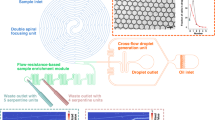
Abstract
Droplet microfluidics has become an indispensable technology to encapsulate cells of interest in a monodispersed aqueous compartment for single-cell analysis. In addition, the confinement of cells in picoliter droplets offers high-throughput, single-cell resolution, and high signal-to-noise ratio for various cellular assays that unmasks cellular heterogeneity from a bulk population. Particularly, co-encapsulation of two distinct cells in a droplet is critically important for studying cell–cell interaction, transcriptomics, genomics, antibody, and drug screening. However, the co-encapsulation of one type A cell and one type B cell per single droplet, termed 1–1–1 encapsulation, has been dictated by double Poisson distribution due to the intrinsic random dispersion of cells, which yields mostly empty droplets and only up to 13.5% of droplets under optimal conditions. Such low 1–1–1 encapsulation efficiency makes it impractical for biological analyses at scale involving low cell concentrations or a large number of variables. Here, we demonstrate a passive co-encapsulation microfluidic device that leverages close packing of cells by hydrodynamic draining to overcome the double Poisson limitation. The results suggest a significant improvement of the 1–1–1 encapsulation efficiency by over two-fold compared to the double Poisson model. The enhanced encapsulation efficiency of this platform demonstrates great potential for a high-throughput, versatile, and simple platform for cell–cell interaction studies within a confined microenvironment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Single-cell analysis has emerged as an indispensable technique to reveal cellular heterogeneity within a cell population and enable profiling that bridges between phenotype and genotype (Altschuler and Wu 2010; Chen et al. 2019; Chattopadhyay et al. 2014; Shalek et al. 2014; Shi et al. 2012). The field of single-cell analysis has gained tremendous interest from both academia and industry to further investigate the cell–cell variability for an accurate understanding of disease diagnosis and progression (Zheng et al. 2017; Lawson et al. 2018; Dhar et al. 2018). Beyond the interrogation of individual cells, the study of cell–cell communication offers in-depth information on deciphering dynamic and heterogeneous responses upon interacting with another cell (Bogdanowicz and Lu 2013; Bl and Is 2014; Haan et al. 2014; Vu et al. 2017). As a plethora of evidence has shown the significance of heterogeneity within a clonal population, it is imperative to investigate the heterogeneity of cellular function and phenotype upon cell–cell interaction. In cancer biology, the cellular interaction between cancer stem cells and other cells within the tumor microenvironment has a major impact on cancer progression, survival, and lineage (Bl and Is 2014). For instance, natural killer (NK) cell is a type of immune cells that can kill tumor cells, such as K562 cells, by inducing apoptosis upon interaction. The heterogeneous membrane marker expression of K562 can directly affects the cytotoxicity and proliferation of NK cells (Streltsova et al. 2019; Kweon et al. 2019). This heterotypic cell–cell interaction within a confined microenvironment would facilitate the development of novel biomedical strategies such as immunotherapy or cell-based therapeutics for disease treatment (Agrawal et al. 1998; Shelton et al. 2021; Zalfa and Paust 2021).
The conventional method for studying cell–cell interactions involves bulk co-culturing in a flask either with or without cell–cell contact. Although the co-culturing system could achieve heterotypic or homotypic interactions by mixing different cell suspensions prior to loading into a monolayer model, the number of interacting cells is difficult to be deterministically controlled (Bogdanowicz and Lu 2013). Moreover, the mixed cells share the same environment, and cannot prevent cross-interference from the neighboring interacting cells. Bulk co-culturing also compromises sensitivity in detecting secretion from single cells due to high background noise and diffusion of secreted molecules into bulk medium. With these limitations, the analysis of single-cell upon cell–cell interaction becomes challenging.
In recent years, droplet microfluidics has been playing a catalytic role in the field of single-cell analysis spanning from genomic sequencing (Macosko et al. 2015; Liu et al. 2019), cytotoxicity screening (Brouzes et al. 2009), directed evolution of cell therapy (Carlo 2019), antibody discovery (Mazutis et al. 2013; Shembekar et al. 2018), and monitoring of dynamic cell–cell interactions (Sarkar et al. 2016, 2017; Konry et al. 2013). The compartmentalization of individual cells or cell pairs in monodispersed aqueous droplets that are suspended in an immiscible oil carrier fluid serves as micro-reactors and circumvents cross-interference from neighboring droplets. Droplets can be individually manipulated and interrogated for complex biological assays. Owing to the drastic reduction of volumetric confinement of cells in droplets, the secreted molecules from a single cell can quickly reach a detectable concentration, which improves the signal-to-noise ratio in comparison to the bulk co-culturing method (Konry et al. 2011, 2016).
