting from FBS. Based on the average spot volume ratio, spots whose relative expression was constant for all the MV samples (coefficient of variation <50%) and/or the spots with higher normalized volume were selected for further analysis.
Extract of MV proteins
MV samples were suspended in PBS, and then, proteins were precipitated by adding ice-cold acetone to a final concentration of 90% (v/v), kept 1 h at −20 °C and recovered by centrifugation at 21,000×g for 10 min at 4 °C. The dried pellets were analyzed by mass spectrometry.
Disc SDS-PAGE isolation
Predominant and high molecular weight proteins can suppress ionization and hinder the LC–MS detection of low-abundance/low molecular weight peptides. One other sample of MVs was separated using an improved Disc SDS-PAGE according to Li et al. [22]. This allows the elimination of high molecular weight proteins (>30 kDa) from the sample. Briefly, three polyacrylamide gels [0.5 cm resolving gel (16.5% T, 6% C), 0.7 cm spacer gel (10% T, 3% C) and 0.5 cm stacking gel (4% T, 3% C)] were sequentially poured into a glass tube that was placed between anode and cathode cells. Sample (270 µg) was added to the stacking gel and allowed to migrate for 1 h 30 at 60 V. The resolving gel (containing low molecular weight proteins) was then fixed in 30% ethanol/10% acetic acid and analyzed by mass spectrometry.
Protein identification using mass spectrometry
Selected samples were analyzed using the Proteomics facility of the Eastern Quebec Genomics Center (Quebec, Canada). Tryptic digestion was performed on a MassPrep liquid handling robot (Waters, Milford, MA). Peptide samples were then separated by online reversed-phase nanoscale capillary liquid chromatography and analyzed by electrospray mass spectrometry (ES MS/MS). All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.4.1) and X! Tandem [The GPM, thegpm.org; version CYCLONE (2010.12.01.1)]. Mascot was set up to search the UR100_12_05_Bos_taurus_9913 database (160131 entries) assuming the digestion enzyme trypsin. X! Tandem was set up to search the UR100_12_05_Homo_sapiens_9606 database (128347 entries) also assuming trypsin. Mascot and X! Tandem were searched with a fragment ion mass tolerance of 0.100 Da for Disc SDS-PAGE sample and 0.50 Da for other samples. Carbamidomethyl of cysteine was specified in Mascot and X! Tandem as a fixed modification. Dehydration of the N-terminus, Glu → pyro-Glu of the N-terminus, ammonia loss of the N-terminus, Gln → pyro-Glu of the N-terminus, deamidation of asparagine and glutamine and oxidation of methionine were specified in X! Tandem as variable modifications.
Scaffold (version Scaffold-3.6.3, Scaffold Proteome Software, RRID:SCR_014345) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm [23, 24] and contained at least two identified peptides. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Data analysis
DAVID 6.7 (Database for Annotation Visualization and Integrated Discovery, RRID:SCR_003033; http://david.abcc.ncifcrf.gov/) and FunRich (a Functional Enrichment analysis tool, http://www.funrich.org/, RRID:SCR_014467) were used to perform analysis.
To analyze the biological functions of MV proteins, a functional annotation clustering tool from the Web-based DAVID database [25] was used. For any protein list, DAVID tools are able to identify enriched biological terms, particularly GO terms, discover enriched functional-related protein groups, visualize proteins on BioCarta and KEGG pathway maps, etc. A biological process or pathway was considered to be significantly enriched in MV proteins if it contained a minimum of three proteins per category featuring Fisher’s exact p value less than 0.05. Human proteome was used as the background data set.
Functional enrichment analysis of the proteins identified in MVs was also performed using FunRich, (http://www.funrich.org), which integrates heterogeneous genomic and proteomic resources (>1.5 million annotations). Enrichment analysis was performed for GO terms, biological pathways, protein domains, site of expression, transcription factors and disease terms using the FunRich database as the background dataset [26]. The statistical significance of the enrichment was assessed by the corrected p value (Benjamini–Hochberg method).
HMEC growth assay
HMECs (3 × 104 cells per well) were seeded in a gelatin-coated 24-well plate and cultured in EGM™-2MV for 24 h. After 24 h, four wells were trypsinized to count the exact number of HMECs in each well. The medium was then changed to EBM-2 basal medium supplemented with 10% EGM™-2MV (basal medium with minimum factors to keep cells alive but not proliferative) with MVs or controls (positive control: basal medium with 5% FBS that stimulate cell growth, negative control: basal medium only) and kept for 6 days with medium change every 2 days. Cells were then trypsinized and counted using a Coulter® counter and expressed as a ratio to the number of cells at J0. MV samples were tested on two different HMEC populations.
