Abstract
While the degree of cerebellar tonsillar descent is considered the primary radiologic marker of Chiari malformation type I (CMI), biomechanical forces acting on the brain tissue in CMI subjects are less studied and poorly understood. In this study, regional brain tissue displacement and principal strains in 43 CMI subjects and 25 controls were quantified using a magnetic resonance imaging (MRI) methodology known as displacement encoding with stimulated echoes (DENSE). Measurements from MRI were obtained for seven different brain regions—the brainstem, cerebellum, cingulate gyrus, corpus callosum, frontal lobe, occipital lobe, and parietal lobe. Mean displacements in the cerebellum and brainstem were found to be 106 and 64% higher, respectively, for CMI subjects than controls (p < .001). Mean compression and extension strains in the cerebellum were 52 and 50% higher, respectively, in CMI subjects (p < .001). Brainstem mean extension strain was 41% higher in CMI subjects (p < .001), but no significant difference in compression strain was observed. The other brain structures revealed no significant differences between CMI and controls. These findings demonstrate that brain tissue displacement and strain in the cerebellum and brainstem might represent two new biomarkers to distinguish between CMI subjects and controls.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
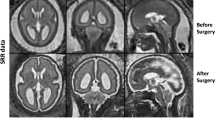
Chiari malformation type I (CMI) is a neurological disorder that is typically diagnosed by a descent of the cerebellar tonsils greater than 5 mm below the foramen magnum (see Fig. 1).25 While tonsillar position (TP) has been used as the primary evidence of CMI, recent radiological studies have shown that TP is limited as a reliable indicator of CMI symptomatology.17,25,28,41 For example, Strahle et al. reported that for the cohort of CMI subjects they evaluated, there were ten times more asymptomatic subjects with a TP greater than 5 mm that symptomatic subjects.37 Also, Smith et al. found that approximately 1–2% of subjects having TP > 5 mm remain asymptomatic.34 Khalsa et al. matched CMI patients by TP and found no significant differences between symptomatic and asymptomatic subjects when comparing two-dimensional or three-dimensional morphometric parameters.41 Moreover, in a survey of 63 experts with a collective surgical experience of more than 15,000 CMI cases, over 85% recommended rejecting the 5-mm rule currently used as the basis for CMI diagnosis.7 Additionally, several studies reported only a weak correlation between TP and CMI symptomatology.17,25,28,41 Due to inconsistent findings regarding the relationship between TP and CMI symptomatology, additional studies have been conducted to identify alternative morphometric diagnostic criteria that are specific to CMI.14,18,26,28 The disconnect between TP and CMI symptomatology is believed to result from variability in the static morphometric measures found in the general population and people with asymptomatic CMI.13,24,34,37 Thus, exploring the brain’s dynamic properties may provide a unique quantitative measure to better understand the effect of biomechanical forces acting on the brain parenchyma, as these forces may be more indicative of CMI symptomatology than TP.
Phase-contrast magnetic resonance imaging (PCMRI) is a dynamic measure that has been used in several studies to characterize brain tissue and cerebrospinal fluid (CSF) dynamics in CMI subjects.4,5,16,21,30,40 It has been speculated that the abnormal CSF flow dynamics observed in CMI patients could result in altered neural tissue biomechanics at the cranio-cervical junction.9,10,22,23 Due to the altered soft tissue properties in the brain, studies have revealed that subjects with CMI exhibit an increase in neural tissue motion.4,5,11,16,21,22,30,40 For example, Pujol et al. determined the movement of the cerebellar tonsils in subjects with CMI and reported a significantly higher motion in CMI subjects as compared to controls.30 They also reported an abnormal deformation of the cerebellar tonsils—which resulted in the obstruction of the CSF flow from the cranial cavity into the spine. Furthermore, in subjects with cough-strain headaches, the amplitude of tonsillar motion was found to be greater as compared to patients without cough-strain headaches.30 In another study, Wolpert et al. determined the movement of the medulla, tonsils, upper cervical cord, and the posterior fossa cerebrospinal fluid pathways in both CMI subjects and normal subjects. They found a tenfold increase in the velocity of the tonsils in CMI subjects compared to normal subjects. Additionally, a significant increase in velocity was also identified in the medulla and upper cervical cord in CMI subjects when compared to normal subjects.40 Hofmann et al. evaluated volumetric motion data of the spinal cord and CSF using axial PCMRI images between 18 subjects with CMI and 18 healthy controls. Out of the 18 CMI subjects, 14 of them had syringomyelia. They found an increase in systolic downward displacement of the spinal cord in patients with syrinx as compared to patients without a syrinx. They attributed the systolic downward displacement of the spinal cord to obstruction of the foramen magnum in subjects with CMI.16 Even though PCMRI has been used extensively to quantify cardiac-induced brain motion, PCMRI-based quantification of brain tissue motion involves integrating the velocity field over the cardiac cycle—which could potentially result in the accumulation of errors in each cardiac phase.36
Other studies have quantified the displacement of neural tissues in CMI subjects by tracking anatomical landmarks in the cerebellum and brainstem.11,12,22 Terem et al. used an amplified MRI methodology to detect brain tissue motion. As part of the study, using a free form deformation of the amplified MRI output, the group obtained displacement maps of a single subject with Chiari malformation. The results from this study demonstrated that CMI subjects exhibited a higher downward tissue displacement in the brainstem and craniocervical junction.38
Displacement encoding with stimulated echoes (DENSE) is a dynamic MRI technique that was initially developed to characterize myocardial tissue biomechanics.3 DENSE MRI is advantageous over PCMRI because it encodes displacement directly into the phase of the image without requiring error-accumulating path integrations.3,43 DENSE MRI also has the capability of encoding displacements smaller than the spatial resolution of the acquired images (peak displacement of less than 200 µm).1,2,29,33,42 DENSE MRI has been widely employed to characterize myocardial and vascular tissue biomechanics.19,20,39,42,43 Soellinger et al. and Zhong et al. demonstrated that it could also be used to evaluate cardiac-induced brain tissue motion.35,42 Other researchers have used the DENSE methodology to quantify neural tissue motion in healthy subjects, where displacement as low as 100 µm was detected.1,2,29,33 Additionally, using a tissue phantom undergoing cyclicical motion, Nwotchouang et al. determined the accuracy of the DENSE MRI protocol for quantifying cardiac-induced brain tissue motion.27 These studies have shown the ability of DENSE MRI to quantify cardiac-induced brain tissue motion.