Although the workflow for encapsulating cells in droplets is relatively straightforward, single-cell encapsulation suffers an inherent fundamental challenge in its ability to control the number of cells per droplet. The randomly dispersed cell aqueous suspension is typically diluted prior to loading into droplets; thus, cell encapsulation statistics is often dictated by Poisson statistics (Fig. 1a). Therefore, the distribution of the number of cells per droplet is governed by
where \(k\) is the number of cells in a droplet and \(\lambda\) is the average number of cells per droplet assuming complete random dispersion. Furthermore, the value of \(\lambda\) is directly proportional to the frequency of each cell entering the droplet generation junction and inversely dependent on the frequency of droplet generation rate (Eq. 2).
a Poisson distribution of single-cell encapsulation in droplets. b–d Distribution of double Poisson statistics for 1–1–1 encapsulation. b The probability to co-encapsulate two distinct cells in a droplet with respect to both \({\lambda }_{A}\) average number of cells type A per droplet and \({\lambda }_{B}\) cell type B per droplet. c The probability of 1–1–1 encapsulation efficiency with respect to both average number of cell A and B less than one \(({\lambda }_{A} and {\lambda }_{B}<1)\) and dictated by the minimum of \({\lambda }_{A} and {\lambda }_{B}<1\). d The 1–1–1 encapsulation efficiency that follows double Poisson statistics exhibits a sigmoidal relationship against the minimum of \({\lambda }_{A} \mathrm{and} {\lambda }_{B}=1\)
The change in \(\lambda\) produces a different distribution of cell occupancy per droplet, where the number of droplets containing exactly one cell is maximized at 36.8% when \(\lambda\) is equal to one. Moreover, the fraction of droplets containing more than one cell becomes insignificant if the average number of cells per droplet is low (\(\lambda <0.3)\). However, the fraction of empty droplets is then accounted for a significant majority of the droplet population that reduces throughput, wastes precious reagents, and complicates the subsequent analytical processes.
This stochastic encapsulation will lead to even more cumbersome and inefficient experiments in co-encapsulating one cell type A and one cell type B in one droplet, termed 1–1–1 encapsulation. Consider a random co-encapsulation of two distinct cells A and B with their respective cell density of \({\lambda }_{A}\) and \({\lambda }_{B}\), the co-encapsulation efficiency is governed by double Poisson statistics (Eq. 3).
For 1–1–1 encapsulation efficiency, the Poisson limited single-cell encapsulation further reduces the probability of co-encapsulating two distinct cells per droplet to at most 13.5% under the optimal condition, whereas most droplets contain either no cells or incorrect cell pairing (Fig. 1b). When compared with the common cell loading density that is typically used in FACS (\({\lambda }_{A}={\lambda }_{B}=0.3\)), the 1–1–1 encapsulation efficiency can be only achieved as low as 4.9%, and over half of the generated droplets (55%) contain no cells. Specifically, in cases where both cell densities are below one, the minimum of \({\lambda }_{A}\) and \({\lambda }_{B}\) dictates the maximum achievable 1–1–1 encapsulation efficiency and suggests a partially linear relationship (Fig. 1c). Thus, the theoretical 1–1–1 encapsulation statistics with respect to the minimum of \({\lambda }_{A}\) and \({\lambda }_{B}\) illustrated in Fig. 1d depicts a sigmoidal relationship for \(\lambda\) between zero and one.
To circumvent this Poisson limitation, both active and passive microfluidic techniques have been devised to improve the encapsulation process in the aspect of cell ordering or manipulation. Collectively, both existing techniques suffer either from high system complexity, low throughput, limited versatility, or poor encapsulation efficiency when encapsulating cells with low concentration. Active technique involves acoustic (Collins et al. 2013), optical (Mingyan He et al. 2005), or magnetic force (Chen et al. 2013) to manipulate cells leading into the droplet generation region and actively generates droplets for cell encapsulation. The use of external force requires electrodes, actuators, or lasers to execute the operation, thus increasing the complexity of droplet generation process. These active methods also have a limited droplet generation rate that is orders of magnitude lower than passive encapsulation. In contrast, passive encapsulation merely relies on hydrodynamic effect and channel features, including inertial (Edd et al. 2008), viscoelastic effect (Shahrivar and Giudice 2021), pinched flow fractionation (Ramji et al. 2014), and gravitational field (Gk and As 2012). Specifically, passive techniques that rely on inertial or Dean force to achieve two ordered particle trains prior to co-encapsulation have been demonstrated with improved 1–1–1 encapsulation efficiency. However, the performance of these passive methods is critically dependent on the flow rates and the properties of particulates and fluids with the prerequisite of high cell loading density (\(\lambda >1)\).
As the 1–1–1 encapsulation efficiency remains a significant technical challenge, we present a passive compartmentalization platform that leverages hydrodynamic draining for close packing of cells to overcome the limitation on double Poisson co-encapsulation efficiency. The presented draining technique in achieving close packing of cells offer simplicity and adaptability, which could increase platform compatibility for various reagents and lower cell loading density. In contrast to other passive methods, this platform is applicable to increase 1–1–1 encapsulation efficiency with the pairing of two different types of cells or with 1-cell and 1-bead encapsulated in a droplet.
2 Materials and methods
2.1 Microfluidic device design
The 1–1–1 co-encapsulation microfluidic device consists of four inlets and two outlets as illustrated in Fig. 2. Three of the four inlets are used for the perfusion of aqueous phases, including two for separate cell loading and one for sheath flow, whereas the remaining inlet is for the perfusion of continuous oil phase. The two outlets serve as droplet collection and aqueous draining for the cell loading streams, respectively. Both the aqueous and oil phases are delivered into the microfluidic device at a constant volumetric flow rate, where the draining outlet withdraws the aqueous phase at a constant volumetric flow rate.