Cell migration assay
HMECs were seeded in a gelatin-coated 12-well plate and cultured in EGM™-2MV until confluence. The medium was then changed to EBM-2 basal medium supplemented with 10% EGM™-2MV and 0.01 mg/ml mitomycin C (MMC) per well (Bio Basic, Markham, ON, Canada). Three hours later, the medium was changed to EBM-2 basal medium supplemented with 10% EGM™-2MV with or without MVs at the indicated concentrations and a scratch test was performed using a 200-µl tip. Phase contrast images (magnification of 4×) were recorded on a digital camera after scratch was performed (T0) and after 24 h of incubation (T24). The cell migration was measured using ImageJ software and expressed as the difference in the wound closure between T0 and T24. MVs samples were tested on two different HMEC populations.
Tube formation assay
HMECs (5 × 104 cells per well) were seeded onto Matrigel®-reduced growth factors (Corning, Bedford, MA, USA)-coated wells in a 24-well plate and cultured in EBM-2 basal medium supplemented with 10% EGM™-2MV in the presence of controls or MVs at the indicated concentrations. Phase contrast images (magnification of 4×) were recorded on a digital camera after incubation for 24 h. The number and total length of the tube structures were measured using ImageJ software and the results expressed as the length and numbers of the network structure after 24 h. MV samples were tested on two different HMEC populations.
Myofibroblast transduction
pLenti6.3-DsRed was made from the ligation of a DsRed amplicon (pDsRed-Express-N1, Clontech) into pLenti6.3V5-TOPO vector (Invitrogen). Two WMyo populations were transduced with lentiviral vectors which have been produced with the previously assembled plasmid and the protocol described in [27]. Fluorescent WMyos were then cultured during 48 h with DMEM + 20% MV-free FBS, and MVs were isolated as described above. Fluorescence of MVs was evaluated using flow cytometry (FACScan Becton Dickenson) and compared to MVs isolated from WMyo. Two HMEC populations at 50% confluency were treated in duplicate with two preparations of MVs isolated from transduced and not transduced WMyo. Quantification of cell fluorescence was performed using flow cytometry after 24 h.
Statistical analysis
Statistical differences were computed by analysis of variance followed by the one-way ANOVA test [28]. The results were considered significant when the p value was <0.05. All data presented are the means (errors bars: ± standard deviation). Difference between two curves obtained following flow cytometry evaluation has been evaluated using Kolmogorov–Smirnov statistics.
Results
Microvesicle and exosome production by WMyos in the presence of serum
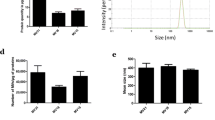
Both MVs production and exosome production have being quantified using differential centrifugations followed by the quantification of proteins. As previously shown, MVs were only detected when WMyos were cultured with serum. In contrast, exosome production was undetectable in the presence or absence of serum (Fig. 1).
Serum stimulated the release of MVs by WMyos but not the release of exosomes. Two different populations of WMyos were incubated for 48 h with or without 20% serum (FBS). Using differential centrifugation, MV (left) and exosome (right) production was then quantified using spectrophotometric measurement of protein content
Protein composition of MVs
To determine the protein composition of MVs released by WMyos, the 2D-DIGE method was used to compare proteins between six different MV preparations. A total of 122 protein spots whose relative expression was constant upon all the MV samples (coefficient of variation <50%) or/and the spots whose relative expression level was the highest were analyzed using mass spectrometry, which allowed 251 human proteins to be identified. This method was focused on the selection of spots according to specific criteria. To be sure that all important proteins were detected, MV proteins from the general extract (total protein extraction) or from the MV sample where high molecular weight proteins (Disc SDS-PAGE isolation) were discarded were also analyzed. Mass spectrometry analysis of the total proteins extraction sample detected 179 human proteins while analysis of the Disc SDS-PAGE sample detected 105 human proteins. A Venn diagram (Fig. 2) showed the repartition of the detected protein numbers according to each method; the 2D-DIGE method was the most powerful one as 64.0% of all the detected proteins were detected with this method. The use of a total protein extract allowed the detection of 45.7% of proteins, whereas in the low molecular weight protein extract 26.8% of the proteins were detected. In this sample, the majority of detected proteins had a molecular weight inferior to 30 kDa. Altogether, 392 human proteins were detected in MV samples (Table 1S).