The goal of the current study was to quantify regional brain tissue displacement and principal strains in CMI subjects and healthy controls. We focused on seven brain regions: the brainstem, cerebellum, cingulate gyrus, corpus callosum, frontal lobe, occipital lobe, and parietal lobe. Differences in regional tissue displacement and principal strains in these regions were determined. The effects of age and body mass index (BMI) on brain tissue displacement and principal strains were also evaluated. Due to the abnormal CSF dynamics and altered neural tissue properties reported in CMI subjects, we hypothesized that neural tissue displacement and principal strain is significantly higher in CMI subjects as compared to healthy controls.
Materials and Methods
Participants
The institutional review boards at The University of Akron and Emory University approved this study, and all subjects provided written informed consent. All subjects were scanned at the Center for Systems Imaging at the Emory University School of Medicine between January 2017 and February 2020. This study included 43 adults with CMI (36 females and seven males; age 37 ± 11 years (mean ± standard deviation); BMI 31 ± 9 kg m−2) and 25 healthy adults (17 females and eight males; age 25 ± 4 years; BMI 22 ± 2 kg m−2). In addition to the DENSE MR images, common clinical symptoms presented by CMI subjects were also collected prior to posterior cranial fossa decompression surgery. These symptoms were collected by a single neurosurgeon (DLB) at Emory University Hospital. The list of clinical symptoms and the number of CMI subjects with specific symptoms are shown in Table 1.
Imaging Protocol
Sagittal T1 and T2 weighted images covering the brain were acquired for all subjects on a PrismaFit 3T MRI scanner (Siemens Healthcare, Erlangen, Germany) using a 20-channel head coil. DENSE scans were acquired in the mid-sagittal orientation using a two-dimensional spiral cine technique employing peripheral pulse unit-gating (Fig. 2).20 Two sets of DENSE images were acquired in each scan, one with displacement encoded in the anterior-posterior direction and one set with displacement encoded in the cranial–caudal direction. The two sets of images were acquired sequentially within the same scan sequence. DENSE MRI acquisition was initialized with a tagging pulse at the peak of the peripheral pulse unit signal, and a total of 13–29 frames were acquired over the cardiac cycle depending on the subject’s heart rate. The total scan time for each subject was dependent on the subject’s heart rate (For a subject with a heart rate of 60 bpm, the scan time was 1.5 min for each direction). The following imaging parameters were used for DENSE acquisition: The flip angle = 15 degrees, temporal resolution/frame = 34 milliseconds, encoding frequency = 0.6 cycles/mm, spiral interleaves per heartbeat = 2, total spiral interleaves per image = 192, field of view = 256 × 256, reconstruction matrix = 256 × 256, pixel size = 0.86–0.94 × 0.86–0.94 mm, and slice thickness = 8 mm.
Example of midsagittal DENSE MR image sets: (a) magnitude image, (b) phase image with displacement encoded in the anterior-posterior direction, and (c) phase image with displacement encoded in the cranial-caudal direction. Black arrows show the displacement encoding direction. Post trigger delay was 34 milliseconds.
Post-processing of DENSE Scans
A custom program developed in MATLAB (MathWorks, Natick, MA) was used to post-process the DENSE scans. The procedures used to obtain the displacement and principal strains were similar to those described in Pahlavian et al.29 The magnitude image was used to identify the seven brain regions (Fig. 3). DENSE phase information in the anterior–posterior and cranial–caudal directions were converted into displacements in millimeters using the encoding frequency. DENSE-measured displacements were calculated after phase unwrapping (if necessary), noise filtering, and selection of the brain regions (shown in Fig. 3). Noise filtering was achieved with the filter2 function in MATLAB 2018b. Filter2 applies a finite impulse response filter to the displacement measurement. Values for the normalized cut off frequency and filter order were 0.15 and 16, respectively, as recommended by Pahlavian et al.29 On the magnitude images obtained with DENSE, the seven brain regions were created using an interactive paintbrush tool in MATLAB. Pixelwise, two-dimensional Eulerian displacement maps were created for each phase image for the selected brain regions.
Mean Displacement and Mean Principal Strains
Due to a reduction in the signal-to-noise ratio at the end of the cardiac cycle caused by T1 decay, only the first two-thirds of each cardiac cycle was used to calculate the mean displacement.29 For each voxel, the magnitude displacement was computed using the measured displacements from the anterior–posterior (AP) and cranial–caudal (CC) displacement maps:
To quantify the peak-to-peak tissue displacement, the minimum displacement was subtracted from the maximum displacement over the cardiac cycle for each voxel. For each anatomical structure, the mean displacement was calculated by spatially averaging the peak tissue displacements.
To determine the principal strains, Eulerian displacement maps were employed. For each brain region, a structured quadrilateral grid was generated. The locations of the voxels in the un-deformed and deformed coordinates were used to compute the two-dimensional deformation gradient tensor F, which was then used to compute the right Cauchy–Green deformation strain tensor C:
Next, the Lagrangian strain tensor E was computed:
where I is the identity matrix.
The principal compression strain was defined as the smallest negative eigenvalue of E, while the principal extension strain was defined as the largest positive eigenvalue of E. Compression or extension of each brain region describes the shortening or elongation of the tissue, respectively. The mean compression strain and mean extension strain for each brain structure were determined by spatially averaging the peak compression and extension strains over the cardiac cycle, respectively, for each of the seven brain structures.