Design principle of passive 1–1–1 encapsulation device. a Schematic of the passive 1–1–1 encapsulation device that consists of four inlets (two cell suspensions, one sheath flow, and one oil) and two outlets (drainage and collection). b Close-up schematic of the 1–1–1 encapsulation device of the drainage and droplet generation junction. The single-filed close packed cell trains merge with sheath flow at the main channel and collectively enter the high shear droplet generation junction to form aqueous droplets upon contacting the immiscible carrier oil. c An initial distance L between the leading and trailing cell enters the drainage junction. The effect of draining causes close packing of cells that results in the initial cell–cell distance L shortening by a distance of \(\updelta\) until the trailing cell also enters the drainage region
The flow of each cell suspension solution, \({Q}_{cell}\), enters the device and passes through a narrow channel with a width of 20 μm and a height of 40 μm. These channel dimensions facilitate the self-assembly of a randomly dispersed cell suspension into a centered single-cell train. Each cell train subsequently moves toward the drainage junction for close packing. As two cells with initial distance L within a cell train subsequently enters the junction, the flow stream \({Q}_{\mathrm{cell}}\) splits into three directions that flows toward both sides of the draining channels \({Q}_{\mathrm{drain}}\) and downstream for encapsulation \({Q}_{\mathrm{cell},\mathrm{ drain}}\). The volumetric flow rates before and after are equal due to the conservation of mass, which can be described as:
Given the volumetric flow rate is the product of channel cross-sectional area A and velocity v or \(Q=Av\), the Eq. 4 can be described as:
where the cell stream channel cross-sectional area \({A}_{\mathrm{cell}}\) is the same for before and after drainage junction. With absence of draining, the velocity of a cell is the same for before and after drainage, where \({v}_{\mathrm{cell}}={v}_{\mathrm{cell},\mathrm{drain}}\). When \({v}_{\mathrm{drain}}>0\), the velocity of a cell decreases after the drainage junction due to \({v}_{\mathrm{cell}}-{v}_{\mathrm{cell},\mathrm{drain}}>0\). While a fraction of \({Q}_{\mathrm{cell}}\) is diverted into two draining channels, the velocity of the leading cell decreases as the trailing cell catches up. At a given drain rate, this transient discrepancy of velocity between the leading and trailing cell results in the initial cell–cell distance L shortening by a distance of \(\delta\) until the trailing cell also enters the drainage junction, which leads to close packing of cells with a cell–cell spacing of \(L-\delta\) as the cells exit the drainage region. The shorten distance δ increases as the drain rate increases that further reduces the cell–cell spacing. The shortened cell–cell spacing effect from drainage junction improves the cell loading process by attaining higher on-chip cell density prior to entering the droplet generation junction, which plays a vital role in enhancing 1–1–1 encapsulation efficiency.
Each drainage junction contains a pair of pillar arrays with 10 μm gap that are positioned on both sides of the channel at a 45º backward angle with respect to the flowing direction. The bilateral draining flows withdraw a fraction of the solution of cell suspension while retaining cells inside the channel. All draining flows are designed to maintain equal hydrostatic pressures and flow rates, where the bifurcation channel design with equidistance from the draining outlet to the four draining channels is employed to ensure equivalent channel resistance. The balanced pressure on both sides of the draining junction is imperative to mitigate cells from escaping the channel to prevent adverse cell loss. In addition, the backward drainage angle of the pillar array acts as a filter and induces microvortices to pivot any escaped cells back into the channel. See the high-speed video recording of the drainage junction in the supporting information. (Movies S1, Supporting Information). The process of draining a fraction of \({Q}_{cell}\) does not only increasing on-chip cell density but also effectively reduces the longitudinal spacing between cells. Due to the increased cell density and shortened cell–cell spacing, the \(\lambda\) value of its respective cell type also increases which results in improving the probability of 1–1–1 encapsulation and attenuating the number of empty droplets.
Both cell trains with shortened cell spacing merge with the sheath flow, where it focuses both cell trains to its respective side of the channel. Given the nature of laminar flow at a low Reynolds number in microfluidic devices, the sheath flow acts as a divider to separate two cell trains to prevent premature interaction before co-encapsulation. In addition, bioassay reagent of interest could also be used as the sheath flow to warrant a temporal control of a reaction. The sheath flow and two streams of cell trains collectively interface with the continuous oil phase at the nozzle to form droplets. The immiscible oil phase symmetrically exerts interfacial shear at the aqueous stream to form droplets at a rate that is optimized for the rate of the cell arriving frequency. The flow rate control of \({Q}_{Cell}\) and the regime of droplet generation are discussed in the Results and Discussion section.
2.2 Device fabrication
The microfluidic devices were fabricated in polydimethylsiloxane (PDMS, Sylgard 184, Dow Corning) following standard soft lithography. The channel geometry was designed in computer-aided design software and printed on mylar masks. The silicon wafer substrates were spin-coated with SU-8 2050 negative photoresist (MicroChem) and subsequently patterned by ultraviolet exposure to form master molds. Polydimethylsiloxane (PDMS) elastomer was mixed with curing agent at a 10:1 ratio and degassed before pouring onto the master mold, followed by curing at 65 °C overnight for complete crosslinking. The PDMS molded imprints and glass microscope slides were oxygen plasma treated (Harrick Plasma Inc) for 2 min and bonded together to form a permanent seal. The devices were baked in an oven at 120 °C overnight to secure their natural hydrophobicity.