Functional analyses of MV proteins
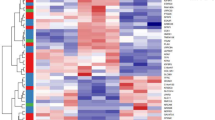
These analyses of MV proteins were performed using two complementary bioinformatic tools, DAVID, which was used here to group MV proteins into functional clusters, and FunRich, which performed enrichment analysis based on a higher number of annotations collected from various databases. A functional analysis of biological pathways was performed on the full list of MV proteins using the DAVID clustering tool. Consequently, 65 clusters with enrichment scores >1.5 related to cellular and biological processes were identified. The cluster with the highest score (enrichment score 22.09) was enriched in proteins of membrane and cytoplasm vesicles (Table 1). MV proteins formed further clusters involved in cell motility (enrichment score 4.26), programmed cell death (apoptosis, enrichment score 4.03), extracellular matrix organization (enrichment score 3.32), cell junction and adhesion (enrichment score 2.95), and blood vessel development and angiogenesis (enrichment score 2.66) (Table 1). The other clusters mainly comprised MV proteins involved in cytoskeletal and cell maintenance pathways (Table 2S). It should be noted that the detected MV proteins did not form clusters involved in inflammation, hemostasis and coagulation, which have usually been reported to involve MVs [18, 29].
Functional enrichment analysis of the GO term “Cellular component” using FunRich showed that the proteins identified in MVs were strongly and significantly enriched in proteins found in exosomes (74% of the detected proteins, p = 8 × 10−143), cytoplasm (72%, p = 10−34), lysosome (56.4%, p = 1.6 × 10−93) and nucleus (49.7%, p = 9 × 10−4) (Table 2). This reflects the process of MV formation: MVs bud from cell membranes, trapping cytosolic proteins. It might also be due to the fact that some proteins exist in different locations. The analysis based on the GO term “Biological Process” showed that MV proteins were significantly enriched in the following annotations “protein metabolism” (21%, p = 5.4 × 10−16) and “cell growth and/or maintenance” (20%, p = 8.5 × 10−18). Regarding the biological pathways (Table 3), MVs were significantly enriched in proteins involved in “metabolism” (30%, p = 9.3 × 10−11), “integrin family cell surface interactions” (33%, p = 0.002), specifically in “beta1 integrin cell surface interactions” (32%, p = 0.03), in “proteoglycan syndecan-mediated signaling events” (31%, p = 0.01), particularly syndecan-1, and in “signaling events mediated by VEGFR1 and VEGFR2 involved in angiogenesis (29%, p = 0.02)” and several growth factors and their receptors (e.g., endothelin, EGFR, GMCSF, IGF1, PDGF/PDGFR) (28–30%, p < 0.035) (Table 3).
Angiogenic properties of MVs
Fluorescent MVs were added in culture medium of HMEC to determine the capacity of HMECs to uptake MVs. MVs isolated from DsRed-transduced WMyo contained the fluorescent marker expressed in the WMyo cytoplasm (Fig. 3a, b). When added in the HMEC medium, the cells became statistically fluorescent (Fig. 3c, d).
MVs produced from DsRed-transduced WMyo contained DsRed and transferred fluorescent staining to HMEC when added on their culture medium. a, b Flow cytometry analysis of MVs produced by WMyo 18 (a) or DsRed-transduced WMyo18 (b); c, d flow cytometry analysis of HMEC stimulated by MVs produced by WMyo18 (light line) or DsRed-transduced WMyo18 (black line) (c) or WMyo21 (d) *p < 0.001
To characterize the angiogenic capacity of MVs, microvascular endothelial cells were cultured and evaluation of cell growth, migration capacity using a scratch test and capillary-like structure formation in Matrigel® in the presence of the MVs was performed. For each experiment, MVs from two different WMyo populations were isolated and tested on two HMEC populations. Representative results obtained with each MV sample were presented.
