Abstract
Aim
To optimize vaccination strategy, evidence on vaccine efficacy against COVID-19 is needed.
Method
The present network meta-analysis uses reconstructed individual patient data from phase III trials on vaccine efficacy (VE), identified through MEDLINE, EMBASE, and Cochrane library (CENTRAL) peer-reviewed and published in English before August 31, 2021. The primary outcome was the VE against confirmed COVID-19 at any time after the first dose as defined in each study. VE was re-estimated using the two-stage approach. Poisson regression models were applied to each trial at the first stage, and the incidence risk ratio (IRR) and their 95% CI were aggregated to allow random-effects network meta-analysis (NMA) at the second stage. VE was expressed as: (1-IRR) × 100. The study protocol is registered in PROSPERO (CRD42020200012).
Results
A total of eight studies, evaluating nine different vaccines were identified and analyzed. Between April 23, 2020 and January 05, 2021, 210,418 participants were recruited in 354 sites worldwide. During a median (IQR) follow-up duration of 69.8 (69.7–70.3) days, 2131 confirmed COVID-19 cases occurred (604; 26.0 per 1000 person–years in vaccine recipients and 1527; 85.9 per 1000 person–years in the control group). The mRNA-1273 vaccine was the most effective (P-score 0.99); at any time after dose 1, incidence reduction for mRNA-1273 ranged from 78% to 98% compared to the other vaccines.
Conclusion
Our results provide evidence for the short-term superiority of mRNA vaccines, especially the mRNA-1273 vaccine in prevention of COVID-19 in different populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mass vaccination campaigns significantly contribute to containing the COVID-19 pandemic. Despite the high circulation of variants of concern, the efficacy of the current vaccines approved worldwide for emergency use authorization (EUA) against COVID-19 in the general population (Dagan et al. 2021; Jara et al. 2021; Kissling et al. 2021) remains similar to that reported in clinical trials (Baden et al. 2021; Polack et al. 2020; Voysey et al. 2021; Logunov et al. 2021; Sadoff et al. 2021; Al Kaabi et al. 2021; Heath et al. 2021; Tanriover et al. 2021). To optimize the vaccination strategy and guide public health recommendations, evidence on the most effective and safe vaccine against COVID-19 is needed. Previous meta-analysis on COVID-19 vaccine efficacy had focused on aggregate data, which were limited to evaluate the dynamic over time of the vaccine efficacy (Harder et al. 2021; Sharif et al. 2021). To address this issue, we initiated in July 2020 a network meta-analysis protocol (PROSPERO registration: CRD42020200012) to compare and rank the efficacy of COVID-19 vaccines using a reconstructed individual patient data (IPD) from published Kaplan–Meier curves due to heterogeneity of time-point of analysis and statistical methods. Here, we report preliminary results on comparison and ranking efficacies of vaccines against COVID-19.
Methods
Search strategy and selection criteria
We searched through MEDLINE, EMBASE, and Cochrane library (CENTRAL) peer-reviewed phase 3 randomized controlled trials (RCTs) that investigated COVID-19 vaccine efficacy, published in English before August 31, 2021. We included RCTs that compared efficacy against any confirmed cases of COVID-19 using reverse transcriptase polymerase chain reaction (O) at different time-points after the first dose (T), any candidate vaccine approved worldwide for EUA to prevent COVID-19 (I), in healthy adults or patients at high risk for SARS-CoV-2 infection (P), versus placebo or vaccine other than SARS-CoV-2 (C). Randomized studies in which the Kaplan–Meier plot did not report the number of at-risk participants were excluded. Two authors (AD and MCB) identified relevant studies, independently reviewed full texts, and disagreements were resolved by discussion. Data were extracted as described in the PROSPERO protocol, and risk of bias was assessed using the Cochrane Risk of Bias tool-2 (Rob-2).