Maximum Displacement and Maximum Principal Strains
The displacement and strain maps were shown to be heterogeneous within a specific brain structure (Fig. 4). In order to obtain a more homogeneous representation of the maximum tissue displacement and principal strains in each brain region, a 30-mm2 circular ROI was automatically identified in sections with the greatest amount of displacement or principal strains. The 30-mm2 was selected based on our previous findings, where we determined that approximately 80 averaged voxels were required to obtain a reliable displacement measurement with DENSE MRI.27 The maximum displacement, maximum compression strain, and maximum extension strain in each circular area were determined by averaging the peak displacements, compression strains, and extension strains, respectively, within the circular area for all brain regions.
Two-dimensional Eulerian displacement maps of the brainstem and the cerebellum of (a) CMI subject and (b) healthy control subject matched by age. Note: The black circles on the displacement maps indicate examples of the 30-mm2 circular areas that were used to compute the maximum displacement, maximum compression strain, and maximum extension strain in each brain region.
Statistical Analysis
The statistical analysis in this study included three comparisons: all CMI subjects vs. all healthy controls, a subset of younger CMI subjects vs. age-matched healthy controls, and younger CMI subjects vs. older CMI subjects. The statistical analysis was performed using MATLAB 2018b, SAS, and Microsoft Excel. The Shapiro–Wilk test was used to check for normality in the data.
First, a multivariate analysis of variance (MANOVA) was used to determine the global effects of group (CMI vs. control) on the displacement and principal strains across all the seven brain structures (i.e., the brain regions as shown in Fig. 3). Next, all 43 CMI subjects’ data were compared to the data for all 25 healthy controls separately for each of the seven dependent variables. Independent t tests and Wilcoxon rank-sum tests were used to determine differences between the CMI subjects and healthy controls. Due to a large number of comparisons (42 comparisons), we controlled for family-wise errors, and statistical significance was adjusted to p < .001 (0.05/42). To make sure that the effects were not due to age differences, the data for 23 younger CMI subjects (mean age = 30 ± 8 years) and 23 age-matched healthy controls (mean age = 25 ± 4 years) was compared. Finally, data for the 23 younger CMI subjects were compared to data from the 20 older CMI subjects (mean age = 48 ± 6 years).
For the clinical data, brain tissue displacement and principal strain for subjects with and without clinical symptoms were also compared. Additionally, because CMI subjects generally have a high BMI,6 we were unable to match the BMI between the CMI subjects and healthy controls. However, we performed an analysis on all 68 subjects by correlating the displacement and principal strain for the seven brain structures with subject age and BMI. The relationship between BMI, age, brain tissue displacement, and principle strains was evaluated using Spearman’s correlation coefficient.
Results
The first statistical comparison included all participants in both groups: 43 CMI subjects and 25 healthy controls. For the mean displacement, the inferior brain structures (brainstem and cerebellum) demonstrated higher tissue motion than the superior brain structures (i.e., the cingulate gyrus, corpus callosum, frontal lobe, occipital lobe, and parietal lobe; see Fig. 5) for both the CMI and control groups. The brainstem demonstrated the largest mean displacement among the seven brain structures, followed by the cerebellum (see Table 2).
Displacement Differences Between CMI and Controls
To test for the global effects of group (CMI vs. control) on the brain tissue displacement, a MANOVA was performed on the displacement measurement for the seven brain regions. Results show that there was a statistically significant multivariate main effect for group (Wilks Lambda = 0.48, F = 8.7, p < .001). Separate t tests revealed that compared to healthy controls, CMI subjects exhibited a significantly higher brainstem mean displacement (192 ± 60 µm vs. 117 ± 38 µm for controls; p < .001) and cerebellum mean displacement (138 ± 55 µm vs. 67 ± 24 µm for controls; p < .001), as can be seen in Figs. 5 and 6a, and Table 2. No significant differences were found between CMI subjects and healthy controls for mean displacement in the cingulate gyrus (26 ± 11 µm for CMI vs. 23 ± 6 µm for controls; p = .38), corpus callosum (48 ± 18 µm for CMI vs. 46 ± 13 µm for controls; p = .79), frontal lobe (26 ± 13 µm for CMI vs. 19 ± 4 µm for controls; p = .07), occipital lobe (20 ± 7 µm for CMI vs. 17 ± 5 µm for controls; p = .05), or parietal lobe (24 ± 10 µm for CMI vs. 20 ± 5 µm for controls; p = .10) as can be seen from Fig. 5.
The maximum displacement from the circular ROI’s for CMI subjects and healthy controls (Table 2) showed a similar trend as the mean displacement. Significantly higher maximum displacement was found in CMI subjects as compared to healthy controls in the brainstem (300 ± 125 µm for CMI vs. 148 ± 43 µm for controls; p < .001) and cerebellum (369 ± 185 µm for CMI vs. 120 ± 42 µm for controls; p < .001). However, no significant differences were found for the superior brain structures (i.e., cingulate gyrus, corpus callosum, frontal lobe, occipital lobe, and parietal lobe, Table 2).
Principal Strain Differences Between CMI and Controls
Similar to the displacement, a MANOVA was also performed on the strain measurements for the seven brain regions. There was a multivariate main effect for group (Wilks Lambda = 0.49, F = 3.8, p < .001)—again, reflecting an overall effect for strain across all dependent variables. For the separate t tests, the mean compression strain and mean extension strain for the seven brain regions in CMI subjects and healthy controls were also compared. The comparisons revealed cerebellum mean compression strain (0.64 ± 0.28% for CMI vs. 0.42 ± 0.11% for controls; p < .001; see Figs. 6b and 7) and cerebellum mean extension strain (0.66 ± 0.34% for CMI vs. 0.44 ± 0.10% for controls; p < .001; see Table 3) to be significantly higher in CMI subjects than in healthy controls (see Fig. 7). Additionally, brainstem mean extension strain was significantly higher in CMI subjects than in healthy controls (0.84 ± 0.30% in CMI subjects vs. 0.60 ± 0.17% for controls; p < .001; see Figs. 6c and 7). However, brainstem mean compression strain was not significantly different in CMI subjects and healthy controls (p = .52). Moreover, no significant differences were found for the mean compression strain or mean extension strain in the other brain regions—cingulate gyrus, corpus callosum, frontal lobe, occipital lobe, and parietal lobe.