To demonstrate the potential role of MVs on HMEC growth, HMECs were treated for 6 days with two MV concentrations. Figure 4 shows that MVs significantly increased the number of cells when 5 and 10 mg/ml of total MV proteins were used, suggesting that MVs can stimulate the endothelial cell growth during wound healing. Because of the low dose of growth factors in medium, cells stimulated with non-conditioned medium preparation (con-MV) did not show any cell growth (Fig. 4) and the doubling time of HMECs was very high (648 h). However, stimulation with the higher dose of MVs induced a similar or higher growth than EBM2 supplemented with 5% FBS (Con+) and a shorter doubling time of cells (134 h for 10 µg/ml MVs and 130 h for Con+).
MV addition in culture medium stimulated endothelial cell growth. HMEC growth after a 6-day treatment with two concentrations of MVs isolated from two different WMyo-conditioned media [WMyo18 (left) and WMyo21 (right)]; EBM2 with 5% FBS (Con+) or non-conditioned medium preparation (con-MV) (*p < 0.05; **p < 0,01; ***p < 0.001 vs. con-MV)
HMEC migration was evaluated using a scratch test. When MVs were added, the migration rate of HMECs was significantly increased compared with the control (con-MV) (Fig. 5)
HMEC migration rate was increased in the presence of MVs. Photographs allowing to evaluate HMEC migration: a, c after the scratch, b, d after 24 h in the presence of (a, b) medium only or (c, d) 10 µg/ml of MVs derived from WMyo18; e, f HMEC migration rate evaluation after 24 h-treatment with two concentrations of MVs isolated from two different WMyo-conditioned media [WMyo18 (e) and WMyo21 (f)]; EBM2 with 5% FBS (Con+) or non-conditioned medium preparation (con-MV) (*p < 0.05; **p < 0,01; ***p < 0.001 vs. con-MV)
The capability of MVs to stimulate capillary-like tube formation in Matrigel® was evaluated. Both the number and length of capillary-like tubes formed by HMECs were increased in the presence of MVs. The length of the capillary-like structures was similar in the presence of the highest concentration of MVs (10 µg/ml) and in the presence of EBM2 supplemented with 5% FBS (Con+). However, MVs stimulated the number of tubes more than Con+ (Fig. 6).
MVs stimulated capillary-like formation by HMECs in Matrigel®-reduced growth factors. Photographs allowing to evaluate tube formation on Matrigel®: comparison between HMEC with con-MV (a) and HMEC with 10 µg/ml of MVs (b). Bar 0.55 mm. Number (c, d) and length (e, f) of capillary-like structures after 24-h treatment with two concentrations of MVs isolated from two different WMyo-conditioned media [WMyo18 (c–e) and WMyo21 (d–f)]; EBM2 supplemented with 5% FBS (Con+) or non-conditioned medium preparation (con-MV) (*p < 0.05; **p < 0,01; ***p < 0.001 vs. con-MV)
Discussion
Myofibroblasts are cells that are detected during physiological healing [30] or pathological states such as cancer [31] or fibrosis [32]. During normal healing, they play a crucial role in extracellular matrix formation and wound contraction [33]. During the first steps of healing, serum proteins are highly present following capillary lesions and the outflow of blood into the wound. Furthermore, during healing, capillaries are permeable and allow the outflow of many plasma proteins. It has been recently demonstrated that, besides extracellular matrix proteins and cytokines, WMyos can produce MVs when activated by serum or plasma [14]. The fact that these MVs stimulated the growth of these mesenchymal cells [14] suggests that MVs can play a role during healing. We determined the protein composition of MVs using a proteomic approach and then determined the biological processes and pathways these MV proteins are involved in by performing enrichment analysis with the bioinformatic tools DAVID® and FunRich® in order to obtain new clues on the processes regulated by MVs.
In the literature, proteomic approaches to identify components of MVs usually used mass spectrometric analysis of tryptic peptides with or without a previous separation of the samples using one-dimensional polyacrylamide gel electrophoresis (SDS-PAGE). However, the presence of highly abundant proteins in the samples can impede the detection of proteins with low abundance. The 2D-DIGE proteomic techniques are advantageous because of their facility to detect robust quantitative differences/similarities between protein spots. Furthermore, orthogonal techniques for the separation of proteins result in a higher separation capacity than SDS-PAGE [34]. Finally, each spot consists of one of several protein species that can be digested in a low-complexity mixture for LC–MS analysis. Our results showed that this method allowed detection of 251 human proteins that were steadily present in MVs. The two other methods used (Disc SDS-PAGE and no treatment) detected 141 further proteins, whereas 152 proteins were detected only by the 2D-DIGE method.