Vaccine exposure
We considered any candidate vaccine approved worldwide for EUA to prevent COVID-19: BNT162b2 manufactured by Pfizer/BioNTech, is a lipid nanoparticle-formulated, nucleoside-modified RNA encoding the SARS-CoV-2 full-length spike; 30 μg; ChAdOx1 nCoV-19 [AZS1222]: manufactured by AstraZeneca, is a recombinant-deficient chimpanzee adenoviral vector containing the SARS-CoV-2 structural glycoprotein antigen: spike protein; nCoV-19; 2.2–6.5×1010 viral particle (VP); mRNA-1273: manufactured by MODERNA, is a lipid nanoparticle (LNP)-encapsulated modified RNA encoding the perfusion stabilized full-length spike protein of the SARS-CoV-2 virus; 100 μg; WIV04 (5 μg) and HB02 (4 μg): manufactured by Siopharm, are inactivated SARS-CoV-2 strains created from Vero cells with aluminum hydroxide adjuvant; Gam-COVID-Vac: manufactured by the Moscow City Health Department, Russian Direct Investment Fund, Sberbank, and RUSAL, is heterologous prime-boost which combined two vector vaccines based on rAd type 26 (rAd26) and rAd type 5 (rAd5) carrying the gene for SARS-CoV-2 full-length glycoprotein S; Ad26.COV2.S: manufactured by Janssen/Johnson & Johnson, is a replication-incompetent adenovirus type 26 (Ad26) vectored vaccine encoding a stabilized variant of the SARS-CoV-2 S protein (5×1010 VP); NVX-CoV2373: manufactured by Novavax, is a recombinant nanoparticle encoding the full-length spike glycoprotein of the prototype strain plus Matrix-M adjuvant (5 μg of NVX-CoV2373 plus 50 μg of Matrix-M adjuvant); CoronaVac: manufactured by the Turkish Health Institutes Association/Sinovac Research & Development, is inactivated whole-virion SARS-CoV-2 vaccine (3 μg of SARS-CoV-2 virion plus 0.45 mg/ml of aluminum hydroxide).
Outcomes and data synthesis
Vaccine efficacy (VE) against confirmed COVID-19 at any time after the first dose as defined in each study was the primary outcome. Secondary outcomes were VE at different time-points: (i) from randomization to day-21 after dose 1 and (ii) starting 7 days after dose 2. IPD were reconstructed by scanning the published Kaplan–Meier cumulative incidence curves using the WebPlotDigitizer software (Rohatgi 2021), then applying the reconstruction algorithm of Guyot and Colleagues (2012), which uses the magnitudes and locations of steps in the Kaplan–Meier curves, together with the numbers of patients at-risk, to infer the number of events and censorings occurring within each time interval. VE was re-estimated using the one-stage approach using the mixed Cox regression models with trials random-effects to account for difference in the study design and the background risk of COVID-19 during study. VE was expressed as: (1-incidence risk ratio [IRR]) × 100. To choose the preferred regimen, the P-score ranging from 0 (worse vaccine) to 1 (best vaccine) was computed for each vaccine, then the vaccine with a higher P-score was selected as better than each competing vaccine. Heterogeneity and inconsistency were quantified using the global Q test proposed by Rucker (Schwarzer et al. 2015). The Q statistic is the sum of statistic for heterogeneity, which represent the proportion of total variation in study estimates (within-designs), and a statistic for inconsistency (between-designs), which represents the variability of vaccine effect between direct and indirect comparisons at the meta-analytic level. To visualize and identify the nodes of single-design inconsistency, we used a network heat plot. Consistency between direct and indirect comparisons was checked using the so-called node-splitting. Sensitivity analysis was conducted by grouping vaccines according to their type (mRNA, viral vector, inactivated and recombinant protein).