For the maximum compression strain and maximum extension strain obtained from the circular ROI’s, only the cerebellum demonstrated significant differences between CMI subjects and healthy controls. The maximum compression strain (1.61 ± 1.02% in CMI subjects vs. 0.80 ± 0.21% for controls; p < .001; see Table 3) and maximum extension strain (1.59 ± 1.11% in CMI subjects vs. 0.89 ± 0.26% for controls; p < .001; see Table 3) in the cerebellum was significantly higher in CMI subjects than in healthy controls. While no significant difference was observed in the brainstem maximum compression strain (p = .09), a trend towards significance was found for the brainstem maximum extension strain (1.63 ± 1.08% in CMI subjects vs. 1.07 ± 0.41% for controls; p = .005; see Table 3).
Comparison Between Individuals With and Without Clinical Symptoms
As part of this study, brain tissue displacement, and principal strain in subjects with ten commonly reported clinical symptoms were compared to subjects without symptoms (see Table 1). These comparisons were focused on the brainstem and cerebellum—as these were the only two structures that were found to be significantly different between CMI subjects and controls. From our analysis, none of the comparisons were significantly different between CMI subjects with and without clinical symptoms (See Supplementary Table).
Effect of Age and BMI on Measured Displacement and Principal Strains
A comparison was also performed between 23 CMI subjects and 23 age-matched healthy controls; the results were similar to those obtained for the full dataset (see Tables 2 and 3 for detailed results). The displacement and strain values for the younger CMI subjects and the older CMI subjects were also compared. No statistically significant differences were found between the two groups in either the displacement or principal strains for any of the seven brain regions. Additionally, an examination of the correlation between age, BMI, and the different brain indices for all brain regions revealed no relationship between age (r values ranged from 0.05 to 0.26; p > .05) or BMI (r values ranged from −0.01 to 0.26; p > .05) and either the displacement or the principal strains.
Discussion
In this study, regional cardiac-induced brain tissue displacement and principal strains were quantified in seven brain structures in CMI subjects and healthy controls. The results of this study demonstrate that CMI subjects have significantly higher tissue motion in the cerebellum and brainstem compared to healthy controls. Additionally, our results show that neither age nor BMI has an impact on the measured cardiac-induced brain tissue displacement or principal strains.
Differences in Displacement for CMI Subjects and Healthy Controls
The altered brain tissue structure in CMI subjects reported by other researchers14,18,26,28 is evident in the CMI subjects in this study. As compared to healthy controls, we found that mean displacement was 64 and 106% higher, respectively, in the brainstem and cerebellum of CMI subjects—but not in other brain regions. As shown in Fig. 8, the distributions of displacement in the brainstem and cerebellum are non-uniform across the tissue structure—with the higher displacements occurring in the inferior aspects of the brainstem and cerebellum. To quantify the increased tissue motion in the inferior aspects, we considered a smaller circular ROI with an area of 30 mm2 (maximum displacement). The maximum displacement in the circular regions was 103% higher in the brainstem and 208% higher in the cerebellum for CMI subjects than for healthy controls. This suggests that while the tissue displacements across the entire brainstem and cerebellum are higher in CMI subjects, the inferior aspects of these brain regions have the greatest contribution to the increase in displacement observed in CMI subjects. This is likely due to the abnormal CSF flow dynamics and altered neural tissue characteristics around the cranio-cervical junction.9,10,22,23
Previous studies have evaluated brain tissue displacement in healthy subjects using DENSE MRI.1,2,29,33 Zhong et al. and Pahlavian et al. reported a displacement in the brainstem of 150 to 210 µm and displacement in the cerebellum of 70 to 105 µm in healthy subjects.29,42 In the current study, mean displacement in the brainstem and cerebellum for healthy subjects were found to be 117 and 67 µm, respectively. One possible reason for the higher displacements reported in other studies is the sample size: Zhong et al.42 included three healthy subjects, and Pahlavian et al.29 evaluated eight healthy subjects, while we evaluated a total of 25 healthy subjects. Additionally, the triggering method used by Zhong et al. and Pahlavian et al. (Electrocardiography) was different from that used in the current study (peripheral pulse unit). Also, the encoding frequency (0.6 cycles/mm) employed in the current study was higher than that used by Zhong et al. (0.4 cycles/mm) but lower than that used by Pahlavian et al. (1.2–1.5 cycles/mm).
Several research groups that evaluated neural tissue motion using PCMRI reported higher displacement values than the ones reported herein. For example, Lawrence et al. evaluated the spinal cord’s displacement in CMI subjects compared to controls and reported displacements of 530 and 230 µm, respectively.21 Alperin et al. reported the maximum cord displacement in CMI subjects and controls to be 390 and 330 µm, respectively.5 Cousins et al. determined the cerebellar tonsils’ motion in the foramen magnum during the cardiac cycle and reported displacements of 570 and 430 µm, for CMI subjects and healthy controls, respectively.11 It should be noted that the measurement of displacements using PCMRI is more localized—either at the foramen magnum21 or the cerebellar tonsils.11 Hence, no displacement information for tissue structures above the foramen magnum is available. Moreover, the PCMRI technique employed in the cited studies measures the velocity directly and, thus, can only provide an indirect measurement of displacement, as the integration of velocity over the cardiac cycle could potentially accumulate errors in each cardiac phase.36 In contrast, the DENSE MRI technique used in the current study measures the displacement directly from the phase information.