In silico enrichment analysis of MV proteins using both DAVID® and FunRich® databases showed that detected MV proteins are, in the literature, associated with the MV compartment (integrins, lactadherin, annexins, Rabs, mitochondrial proteins) but not with the exosomes (absence of Alix or CD81) [35]. The classification of the proteins likewise determined that they can be linked to signaling pathways triggered by extracellular matrix proteins and mediated by integrins, growth factor receptors and proteoglycans and to extracellular matrix organization. These results reflect the known functions of myofibroblasts in extracellular matrix formation and remodeling during healing [33]. The other cluster that MV proteins have been linked to is cell motility. During healing, cells need to migrate into granulation tissue, the temporary tissue, to repopulate it. Fibroblasts and myofibroblasts then reconstitute the extracellular matrix, whereas endothelial cells need to migrate to reconstitute a dense capillary network.
The DAVID analysis identified that MV proteins can be associated with the angiogenic cluster. This finding is in agreement with the enrichment analysis performed with FunRich, which showed that ~30% of MV proteins are members of the pro-angiogenic VEGF/VEGFR, EGFR and PDGFR pathways and with our previous study [14], which detected VEGF and FGF2 in MVs. Furthermore, we show here that MVs produced by serum-activated wound myofibroblasts can be incorporated into endothelial cells and significantly stimulate microvascular endothelial cells growth, migration rate and capillary-like structure formation, demonstrating that MVs secreted by myofibroblasts can promote angiogenesis.
EVs produced by platelets [36], endothelial [8], mesenchymal [15, 16] or cancer cells [5] have been shown to stimulate angiogenesis. However, these EVs are usually exosomes, which have been isolated by ultracentrifugation and contain markers such as Alix or CD81. Furthermore, they are usually obtained after serum starvation [8, 15, 16]. By contrast, in our experimental conditions, myofibroblasts only produced MVs when they were stimulated by serum [14] while exosome production was very low. It has to be noted, however, that differential centrifugation processing results in enriched MV and enriched exosome preparations but not in pure preparations, the size distribution of both groups overlapping. However, the lack of detection of exosome markers such as Alix protein in MV-enriched preparations supports the fact that the MV-enriched preparation contains very few exosomes. Exosome production by normal mesenchymal cells (NIH3T3) has been detected in the presence of serum, but the number of cells required to detect such production was around 50 times higher than in our experiments [37].
The action of WMyos [27] as well as of MVs [38] from other cells on angiogenesis has been documented in a variety of cell types. MVs may fully or partially fuse with the target cell, allowing for a complete or selective transfer of contents, including membrane and cytosolic proteins, bioactive lipids or even whole cell organelles [2]. The action on endothelial cells has been shown to be linked to several proteins such as endoglin [8], MMP-2 [15], VEGF [6] or PDGF [39] or RNA transfer (miR-150 [40], and miR-31 [16]). Several of these proteins have been detected in MVs produced by WMyos and can be valuable molecules to account for the angiogenic activity of MVs. However, we have identified in this study both experimentally and in silico other pro-angiogenic proteins and receptors, which might be linked with the molecular mechanisms underlying the pro-angiogenic effect of MVs.
Conclusion
This study demonstrates that MVs produced by serum-stimulated myofibroblasts contain proteins contributing to extracellular matrix organization, cell motility, cell junction and adhesion and that promote angiogenesis, probably through several growth factor regulated pathways. MV release is thus an important step in myofibroblast activity during wound healing and is involved in the finely orchestrated mechanism of skin repair.