Results
Study characteristics and risk of bias
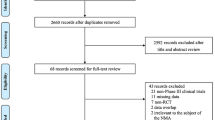
Of 666 retrieved citations, 52 were full-text reviewed, and 8 were included in the quantitative analysis (Fig. 1) (Baden et al. 2021; Polack et al. 2020; Voysey et al. 2021; Logunov et al. 2021; Sadoff et al. 2021; Al Kaabi et al. 2021; Heath et al. 2021; Tanriover et al. 2021). Figure 2 shows the network for efficacy captured by the SARS-CoV-2 vaccines. Reconstructed IPD are shown in Fig. S1; they agree exactly with reported data for each vaccine groups and for each trial. Between April 23, 2020 and January 05, 2021, 210,418 participants were recruited in 354 sites worldwide. Of these participants, 124,099 (59%) were male with a median age ranging from 36.1 to 56 years, 81,521 (38.7%) had a comorbidity, including hypertension, diabetes and obesity, and 8401 (4%) have had a positive PCR or IgG at baseline. During a median (interval inter quartile [IQR]) follow-up duration of 69.8 (69.7–70.3) days, 2131 confirmed COVID-19 cases occurred (604; 26.0 per 1000 person–years in vaccines recipient and 1527; 85.9 per 1000 person–years in the control group). Figure 3 shows the vaccines efficacy compared with controls at different time-points after dose 1. A risk of attrition bias (incomplete outcome data) was detected in some trials (Fig. S2). Furthermore, no evidence of the presence of publication bias was detected (Fig. S3).
Network graph of eligible COVID-19 vaccines comparisons for efficacy. Line width is proportional to the number of trials comparing every pair of vaccine. The size of the circle is proportional to the number of participants assigned to receive the vaccine; BNT162b2 (lipid nanoparticle-formulated, nucleoside-modified RNA encoding the SARS-CoV-2 full-length spike): 30 μg; ChAdOx1 nCoV-19 (AZS1222): recombinant-deficient chimpanzee adenoviral vector containing the SARS-CoV-2 structural glycoprotein antigen (spike protein; nCoV-19): 2.2–6.5×1010 viral particle (VP); mRNA-1273 lipid nanoparticle (LNP)-encapsulated modified RNA encoding the perfusion stabilized full-length spike protein of the SARS-CoV-2 virus): 100 μg; WIV04 (5 μg) and HB02 (4 μg): inactivated SARS-CoV-2 strains created from Vero cells with aluminum hydroxide adjuvant; Gam-COVID-Vac: heterologous prime-boost which combined two vector vaccine based on rAd type 26 (rAd26) and rAd type 5 (rAd5) carrying the gene for SARS-CoV-2 full-length glycoprotein S; Ad26.COV2.S: replication-incompetent adenovirus type 26 (Ad26) vectored vaccine encoding a stabilized variant of the SARS-CoV-2 S protein (5×1010 VP); NVX-CoV2373: recombinant nanoparticle encoding the full-length spike glycoprotein of the prototype strain plus Matrix-M adjuvant (5 μg of NVX-CoV2373 plus 50 μg of Matrix-M adjuvant); CoronaVac: inactivated whole-virion SARS-CoV-2 vaccine (3 μg of SARS-CoV-2 virion plus 0.45 mg/ml of aluminum hydroxide)
Efficacy of vaccines against COVID-19 at different time-points compared with control from reconstructed individual patient data. Vaccine efficacy estimates are provided as 1 minus incidence risk ratio (IRR) expressed as percentage with 95% confidence interval. BNT162b2 (lipid nanoparticle-formulated, nucleoside-modified RNA encoding the SARS-CoV-2 full-length spike): 30 μg; ChAdOx1 nCoV-19 (AZS1222): recombinant-deficient chimpanzee adenoviral vector containing the SARS-CoV-2 structural glycoprotein antigen (spike protein; nCoV-19): 2.2–6.5×1010 viral particle (VP); mRNA-1273 lipid nanoparticle (LNP)-encapsulated modified RNA encoding the perfusion stabilized full-length spike protein of the SARS-CoV-2 virus): 100 μg; WIV04 (5 μg) and HB02 (4 μg): inactivated SARS-CoV-2 strains created from Vero cells with aluminum hydroxide adjuvant; Gam-COVID-Vac: heterologous prime-boost which combined two vector vaccine based on rAd type 26 (rAd26) and rAd type 5 (rAd5) carrying the gene for SARS-CoV-2 full-length glycoprotein S; Ad26.COV2.S: replication-incompetent adenovirus type 26 (Ad26) vectored vaccine encoding a stabilized variant of the SARS-CoV-2 S protein (5×1010 VP); NVX-CoV2373: recombinant nanoparticle encoding the full-length spike glycoprotein of the prototype strain plus Matrix-M adjuvant (5 μg of NVX-CoV2373 plus 50 μg of Matrix-M adjuvant); CoronaVac: inactivated whole-virion SARS-CoV-2 vaccine (3 μg of SARS-CoV-2 virion plus 0.45 mg/ml of aluminum hydroxide)