2.3 Cell culture
Human erythromyeloblastoid leukemia cells K562 (American Type Culture Collection (ATCC)) were used to assess the performance of 1–1–1 encapsulation. K562 cells were cultured in a T-75 cell culture flask using RPMI 1640 medium (Gibco) supplemented with 10% (v/v) fetal bovine serum (Gibco), and 100 U/mL penicillin–streptomycin (Gibco). The cells were cultured in a condition of 5% CO2 humidified incubator at 37 °C and passaged every 2–3 days with a seeding density of 1 \(\times\) 106 cells/mL. In preparation for encapsulation, the aqueous phase solution was prepared in RPMI medium supplemented with 16% Optiprep, 1% BSA, 0.01% Triton X-100, and 10 U/mL of DNase I, which would serve as sheath flow and cell suspension. Subsequently, the cells were resuspended in the aqueous phase at a desired volume fraction, ranging from 2 to 2.5%. For the continuous phase, 2% (w/v) 008-FluoroSurfactant in HFE 7500 oil (Ran Biotechnologies) was used.
2.4 Experimental setup
Cell suspensions, sheath flow solution, and oil were separately loaded into the microfluidic co-encapsulation device using four 1 mL plastic syringes (BD, Breda, Netherlands) and were connected to their respective inlets with PTFE tubing (Cole Parmer, ID 0.022 inch and OD 0.042 inch). A syringe filled with aqueous solution was connected to the drainage outlet with PTFE tubing, in which the absence of air in a syringe would prevent any gas contraction or expansion to ensure a constant drain rate. Lastly, PTFE tubing was connected to the outlet of the device to direct droplets into a collection Eppendorf tube. Both the dispersed phase and the continuous phase are delivered into the microfluidic device at constant volumetric flow rate and individually controlled by syringe pump (Pico Plus; Harvard Apparatus, Inc., MA, USA). Syringe pump as flow control system is desirable due to its consistent volumetric flow delivery and independent of channel resistance. The flow rate of the continuous oil phase was set at 5 µL/min, and the dispersed aqueous phase for cell was set at 3–3.5 µL/min. The sheath flow was initially set at 1 µL/min and could be adjusted to tune the droplet generation rate as needed. The syringe for draining was mounted on a syringe pump with withdrawal mode at a flow rate of 0-2.5 µL/min to remove the aqueous solution.
The microfluidic chip was mounted onto the stage of a Nikon 100-S inverted microscope and monitored using a computer-controlled high-speed Phantom camera V-310 (Vision Research, Wayne, NJ) for image recording. Droplet generation videos were acquired at 10,000 frames per second. A customized MATLAB code was utilized to assess cell–cell longitudinal spacing. To obtain co-encapsulation statistics, we analyzed the high-speed videos frame by frame during cell co-encapsulation using ImageJ, a public domain Java-based image processing software program developed at the National Institutes of Health.
3 Results and discussion
3.1 Droplet generation rate for an effective 1–1–1 encapsulation
The droplet generation rate is a function of flow rates of both dispersed and continuous phase, size dimensions of a nozzle, and interfacial tension. Droplet production processes have a direct impact on the efficiency of 1–1–1 encapsulation of cells, which is critical to understand the rate of droplet generation with respect to its associated parameters. A large discrepancy between droplet generation frequency and cell arrival rate at the nozzle can be detrimental to the overall encapsulation efficiency. Thus, the effect of droplet generation frequency and cell arrival rate are investigated for various flow rates for optimal 1–1–1 encapsulation.
The droplet generation frequency exhibits a positive relationship as the flow rate of dispersed phase increases, as well as the increasing of continuous oil flow rate (Fig. 3a). The results are congruent with the physics theory of droplet generation, where the modes of droplet formation can be determined according to the capillary number Ca.
a Characterization of droplet generation frequency with respect to various aqueous and oil flow rate. b The arrival frequency of cells at the droplet generation nozzle is approximated under the assumption of uniformly dispersed cells. The frequency difference among various cell concentrations increases as the cell flow rate increases
The capillary number Ca is a non-dimensional quantity and is dependent on viscosity \(\mu\) and characteristics velocity U of the continuous phase and surface tension \(\gamma\) of the water–oil interface. With the increasing value of Ca, the mode of droplet generation would be in transition in the order of squeezing, dripping, and jetting regime.