References
Freyssinet JM (2003) Cellular microparticles: what are they bad or good for? J Thromb Haemost 1(7):1655–1662
Mause SF, Weber C (2010) Microparticles: protagonists of a novel communication network for intercellular information exchange. Circul Res 107(9):1047–1057. doi:10.1161/CIRCRESAHA.110.226456
Agouni A, Lagrue-Lak-Hal AH, Ducluzeau PH, Mostefai HA, Draunet-Busson C, Leftheriotis G, Heymes C, Martinez MC, Andriantsitohaina R (2008) Endothelial dysfunction caused by circulating microparticles from patients with metabolic syndrome. Am J Pathol 173(4):1210–1219. doi:10.2353/ajpath.2008.080228
Boilard E, Nigrovic PA, Larabee K, Watts GF, Coblyn JS, Weinblatt ME, Massarotti EM, Remold-O’Donnell E, Farndale RW, Ware J, Lee DM (2010) Platelets amplify inflammation in arthritis via collagen-dependent microparticle production. Science 327(5965):580–583. doi:10.1126/science.1181928
Munster M, Fremder E, Miller V, Ben-Tsedek N, Davidi S, Scherer SJ, Shaked Y (2014) Anti-VEGF-A affects the angiogenic properties of tumor-derived microparticles. PLoS ONE 9(4):e95983. doi:10.1371/journal.pone.0095983
Lemoinne S, Cadoret A, Rautou PE, El Mourabit H, Ratziu V, Corpechot C, Rey C, Bosselut N, Barbu V, Wendum D, Feldmann G, Boulanger C, Henegar C, Housset C, Thabut D (2015) Portal myofibroblasts promote vascular remodeling underlying cirrhosis formation through the release of microparticles. Hepatology 61(3):1041–1055. doi:10.1002/hep.27318
Mao G, Liu Y, Fang X, Liu Y, Fang L, Lin L, Liu X, Wang N (2015) Tumor-derived microRNA-494 promotes angiogenesis in non-small cell lung cancer. Angiogenesis 18(3):373–382. doi:10.1007/s10456-015-9474-5
Belik D, Tsang H, Wharton J, Howard L, Bernabeu C, Wojciak-Stothard B (2016) Endothelium-derived microparticles from chronically thromboembolic pulmonary hypertensive patients facilitate endothelial angiogenesis. J Biomed Sci 23:4. doi:10.1186/s12929-016-0224-9
Tetta C, Bruno S, Fonsato V, Deregibus MC, Camussi G (2011) The role of microvesicles in tissue repair. Organogenesis 7(2):105–115. doi:10.4161/org.7.2.15782
Bastos-Amador P, Royo F, Gonzalez E, Conde-Vancells J, Palomo-Diez L, Borras FE, Falcon-Perez JM (2012) Proteomic analysis of microvesicles from plasma of healthy donors reveals high individual variability. J Proteom 75(12):3574–3584. doi:10.1016/j.jprot.2012.03.054
Bruno S, Grange C, Deregibus MC, Calogero RA, Saviozzi S, Collino F, Morando L, Busca A, Falda M, Bussolati B, Tetta C, Camussi G (2009) Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J Am Soc Nephrol 20(5):1053–1067. doi:10.1681/ASN.2008070798
Moulin V (1995) Growth factors in skin wound healing. Eur J Cell Biol 68:1–7
Moulin V, Castilloux G, Auger FA, Garrel D, O’Connor-McCourt M, Germain L (1998) Modulated response to cytokines of human wound healing myofibroblasts compared to dermal fibroblasts. Exp Cell Res 238:283–293
Moulin VJ, Mayrand D, Messier H, Martinez MC, Lopez-Valle CA, Genest H (2010) Shedding of microparticles by myofibroblasts as mediator of cellular cross-talk during normal wound healing. J Cell Physiol 225:734–740
Beltramo E, Lopatina T, Berrone E, Mazzeo A, Iavello A, Camussi G, Porta M (2014) Extracellular vesicles derived from mesenchymal stem cells induce features of diabetic retinopathy in vitro. Acta Diabetol 51(6):1055–1064. doi:10.1007/s00592-014-0672-1
Kang T, Jones TM, Naddell C, Bacanamwo M, Calvert JW, Thompson WE, Bond VC, Chen YE, Liu D (2016) Adipose-derived stem cells induce angiogenesis via microvesicle transport of miRNA-31. Stem Cells Transl Med 5(4):440–450. doi:10.5966/sctm.2015-0177
Castellana D, Toti F, Freyssinet JM (2010) Membrane microvesicles: macromessengers in cancer disease and progression. Thromb Res 125(Suppl 2):S84–S88. doi:10.1016/S0049-3848(10)70021-9
Distler JH, Huber LC, Gay S, Distler O, Pisetsky DS (2006) Microparticles as mediators of cellular cross-talk in inflammatory disease. Autoimmunity 39(8):683–690. doi:10.1080/08916930601061538
Moulin V, Garrel D, Auger FA, O’Connor-McCourt M, Castilloux G, Germain L (1999) What’s new in human wound healing myofibroblasts? Curr Top Pathol 93:123–133