Brain tissue displacement has been quantified in CMI subjects and controls using a steady-state free procession MRI techniques.12,22,38 With this technique, Leung et al. reported displacements in the cerebellar tonsils of 790 µm in CMI subjects (vs. 300 µm in healthy controls) and displacements in the cervicomedullary junction of 600 µm in CMI subjects (vs. 320 µm in controls).12,22 Dawes et al. used a similar technique and reported a median displacement of 560 µm from the fastigium to the tip of the tonsils, and a mean displacement of 200 µm from the superior aspect of the cerebellum to the fastigium.12 One major drawback of these studies is that the displacement was determined by tracking specific anatomical locations over the entire cardiac cycle. By relying on a single point for the measured displacement, an overall understanding of the displacement over the entire brainstem and cerebellum cannot be determined. By using DENSE MRI, we were able to obtain the displacement over the entire brainstem and cerebellum; as shown in Fig. 8, the distributions of displacements across these two structures are non-uniform—with lower displacements in the superior aspects of the structures and higher displacements in the inferior aspects.
Differences in Principal Strains for CMI Subjects and Healthy Controls
In the current study, we compared the mean compression strain and mean extension strain for CMI subjects and healthy controls. The mean compression strain and mean extension strain in the cerebellum were 52 and 50% higher in CMI subjects than in healthy controls, respectively (p < .001), and the brainstem mean extension strain was 41% higher in CMI subjects than in healthy controls (p < .001). No significant differences were found for the mean compression strain and mean extension strain for the superior brain structures (cingulate gyrus, corpus callosum, frontal lobe, occipital lobe, and parietal lobe). Similar to the displacement results described above, the mean compression strain and mean extension strain were also non-uniformly distributed across the cerebellum and brainstem (see Fig. 8). The non-uniformity is evident by a 100% increase in cerebellar maximum compression strain and a 78% increase in cerebellar maximum extension strain in CMI subjects as compared to healthy controls.
Other researchers have quantified brain tissue strain using DENSE MRI.1,2,29,33 Pahlavian et al.29 determined the principal regional strain in healthy controls and reported cerebellar mean compression strain and mean extension strain of 0.3 and 0.23%, respectively—which are lower than the reported principal strains for the cerebellum for healthy subjects in the current study (where mean compression strain and mean extension strain were 0.42% and 0.44%, respectively). Similarly, higher brainstem mean compression strain and mean extension strain were reported for the current study than were reported by Pahlavian et al. The possible reasons for these discrepancies are the same as those for the differences in displacement. Although Pahlavian et al. reported higher displacement, we reported higher strain values. It is important to note that displacement alone does not cause damage to tissues, but strain does. Moreover, since an increase in displacement does not necessarily lead to an increase in strain (as displacement is not directly proportional to strain), an increase in displacement within a tissue structure may not result in an increase in strain. This is because compression or extension strains are caused by a gradient of displacement across the tissue. Areas in the tissue with a higher gradient of displacement will experience higher compression or extension strains—and vice versa.
Dawes et al. and Leung et al. also quantified brain tissue strain in subjects with CMI.12,22 Leung et al. quantified the strain from the fastigium of the fourth ventricle to the tip of the tonsils and reported a mean strain of about 0.012 and 0.008%, before and after decompression surgery, respectively. Dawes et al. reported a median strain of 1.48% from the fastigium to the tip of the tonsils before decompression surgery, and 0.51% after decompression surgery. Additionally, for the superior aspect of the cerebellum to the fastigium, a median strain of 0.69 and 0.47% was reported before and after decompression surgery, respectively.12 Since regional cerebellar compression or extension strains were not quantified in these studies, we are unable to make a direct comparison with our findings.
Previous studies have shown that subjects with CMI have abnormal CSF flow dynamics, which, in turn, can lead to altered neural tissue characteristics at the cranio-cervical junction.9,10,22,23 Sloots et al. demonstrated that cardiac-induced brain tissue volumetric strain was three times larger than the respiratory-induced volumetric strain.33 Thus, the strain generated in the brain is influenced more by the cardiac cycle than the respiratory cycle. CMI subjects have been reported to experience higher intracranial pressures and severe occipital headaches.8,15 To reduce symptoms, CMI subjects undergo posterior fossa decompression surgery, which helps to restore CSF flow and increases the area in the CSF space inferior to the cerebellum. One technique commonly used by neurosurgeons as a part of this surgery is to remove sections of the occipital bone. From Fig. 8, it can be noticed that the extension and compression strains are more localized in the inferior–posterior section of the cerebellum. The occipital bone—which is located in the posterior aspect of the cerebellum—may prevent the movement of the cerebellum if the cranio-cervical junction is crowded or if the cerebellar tonsils are compressed against the occipital bone, which would not permit adequate CSF flow during each cardiac cycle (as is often seen in CMI subjects). Thus, a gradient of displacement may be created—with higher displacements in the inferior–anterior section of the cerebellum and minimal displacements in the inferior–posterior section (Fig. 8). This gradient of displacement may, in turn, lead to an increase in strain in the cerebellum. This circumstance may explain why lower cerebellum strains are reported for CMI subjects following decompression surgery.12,22
Comparison Between Individuals With and Without Clinical Symptoms
In the current study, we did not find any significant differences in the brain tissue displacement or principal strain between CMI subjects with and without clinical symptoms (p > 0.05). In the literature, there have been conflicting findings relating to the differences in tissue displacement between CMI subjects with and without clinical symptoms. For example, Pujol et al. reported that in subjects with cough-strain headaches, the amplitude of tonsillar motion was greater as compared to patients without cough-strain headaches.30 Also, subjects with Valsalva headaches have been shown to have a higher motion at the cerebellar tonsils as compared to subjects without Valsalva headaches.22 Even though these studies reported a relationship between tissue motion and clinical symptoms, another study found no statistically significant differences in brain motion in subjects with and without Valsalva headaches12—which is in agreement with the results reported herein. While the results of this paper show that we can distinguish between CMI subjects and controls based on both displacement and strain, these measurements did not discriminate between CMI subjects with and without specific symptomatology.