In the dripping regime, the Ca number is high enough, such that the droplet formation is shear dominant with a generation frequency in the order of thousands of droplets per second. Particularly, the droplet generation frequency reaches roughly 7000 droplets per second on the 1–1–1 encapsulation device (Fig. 4a). This high droplet generation frequency would lead to an exceedingly low \(\lambda\) value according to Eq. 2, which results in generating a plenitude of empty droplets. Such a high fraction of empty droplets is challenging to be offset by the number of droplets containing correct pairing, which conversely undermines the efficiency of 1–1–1 co-encapsulation. Despite higher cell loading density or flow rate \({Q}_{cell}\) could increase the frequency of cells arriving at the nozzle, this method can often lead to channel clogging from highly concentrated cell suspension or excessive shear stress at high flow rate. In the squeezing regime when the Ca value is low, the tip of dispersed phase transiently blocks the nozzle and protrudes outward into the continuous phase. The neck of the interface is squeezed from the increased pressure until it breaks off (Fig. 4b). This mode of droplet generation offers a relatively slower generation frequency near or below one thousand droplets per second that is suitable for encapsulating cells with lower cell loading density.
a Droplet generation at squeezing regime, where the liquid–liquid interface makes contact at both sides of the nozzle before breakoff. b Droplet generation at dripping regime, where the droplet formation is shear-dominated by the continuous phase and the liquid–liquid interface separates from the nozzle
For a passive device, the droplet generation rate could not be actively modulated to accommodate any fluctuation of cell arrival rate due to intrinsic random dispersion of cells. However, the droplet generation in squeezing regime can tolerate for the fluctuation of cell spacing without adversely compromising droplet throughput. Assuming cells are uniformly dispersed, the analytical simulation on the arrival rate of cells at droplet generating nozzle demonstrates a positive linear relationship with the flow \({Q}_{\mathrm{cell}}\) for various cell density (Fig. 3b). Consider the cell loading density of \(10\times {10}^{6}\) cells/mL with local cell density fluctuation between 5 to 15 \(\times {10}^{6}\) cells/mL, a \({Q}_{cell}\) flow rate of 8 µL/min can trigger an extensive range of cell arrival frequency in comparison to the one with slower flow rate. Due to the fixed droplet generation rate, this broad range of cell arrival frequency would lead to large number of droplets with incorrect encapsulation. Slower flow rate \({Q}_{\mathrm{cell}}\) is optimal as it presents a narrower range of cell arrival frequency. Furthermore, the cell arrival frequency for high flow rate \({Q}_{\mathrm{cell}}\) can quickly reach over one thousand cells per second that demands a higher droplet generation rate in dripping regime. The droplet generation rate over a thousand droplets per second can lead to a plethora of empty droplets. Thus, a slower flow rate \({Q}_{\mathrm{cell}}\) does not only lead to a narrower range of cell arrival rate despite the fluctuation of cell density but also falls in the same order of magnitude with the rate of droplet generation in squeezing regime. Lastly, the modulation of flow rate in sheath flow enables a fine adjustment of droplet generation rate. As a result, the droplet generation in squeezing regime and slower flow rate \({Q}_{\mathrm{cell}}\) are chosen for the assessment of the 1–1–1 encapsulation platform.
3.2 Self-assembly of cell trains through close packing
The drainage junction consists of pillar arrays on both sides of each cell channel, which enables balanced drain rates and pressures that are exerted on each passing cell to mitigate the occurrence of cells escaping the channel. The pillar array serves as a filter with 10 \(\mu\) m gaps and is slanted at a 45° angle backward with respect to the flow direction. We discovered that this pillar design could induce microvortices to redirect any escaped cells back into the cell channel to further prevent cell loss. A computational fluid dynamic simulation was performed to analyze the velocity flow fields at the drainage junction. Single phase creeping flow module was utilized for the simulation, where the flow rate \({Q}_{\mathrm{cell}}\) was set at 2 μL/min and the total drain rate at 1.25 μL/min. At the drainage junction, the flow \({Q}_{\mathrm{cell}}\) is divided into three different directions, such that the cell suspension solution either flows straight to merge with the sheath flow downstream or diverts to the pillar arrays on both sides. The velocity field indicates that the angle of pillar position facilitates the formation of recirculation at the downstream part of the array (Fig. 5). In addition to the equal drain rates, the recirculation is beneficial to further mitigate cell loss to redirect the escaped cells back into the flowing stream. The drain rates tested in the experiments did not present any significant cell loss from the cell suspension solution. However, exceedingly high drain rate could contribute a detrimental effect on cell loss, where majority of the cell suspension solution would preferentially flow toward the pillar arrays on both sides. This is attributed to the lower channel resistance of the pillar arrays in comparison to that of the cell channel, where the cross-sectional area of the pillar arrays is six-fold of the cell channel. Thus, the optimization of the draining rate and the flow \({Q}_{cell}\) is imperative for achieving an effective close packing of cells.
To evaluate the efficacy of close packing of cells from the drainage junction, a custom-made MATLAB script was developed to analyze the distance of cell spacing. Briefly, slow-motion videos captured by the high-speed Phantom camera were used to analyze each frame. The grayscale frames were converted into binary format, which enables the identification of cells. Subsequently, two regions of interest were selected at the channel entrance and exit of the drainage junction to detect the arrival of cells. When a cell crosses the region-of-interest area, a spike of pixel color will be detected. Peak-to-peak analysis was utilized to obtain the statistics of spacing between cells.