Germain L, Jean A, Auger F, Garrel DR (1994) Human wound healing fibroblasts have greater contractile properties than dermal fibroblasts. J Surg Res 57:268–273
Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25(9):1327–1333. doi:10.1002/elps.200305844
Li T, Dai S, Wang Z, Zhang H (2010) Improved disc SDS-PAGE for extraction of low molecular weight proteins from serum. Electrophoresis 31(6):1090–1096. doi:10.1002/elps.200900423
Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74(20):5383–5392
Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75(17):4646–4658
da Huang W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57. doi:10.1038/nprot.2008.211
Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim A, Bacic A, Hill AF, Stroud DA, Ryan MT, Agbinya JI, Mariadason JM, Burgess AW, Mathivanan S (2015) FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics 15(15):2597–2601. doi:10.1002/pmic.201400515
Mayrand D, Laforce-Lavoie A, Larochelle S, Langlois A, Genest H, Roy M, Moulin V (2012) Angiogenic properties of myofibroblasts isolated from normal human skin wounds. Angiogenesis 15:199–212
Scherrer B (1984) Biostatistique. Morin, G., Montreal
Abid Hussein MN, Boing AN, Biro E, Hoek FJ, Vogel GM, Meuleman DG, Sturk A, Nieuwland R (2007) Phospholipid composition of in vitro endothelial microparticles and their in vivo thrombogenic properties. Thromb Res 121(6):865–871
Gabbiani G (2003) The myofibroblast in wound healing and fibrocontractive diseases. J Pathol 200(4):500–503
de Wever O, Demetter P, Mareel M, Bracke M (2008) Stromal myofibroblasts are drivers of invasive cancer growth. Int J Cancer 123:2229–2238
Ronnov-Jessen L, Van Deurs B, Nielsen M, Petersen OW (1992) Identification, paracrine generation, and possible function of human breast carcinoma myofibroblasts in culture. In Vitro Cell Dev Biol Anim 28A:273–283
Moulin VJ (2016) The role of myofibroblasts in normal skin wound healing. In: Martinez A (ed) Myofibroblasts: origin, function and role in disease. Cell biology research progress. Nova Science Publishers, Hauppauge, NY, pp 1–12
Kreimer S, Belov AM, Ghiran I, Murthy SK, Frank DA, Ivanov AR (2015) Mass-spectrometry-based molecular characterization of extracellular vesicles: lipidomics and proteomics. J Proteome Res 14(6):2367–2384. doi:10.1021/pr501279t
Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW, Thery C (2014) Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 3:26913. doi:10.3402/jev.v3.26913
Brill A, Dashevsky O, Rivo J, Gozal Y, Varon D (2005) Platelet-derived microparticles induce angiogenesis and stimulate post-ischemic revascularization. Cardiovasc Res 67(1):30–38
Ji H, Erfani N, Tauro BJ, Kapp EA, Zhu HJ, Moritz RL, Lim JW, Simpson RJ (2008) Difference gel electrophoresis analysis of Ras-transformed fibroblast cell-derived exosomes. Electrophoresis 29(12):2660–2671. doi:10.1002/elps.200800015
Shai E, Varon D (2011) Development, cell differentiation, angiogenesis–microparticles and their roles in angiogenesis. Atertioscler Thromb Vasc Biol 31(1):10–14. doi:10.1161/ATVBAHA.109.200980
Lopatina T, Bruno S, Tetta C, Kalinina N, Porta M, Camussi G (2014) Platelet-derived growth factor regulates the secretion of extracellular vesicles by adipose mesenchymal stem cells and enhances their angiogenic potential. Cell Commun Signal (CCS) 12:26. doi:10.1186/1478-811X-12-26
Li J, Zhang Y, Liu Y, Dai X, Li W, Cai X, Yin Y, Wang Q, Xue Y, Wang C, Li D, Hou D, Jiang X, Zhang J, Zen K, Chen X, Zhang CY (2013) Microvesicle-mediated transfer of microRNA-150 from monocytes to endothelial cells promotes angiogenesis. J Biol Chem 288(32):23586–23596. doi:10.1074/jbc.M113.489302
Acknowledgements
This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) (RGPIN-2014-04404); Le Réseau de Thérapie Cellulaire et Tissulaire du FRQS (ThéCell). MM was recipient of a Soeurs Mallet fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Merjaneh, M., Langlois, A., Larochelle, S. et al. Pro-angiogenic capacities of microvesicles produced by skin wound myofibroblasts. Angiogenesis 20, 385–398 (2017). https://doi.org/10.1007/s10456-017-9554-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10456-017-9554-9