Effects of Age and BMI
The effects of age and BMI on the measured brain indices were also evaluated in this study. To determine the effect of age on the brain tissue indices, three methods were used. First, 23 younger CMI subjects were compared to 23 age-matched healthy controls; the results were similar to that of the full dataset. A second comparison showed that the brain indices for the younger CMI subjects were not significantly different from those in a group of older CMI subjects. A correlation study revealed no relationship between age and any of the brain indices. These results suggest that age had no impact on our final results. An additional correlation study using data for all study participants revealed no relationship between BMI and brain tissue displacement or principal strains. Thus, we concluded that age and BMI most likely had no effect on the measured brain tissue displacement or principal strains.
Limitations
There were limitations to this study. The displacements in this study were quantified on a two-dimensional sagittal plane—without consideration of the through-plane direction. Previous studies have shown that the through-plane tissue displacement is relatively small and should have no impact on the measured displacement.29 Even though the through-plane displacement may result in a small amount of strain, our current technique, as designed, is unable to quantify this brain tissue strain. Future work will focus on improving the current post-processing methodology of evaluating strain, taking into consideration the through-plane brain tissue displacement. Furthermore, three-dimensional measurements were not performed in the current study. Previous studies have reported that the maximum principal strain orientation of the brainstem and cerebellum was found at the sagittal plane in a volunteer.31 Thus, the current study was focused on measurements obtained on the sagittal plane. Additionally, the cine DENSE sequence employed in the current, as designed, was unable to obtain through-plane displacements without imaging artifacts for encoding values optimized for quantifying small brain tissue motion. Future work will focus on improving the sequence to better the quality of the through-plane displacement measurement. Also, the encoding frequency was not optimized for maximum sensitivity to detect displacement. The clinical images employed in the current study used an encoding frequency of 0.6 cycles/mm to avoid phase wraps.
Posture may also play a role in the position or structure of the brain tissue, as reported in previous studies.32 However, in the current study, all subjects underwent scanning in the same orientation. Additionally, blurring effects can be noticed due to the prospective triggering caused by artifacts from heart rate variations. Because the brain regions in our study were manually created, there was a possibility that voxel averaging might affect the measurement of the displacement and principal strains. To reduce the possibility of adding unwanted structures to the outline of the brain region, an area that was between two to three voxels wide around the entire boundary of each brain region was excluded from the analysis. In the current study, only two-thirds of the cardiac cycle was employed during the analysis. It is important to note that using only two-thirds of the cardiac cycle may underestimate the measured average displacement by up to 12 µm (Table 4). Even though the differences in the displacement between the two methods was small, a paired t test between the methods revealed significant differences (Table 4).
As described in the methods section, the mean displacement in the current study was determined by subtracting the minimum displacement from the maximum displacement for each voxel. However, in a case where a region-averaged displacement curve is first determined before the minimum displacement is subtracted from the maximum displacement, the region-averaged method may underestimate the mean displacement by up to 12 µm (Table 5). While differences were found between the displacement obtained between the two methods, as shown in Table 5, a paired t test analysis between the two methods revealed no statistically significant differences (Table 5).
In summary, we quantified regional tissue displacement and principal strains in seven brain structures. Cardiac-induced brain tissue displacement and principal strains were significantly higher in the cerebellum and brainstem of CMI subjects compared to healthy controls, and inferior brain structures showed higher displacements than superior brain structures. The displacements and principal strains were non-uniformly distributed in the brainstem and cerebellum—with higher displacements and principal strains reported in the inferior aspects than in superior aspects. The results reported herein confirm our hypothesis that neural tissue displacement and principal strains are significantly higher in CMI subjects when compared to healthy controls. Thus, the different brain indices reported in this study may serve as additional diagnostic criteria for CMI.
References
Adams, A. L., H. J. Kuijf, M. A. Viergever, P. R. Luijten, and J. J. M. Zwanenburg. Quantifying cardiac-induced brain tissue expansion using DENSE. NMR Biomed. 32(2):e4050, 2019.
Adams, A. L., M. A. Viergever, P. R. Luijten, and J. J. M. Zwanenburg. Validating faster DENSE measurements of cardiac-induced brain tissue expansion as a potential tool for investigating cerebral microvascular pulsations. Neuroimage 208:116466, 2020.
Aletras, A. H., S. Ding, R. S. Balaban, and H. Wen. DENSE: displacement encoding with stimulated echoes in cardiac functional MRI. J. Magn. Reson. 137(1):247–252, 1999.
Alperin, N., J. R. Loftus, C. J. Oliu, A. M. Bagci, S. H. Lee, B. Ertl-Wagner, B. Green, and R. Sekula. Magnetic resonance imaging measures of posterior cranial fossa morphology and cerebrospinal fluid physiology in Chiari malformation type I. Neurosurgery 75(5):515–522, 2014; (discussion 522).
Alperin, N., A. Sivaramakrishnan, and T. Lichtor. Magnetic resonance imaging-based measurements of cerebrospinal fluid and blood flow as indicators of intracranial compliance in patients with Chiari malformation. J. Neurosurg. 103(1):46–52, 2005.
Arnautovic, K. I., D. Muzevic, B. Splavski, and F. A. Boop. Association of increased body mass index with Chiari malformation Type I and syrinx formation in adults. J. Neurosurg. 119(4):1058–1067, 2013.
Bolognese, P. A., A. Brodbelt, A. B. Bloom, and R. W. Kula. Chiari I malformation: opinions on diagnostic trends and controversies from a panel of 63 international experts. World Neurosurg. 130:e9–e16, 2019.