For cell spacing analysis, a suspension of K562 cells was delivered into the device at a constant flow rate of 3 µL/min, and three different draining flow rates (0, 1.25, and 2.5 µL/min) were examined. No significant difference was observed in cell spacing before and after the drainage junction in the absence of draining (Fig. 6a). Each passing cell flows at the same velocity across the drainage junction as no volume of cell suspension is removed, thus the lack of velocity difference between cells results in \(\updelta\) to be zero. Conversely, the presence of drain rate removes a fraction of cell suspension volume that leads to differential in the velocity of flowing cell before and after the drainage. This velocity difference between the leading and trailing cells within a cell train leads to the positive value of \(\updelta\) that shortens the cell–cell distance. In addition to both drain rates of 1.25 and 2.5 µL/min showed a substantially shortened cell spacing after drainage junction (Fig. 6b, c), the value of \(\updelta\) is also dependent on the level of drain rate. As high volume is being removed in the drain rate of 2.5 µL/min, the velocity of a leading cell would be lower in comparison to that of a drain rate of 1.25 µL/min, which increases the time for a trailing cell to catch up and increases the value of \(\updelta\) for shortening cell–cell spacing. Consequently, the drain rate of 2.5 µL/min indicates nearly half of cell–cell spacing reduction across a drainage junction, where there is 21% reduction for drain rate of 1.25 µL/min. According to Fig. 6d, an average cell–cell spacing before drainage junction is around 200 µm, which is equivalent of a cell arrival rate of 300 cells per second. The drain rate of 2.5 µL/min effectively enhances the on-chip cell density that results in a 21% increase of cell arrival rate. Thus, the drainage junction can effectively close pack a randomly dispersed cell train to a level that minimizes the number of empty droplets when the droplet generation rate is fixed.
Cell spacing assessment on drainage effect using 3 µL/min cell flow rate of K562 cells in 10 million/mL density and various drain rates. a With no draining, the cell spacing does not show any statistical difference in before and after drainage junction (p > 0.1). b The drain rate of 1.25 µL/min demonstrated a significant shift in cell spacing after the drainage junction (p < 0.001). c The cell spacing distance was significantly shortened under the drain rate of 2.5 µL/min (p < 0.001). d The average cell–cell spacing under various drain rates 0, 1.25, and 2.5 µL/min (N = 3)
Although the drainage junction reduces the cell–cell distance of \(\delta\), the intrinsic random cell spacing persists with decreased range of dispersion. With a more regulated cell–cell spacing, the frequency of droplet generation is crucial to accommodate the random dispersed cell train. As the cell arrival rate after drainage junction is roughly 400 cells per second, the droplet generation rate is set at a rate of a thousand droplets per second. At this droplet generation rate, it can accommodate the randomness of cell–cell spacing that minimizes the fraction of droplets with more than 2 cells of the same cell type. Therefore, the increase of cell arrival rate and narrower range of cell–cell spacing contribute to the improvement of 1–1–1 co-encapsulation efficiency.
3.3 Assessment of 1–1–1 efficiency
The 1–1–1 encapsulation device was applied to co-compartmentalize K562 cells with two different densities to demonstrate performance (Fig. 7a). Each flow rate of cell suspension solution was set to 3 µL/min with a draining rate of 2.5 µL/min, whereas the sheath flow and continuous oil were, respectively flowing at 1 µL/min and 5 µL/min. See the high-speed video recording of the co-encapsulation in the supporting information. (Movies S2, Supporting Information). The droplet generation frequency was maintained at around 980 droplets per second, which resulted in the expected number of cells per droplet to have a \(\lambda\) value of 0.4 for the given cell loading densities. The encapsulation probability for both cell channels is illustrated in Fig. 7b in comparison with its theoretical single-cell Poisson distribution. The increase in cell arrival rate led to the improvement of single-cell encapsulation efficiency to be as much as 43% with respect to Poisson statistics. The close packing of cells decreases the overall cell-cell spacing, which reduces 23% on the number of empty droplets for both channels. The enhancement of single-cell encapsulation and reduction on the number of empty droplets directly relate to the improvement on the 1-1-1 encapsulation efficiency.
Encapsulation statistical analysis on 1–1–1 encapsulation device. a Bright-field microscopic image demonstrates the co-encapsulation of cells in monodispersed droplets. b Comparison of the distribution on various number of cells per droplet against Poisson statistics with \(\uplambda =0.4\) (N = 3 with average 210 droplets). c Distribution of experimental co-encapsulation statistics normalized against double Poisson distribution (N = 3 with average 210 droplets)
Owing to the benefits of drainage junction, the 1–1–1 encapsulation efficiency achieved as high as 21% of the droplets contain correct pairing, whereas the double Poisson efficiency is only 12%. The overall co-encapsulation statistics over three separate experiments with cell density of \(\lambda \approx 0.4\) is normalized against the double Poisson (Fig. 7c). The 1–1–1 encapsulation devices were performed to yield an average of over 2.3-fold improvement in 1–1–1 encapsulation efficiency, which exceeds the intrinsic limited double Poisson statistics. One strong assumption behind the Poisson distribution is that cells arrive independently in droplets. This is not the case as soon as there are interactions between cells, for instance, cells adhere to each other that are closely packed as they are delivered into the microfluidic device. Such an increase in 1-1-1 encapsulation efficiency is the result of the packing of cells upon entering the droplet generation junction, which also resulted in the increase of λ values of cell loading density. In addition, the distribution of the number of cells per droplet demonstrates that over 38% reduction in the number of empty droplets, which is consistent with the single-cell encapsulation results in Fig. 7b. The fraction of single-cell cell during co-encapsulation is comparable with the double Poisson statistics, whereas an increment in the fraction of droplets containing three or more cells per droplet was observed. Despite the increase on the number of droplets with multiple cells, it only constitutes only 8% of the total droplets. This observation is the result of close packing of cells prior to encapsulation. As a result, the technique of hydrodynamic draining to achieve close packing of cell can overcome the double Poisson limitation with over 2.3-fold improvement in 1-1-1 encapsulation efficiency, which indicates the robustness of the performance.
4 Conclusion
In this work, we presented a passive microfluidic device that utilizes close packing of cells through draining of aqueous phase and hydrodynamic sheath flow for single-file cell trains prior to droplet generation. With these passive techniques to achieve the ordering of cell trains, the cell–cell distance has been substantially shortened to reduce the fraction of empty droplets. Another notable feature of this technique is that it could perform high-efficiency co-encapsulation at low cell concentrations (\(\lambda <1\)), and this platform is all passive, controlled only by the flow rates of the two phases, which does not require complex channel structures or active elements. Thus, the resulting 1–1–1 co-encapsulation efficiency surpasses the double Poisson limitation by over two-fold improvement for pairing two separate cells in droplets at 1 kHz rate. Furthermore, the improved encapsulation efficiency in single cell and cell pairing from this platform would be particularly beneficial for cell–cell pairing analysis by providing an abundance of useable droplets for various control groups during cellular analysis. One limitation of this technique is the intrinsic random dispersion of cells where the fluctuation of cell density is too large. Extensive variation in cell spacing, particularly in between cell trains, could be detrimental to the overall encapsulation efficiency. Progressive draining with a serial drainage junction holds a great potential to further improve the uniformity of cell-cell spacing. Different from other passive techniques such as inertial microfluidics, the 1–1–1 encapsulation platform presented here does not rely on the intrinsic properties of cells, such as cell size, concentration, and stiffness, or rheology of fluids to attain improvement of co-encapsulation efficiency. Therefore, the platform minimizes the dependency on the properties of cells and fluids without compromising throughput, versatility, and simplicity of the encapsulation of cell pairs in droplets. This simple, passive, and promising 1–1–1 co-encapsulation platform has the potential for a broader range of applications in single-cell or cell pairing analysis and is compatible with barcoded-based genomic analysis.
References
Agrawal B, Gendler SJ, Longenecker BM (1998) The biological role of mucins in cellular interactions and immune regulation: prospects for cancer immunotherapy. Mol Med Today 4(9):397–403
Altschuler SJ, Wu LF (2010) Cellular heterogeneity: do differences make a difference? Cell 141(4):559–563
Bl B, Is J (2014) Cell–cell communication in the tumor microenvironment, carcinogenesis, and anticancer treatment. Cell Physiol Biochem 34(2):213–43. Available from: https://pubmed.ncbi.nlm.nih.gov/25034869/
Bogdanowicz DR, Lu HH (2013) Studying cell-cell communication in co-culture. Biotechnol J 8(4):395. Available from: /pmc/articles/PMC4230534/
Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB et al (2009) Droplet microfluidic technology for single-cell high-throughput screening. Proc Natl Acad Sci USA 106(34):14195–200. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2732882&tool=pmcentrez&rendertype=abstract
Chattopadhyay PK, Gierahn TM, Roederer M, Love JC (2014) Single-cell technologies for monitoring immune systems. Nat Immunol 15:2
Chen Z, Chen JJ, Fan R (2019) Single-cell protein secretion detection and profiling. Annu Rev Anal Chem 12:431–449
Chen A, Byvank T, Chang WJ, Bharde A, Vieira G, Miller BL et al (2013) On-chip magnetic separation and encapsulation of cells in droplets. Lab on a Chip 13(6):1172–81. Available from: https://pubs.rsc.org/en/content/articlehtml/2013/lc/c2lc41201b
Collins DJ, Alan T, Helmerson K, Neild A (2013) Surface acoustic waves for on-demand production of picoliter droplets and particle encapsulation. Lab on a Chip 13(16):3225–31. Available from: https://pubs.rsc.org/en/content/articlehtml/2013/lc/c3lc50372k
den Haan JMM, Arens R, van Zelm MC (2014) The activation of the adaptive immune system: cross-talk between antigen-presenting cells, T cells and B cells. Immunol Lett 162(2):103–112. https://doi.org/10.1016/j.imlet.2014.10.011
Dhar M, Lam JN, Walser T, Dubinett SM, Rettig MB, di Carlo D (2018) Functional profiling of circulating tumor cells with an integrated vortex capture and single-cell protease activity assay. Proc Natl Acad Sci 115(40):9986–9991
di Carlo D (2019) Technologies for the directed evolution of cell therapies. SLAS Technol 24(4):359–372
Edd JF, di Carlo D, Humphry KJ, Köster S, Irimia D, Weitz DA et al (2008) Controlled encapsulation of single-cells into monodisperse picolitre drops. Lab Chip 8(8):1262–1264
Gk K, As B (2012) Field-free particle focusing in microfluidic plugs. Biomicrofluidics 6(2). Available from: https://pubmed.ncbi.nlm.nih.gov/22655011/