Chavez, A., M. Roguski, A. Killeen, C. Heilman, and S. Hwang. Comparison of operative and non-operative outcomes based on surgical selection criteria for patients with Chiari I malformations. J. Clin. Neurosci. 21(12):2201–2206, 2014.
Clarke, E. C., D. F. Fletcher, M. A. Stoodley, and L. E. Bilston. Computational fluid dynamics modelling of cerebrospinal fluid pressure in Chiari malformation and syringomyelia. J. Biomech. 46(11):1801–1809, 2013.
Clarke, E. C., M. A. Stoodley, and L. E. Bilston. Changes in temporal flow characteristics of CSF in Chiari malformation type I with and without syringomyelia: implications for theory of syrinx development. J. Neurosurg. 118(5):1135–1140, 2013.
Cousins, J., and V. Haughton. Motion of the cerebellar tonsils in the foramen magnum during the cardiac cycle. AJNR Am. J. Neuroradiol. 30(8):1587–1588, 2009.
Dawes, B. H., R. A. Lloyd, J. M. Rogers, J. S. Magnussen, L. E. Bilston, and M. A. Stoodley. Cerebellar tissue strain in Chiari malformation with headache. World Neurosurg. 134:e74–e81, 2019.
Elster, A. D., and M. Chen. Chiari I malformations: clinical and radiologic reappraisal. Radiology 183(2):347–353, 1992.
Eppelheimer, M. S., J. R. Houston, J. R. Bapuraj, R. Labuda, D. M. Loth, A. M. Braun, N. J. Allen, S. Heidari Pahlavian, D. Biswas, A. Urbizu, B. A. Martin, C. O. Maher, P. A. Allen, and F. Loth. A retrospective 2D morphometric analysis of adult female chiari type I patients with commonly reported and related conditions. Front Neuroanat. 12:2, 2018.
Fukuoka, T., Y. Nishimura, M. Hara, S. Haimoto, K. Eguchi, S. Yoshikawa, T. Wakabayashi, and H. J. Ginsberg. Chiari type 1 malformation-induced intracranial hypertension with diffuse brain edema treated with foramen magnum decompression: a case report. NMC Case Rep. J. 4(4):115–120, 2017.
Hofmann, E., M. Warmuth-Metz, M. Bendszus, and L. Solymosi. Phase-contrast MR imaging of the cervical CSF and spinal cord: volumetric motion analysis in patients with Chiari I malformation. Am. J. Neuroradiol. 21(1):151, 2000.
Houston, J. R., P. A. Allen, J. M. Rogers, M.-C. Lien, N. J. Allen, M. L. Hughes, J. R. Bapuraj, M. S. Eppelheimer, F. Loth, M. A. Stoodley, S. J. Vorster, and M. G. Luciano. Type I Chiari malformation, RBANS performance, and brain morphology: connecting the dots on cognition and macrolevel brain structure. Neuropsychology 33(5):725–738, 2019.
Houston, J. R., M. S. Eppelheimer, S. H. Pahlavian, D. Biswas, A. Urbizu, B. A. Martin, J. R. Bapuraj, M. Luciano, P. A. Allen, and F. Loth. A morphometric assessment of type I Chiari malformation above the McRae line: a retrospective case-control study in 302 adult female subjects. J. Neuroradiol. 45(1):23–31, 2018.
Iffrig, E., J. S. Wilson, X. Zhong, and J. N. Oshinski. Demonstration of circumferential heterogeneity in displacement and strain in the abdominal aortic wall by spiral cine DENSE MRI. J. Magn. Reson. Imaging 49(3):731–743, 2019.
Kim, D., W. D. Gilson, C. M. Kramer, and F. H. Epstein. Myocardial tissue tracking with two-dimensional cine displacement-encoded MR imaging: development and initial evaluation. Radiology 230(3):862–871, 2004.
Lawrence, B. J., M. Luciano, J. Tew, R. G. Ellenbogen, J. N. Oshinski, F. Loth, A. P. Culley, and B. A. Martin. Cardiac-related spinal cord tissue motion at the foramen magnum is increased in patients with type I Chiari malformation and decreases postdecompression surgery. World Neurosurg. 116:e298–e307, 2018.
Leung, V., J. S. Magnussen, M. A. Stoodley, and L. E. Bilston. Cerebellar and hindbrain motion in Chiari malformation with and without syringomyelia. J. Neurosurg. Spine 24(4):546–555, 2016.
Lloyd, R. A., D. F. Fletcher, E. C. Clarke, and L. E. Bilston. Chiari malformation may increase perivascular cerebrospinal fluid flow into the spinal cord: a subject-specific computational modelling study. J. Biomech. 65:185–193, 2017.
Meadows, J., M. Kraut, M. Guarnieri, R. I. Haroun, and B. S. Carson. Asymptomatic Chiari Type I malformations identified on magnetic resonance imaging. J. Neurosurg. 92(6):920–926, 2000.
Milhorat, T. H., M. W. Chou, E. M. Trinidad, R. W. Kula, M. Mandell, C. Wolpert, and M. C. Speer. Chiari I malformation redefined: clinical and radiographic findings for 364 symptomatic patients. Neurosurgery 44(5):1005–1017, 1999.
Nwotchouang, B. S. T., M. S. Eppelheimer, P. Bishop, D. Biswas, J. M. Andronowski, J. R. Bapuraj, D. Frim, R. Labuda, R. Amini, and F. Loth. Three-dimensional CT morphometric image analysis of the clivus and sphenoid sinus in Chiari malformation type I. Ann. Biomed. Eng. 47(11):2284–2295, 2019.
Nwotchouang, B. S. T., M. S. Eppelheimer, D. Biswas, S. H. Pahlavian, X. Zhong, J. N. Oshinski, D. L. Barrow, R. Amini, and F. Loth. Accuracy of cardiac-induced brain motion measurement using displacement-encoding with stimulated echoes (DENSE) magnetic resonance imaging (MRI): a phantom study. Magn. Reson. Med. 2020. https://doi.org/10.1002/mrm.28490.