Konry T, Dominguez-Villar M, Baecher-Allan C, Hafler DA, Yarmush ML (2011) Droplet-based microfluidic platforms for single T cell secretion analysis of IL-10 cytokine. Biosensors Bioelectron 26(5):2707–2710. https://doi.org/10.1016/j.bios.2010.09.006
Konry T, Golberg A, Yarmush M (2013) Live single cell functional phenotyping in droplet nano-liter reactors. Sci Rep 3:1–5
Konry T, Sarkar S, Sabhachandani P, Cohen N (2016) Innovative tools and technology for analysis of single cells and cell-cell interaction. Annu Rev Biomed Eng 18(1):259–284
Kweon S, Phan MTT, Chun S, Yu HB, Kim J, Kim S et al (2019) Expansion of human NK cells using K562 cells expressing OX40 ligand and short exposure to IL-21. Front Immunol 10:879
Lawson DA, Kessenbrock K, Davis RT, Pervolarakis N, Werb Z (2018) Tumour heterogeneity and metastasis at single-cell resolution. Nat Cell Biol 20(12):1349–1360. https://doi.org/10.1038/s41556-018-0236-7
Liu L, Dalal CK, Heineike BM, Abate AR (2019) High throughput gene expression profiling of yeast colonies with microgel-culture Drop-seq. Lab Chip 19(10):1838–1849
Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M et al (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161(5):1202–14. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26000488.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4481139
Mazutis L, Gilbert J, Ung WL, Weitz DA, Griffiths AD, Heyman JA (2013) Single-cell analysis and sorting using droplet-based microfluidics. Nat Protoc 8(5):870–891
Mingyan He J, Edgar S, Jeffries GDM, Lorenz RM, Patrick Shelby J, Chiu DT (2005) Selective encapsulation of single cells and subcellular organelles into picoliter- and femtoliter-volume droplets. Anal Chem 77(6):1539–1544. https://doi.org/10.1021/ac0480850
Ramji R, Wang M, Bhagat AAS, Weng DTS, Thakor NV, Lim CT et al (2014) Single cell kinase signaling assay using pinched flow coupled droplet microfluidics. Biomicrofluidics 8(3):3–11. https://doi.org/10.1063/1.4878635
Sarkar S, Sabhachandani P, Stroopinsky D, Palmer K, Cohen N, Rosenblatt J et al (2016) Dynamic analysis of immune and cancer cell interactions at single cell level in microfluidic droplets. Biomicrofluidics. https://doi.org/10.1063/1.4964716
Sarkar S, Sabhachandani P, Ravi D, Potdar S, Purvey S, Beheshti A et al (2017) Dynamic analysis of human natural killer cell response at single-cell resolution in B-cell non-hodgkin lymphoma. Front Immunol 8:1–13
Shahrivar K, del Giudice F (2021) Controlled viscoelastic particle encapsulation in microfluidic devices. Soft Matter 17(35):8068–8077
Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D et al (2014) Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510(7505):363–369
Shelton SE, Nguyen HT, Barbie DA, Kamm RD (2021) Engineering approaches for studying immune-tumor cell interactions and immunotherapy. Science 24(1):101985
Shembekar N, Hu H, Eustace D, Merten CA (2018) Single-cell droplet microfluidic screening for antibodies specifically binding to target cells. Cell Rep 22(8):2206–2215. https://doi.org/10.1016/j.celrep.2018.01.071
Shi Q, Qin L, Wei W, Geng F, Fan R, Shik Shin Y et al (2012) Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc Natl Acad Sci 109(2):419–24. Available from: http://www.pnas.org/cgi/doi/https://doi.org/10.1073/pnas.1110865109
Streltsova MA, Erokhina SA, Kanevskiy LM, Grechikhina M v., Kobyzeva PA, Lee DA et al (2019) Recurrent stimulation of natural killer cell clones with k562 expressing membrane-bound interleukin-21 affects their phenotype, interferon-γ production, and lifespan. Int J Mol Sci 20:443 [Internet]. 2019 Jan 21 [cited 2022 Aug 17];20(2):443. Available from: https://www.mdpi.com/1422-0067/20/2/443/htm
Vu TQ, de Castro RMB, Qin L (2017) Bridging the gap: microfluidic devices for short and long distance cell-cell communication. Lab Chip 17(6):1009–1023
Zalfa C, Paust S (2021) Natural killer cell interactions with myeloid derived suppressor cells in the tumor microenvironment and implications for cancer immunotherapy. Front Immunol 5(12):565
Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R et al (2017) Massively parallel digital transcriptional profiling of single cells. Nat Commun 8:1 [Internet]. 2017 Jan 16 [cited 2021 Sep 7];8(1):1–12. Available from: https://www.nature.com/articles/ncomms14049
Funding
The authors acknowledge support from the National Science Foundation and industrial members of the Center for Advanced Design and Manufacturing of Integrated Microfluidics (NSF I/UCRC award no. IIP 1841509).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (MP4 15463 KB)
Supplementary file2 (MP4 127073 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Luo, X., Lee, A.P. Overcoming double Poisson limitation for co-encapsulation in droplets through hydrodynamic close packing of cells. Microfluid Nanofluid 27, 3 (2023). https://doi.org/10.1007/s10404-022-02600-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10404-022-02600-9