Nwotchouang, B. S. T., M. S. Eppelheimer, A. Ibrahimy, J. R. Houston, D. Biswas, R. Labuda, J. R. Bapuraj, P. A. Allen, D. Frim, and F. Loth. Clivus length distinguishes between asymptomatic healthy controls and symptomatic adult women with Chiari malformation type I. Neuroradiology 62:1389–1400, 2020.
Pahlavian, S. H., J. Oshinski, X. Zhong, F. Loth, and R. Amini. Regional quantification of brain tissue strain using displacement-encoding with stimulated echoes magnetic resonance imaging. J. Biomech. Eng. 2018. https://doi.org/10.1115/1.4040227.
Pujol, J., C. Roig, A. Capdevila, A. Pou, J. L. Marti-Vilalta, J. Kulisevsky, A. Escartin, and G. Zannoli. Motion of the cerebellar tonsils in Chiari type I malformation studied by cine phase-contrast MRI. Neurology 45(9):1746–1753, 1995.
Reese, T. G., D. A. Feinberg, J. Dou, and V. J. Wedeen. Phase contrast MRI of myocardial 3D strain by encoding contiguous slices in a single shot. Magn. Reson. Med. 47(4):665–676, 2002.
Roberts, D. A.-O., D. A.-O. Asemani, P. A.-O. Nietert, M. A.-O. Eckert, D. A.-O. Inglesby, J. A.-O. Bloomberg, M. A.-O. George, and T. A.-O. Brown. Prolonged microgravity affects human brain structure and function. AJNR Am. J. Neuroradiol. 40(11):1878–1885, 2019.
Sloots, J. J., G. J. Biessels, and J. J. M. Zwanenburg. Cardiac and respiration-induced brain deformations in humans quantified with high-field MRI. Neuroimage 210:116581, 2020.
Smith, B. W., J. Strahle, J. R. Bapuraj, K. M. Muraszko, H. J. Garton, and C. O. Maher. Distribution of cerebellar tonsil position: implications for understanding Chiari malformation. J. Neurosurg. 119(3):812–819, 2013.
Soellinger, M., A. K. Rutz, S. Kozerke, and P. Boesiger. 3D cine displacement-encoded MRI of pulsatile brain motion. Magn. Reson. Med. 61(1):153–162, 2009.
Spottiswoode, B. S., X. Zhong, A. T. Hess, C. M. Kramer, E. M. Meintjes, B. M. Mayosi, and F. H. Epstein. Tracking myocardial motion from cine DENSE images using spatiotemporal phase unwrapping and temporal fitting. IEEE Trans. Med. Imaging 26(1):15–30, 2007.
Strahle, J., K. M. Muraszko, J. Kapurch, J. R. Bapuraj, H. J. Garton, and C. O. Maher. Natural history of Chiari malformation Type I following decision for conservative treatment. J. Neurosurg. Pediatr. 8(2):214–221, 2011.
Terem, I., W. W. Ni, M. Goubran, M. S. Rahimi, G. Zaharchuk, K. W. Yeom, M. E. Moseley, M. Kurt, and S. J. Holdsworth. Revealing sub-voxel motions of brain tissue using phase-based amplified MRI (aMRI). Magn. Reson. Med. 80(6):2549–2559, 2018.
Wehner, G. J., L. Jing, C. M. Haggerty, J. D. Suever, J. Chen, S. M. Hamlet, J. A. Feindt, W. Dimitri-Mojsejenko, M. A. Fogel, and B. K. Fornwalt. Comparison of left ventricular strains and torsion derived from feature tracking and DENSE CMR. J. Cardiovasc. Magn. Reson. 20(1):63, 2018.
Wolpert, S. M., R. A. Bhadelia, A. R. Bogdan, and A. R. Cohen. Chiari I malformations: assessment with phase-contrast velocity MR. Am. J. Neuroradiol. 15(7):1299–1308, 1994.
Khalsa, S. S. S., N. Geh, B. A. Martin, P. A. Allen, J. Strahle, F. Loth, D. Habtzghi, A. Urbizu Serrano, D. McQuaide, H. J. L. Garton, K. M. Muraszko, and C. O. Maher. Morphometric and volumetric comparison of 102 children with symptomatic and asymptomatic Chiari malformation Type I. J. Neurosurg. Pediatr. 21(1):65–71, 2018.
Zhong, X., C. H. Meyer, D. J. Schlesinger, J. P. Sheehan, F. H. Epstein, J. M. Larner, S. H. Benedict, P. W. Read, K. Sheng, and J. Cai. Tracking brain motion during the cardiac cycle using spiral cine-DENSE MRI. Med. Phys. 36(8):3413–3419, 2009.
Zhong, X., B. S. Spottiswoode, C. H. Meyer, C. M. Kramer, and F. H. Epstein. Imaging three-dimensional myocardial mechanics using navigator-gated volumetric spiral cine DENSE MRI. Magn. Reson. Med. 64(4):1089–1097, 2010.
Acknowledgments
The authors would like to thank Conquer Chiari and the National Institutes of Health, NINDS R15 (Grant No. 1R15NS109957-01A1) for providing funding for this research work. This work was also supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1TR002378. The thoughtful comments of Sheila Pearson at The University of Akron are also acknowledged.
Conflict of interest
All authors declared that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Umberto Morbiducci oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nwotchouang, B.S.T., Eppelheimer, M.S., Pahlavian, S.H. et al. Regional Brain Tissue Displacement and Strain is Elevated in Subjects with Chiari Malformation Type I Compared to Healthy Controls: A Study Using DENSE MRI. Ann Biomed Eng 49, 1462–1476 (2021). https://doi.org/10.1007/s10439-020-02695-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-020-02695-7