At any time after the first dose
mRNA-1273 was the most effective vaccine to reduce incident cases of COVID-19 with a probability of 99.9% (P-score 0.999). Incidence reductions were 61% (95% CI, 33–78%) compared with BNT162b2 (P-score 0.881). The corresponding incidence reductions were 75% (56–85%), 76% (58–87%), 76% (56–87%), 79% (63–88%), 84% (74–90%), and 84% (75–92%) compared with Sputnik V (Gam-COVID-Vac; P-score 0.672), NVX-CoV2373 (P-score 0.617), ChAdOx1 nCov-19 (P-score 0.616), HB02 (P-score 0.521), Ad26.COV2.S (P-score 0.298), and WIV04 (P-score 0.231) vaccines, respectively. Incidence reductions were 87% (77–93%) for mRNA-1273, 67% (48–79%) for BNT162b2, 49% (21–67%) for Sputnik V, 46% (13–66%) for NVX-CoV2373, 46% (9–68%) for ChAdOx1 nCov-19, and 38% (2–61%) for HB02 vaccine compared with CoronaVac (P-score 0.164) recipient (Table S1).
Between randomization to 21-day after dose 1
Sputnik V was the most effective vaccine (P-score 0.937) followed by the ChAdOx1 nCov-19 vaccine (P-score 0.751). Compared with the WIV04 vaccine (P-score 0.529), COVID-19 incidence reduction for Sputnik V was 51% (5–75%). The corresponding incidence reductions were 60% (32–76%), 63% (36–79%), and 66% (21–85%) compared with CoronaVac (P-score 0.388), NVX-CoV2373 (P-score 0.305), and mRNA-1273 vaccines (P-score 0.272), respectively. Incidence reductions were 52% (8–75%) for HB02 (P-score 0.669), 56% (33–71%) for BNT162b2 (P-score 0.740), and 71% (59–80%) for Sputnik V compared with a single dose of Ad26.COV2.S vaccine (P-score 0.072) (Table S2).
Starting 7 days after the second dose
One week after the second dose or 35 days after the single dose of Ad26.COV2.S, the mRNA-1273 vaccine remained the most effective (P-score 0.929) with incidence reductions from 68% to 95% when compared with CoronaVac (P-score 0.574) and other vaccines. The corresponding incidence reductions were 73% to 95% for BNT162b2 (P-score 0.913), and 60% to 92% for NVX-CoV2373 (P-score 0.789) when compared with Sputnik V (P-score 0.463), HB02 (P-score 0.457), ChAdOx1 nCov-19 (P-score 0.402), WIV04 (P-score 0.278), and Ad26.COV2.S (P-score 0.196) vaccines, respectively (Table S3) (Table 1).
Sensitivity analysis, Heterogeneity, and Consistency
After grouping vaccines according to their type, results were similar to those of the main analysis. Except from randomization to 21 days after dose 1, mRNA vaccines were the most effective with an incidence reduction of 57% to 96%, while during this interval, DNA vaccines reduced the COVID-19 incidence by 43% and 46% compared with control and recombinant protein vaccines (Table 2). Because only one single closed loop due to the presence of one direct between-vaccine comparison (HB02 and WIV04) was available in our sample, as shown in Fig. 2, we were unable to compute the global heterogeneity for both primary outcome and secondary outcomes.
Discussion
We provide information on the dynamics of vaccine efficacy at different time-points. Our findings provide evidence of higher short-term efficacy of mRNA vaccines, especially the mRNA-1273 vaccine, in reducing the incidence of COVID-19 at any time-point after dose 1. These findings are consistent with the reported VE of 76% (58–87%) for mRNA-1273 (Puranik et al. 2021), 42% (13–62%) (Puranik et al. 2021), and 88% (85–90%) for BNT162b2, 67% (61–72%) for ChAdOx1 nCov-19 (Lopez Bernal et al. 2021), and 75.7% (69.3–80.8%) for the pooled VE from 17 studies (Harder et al. 2021) against symptomatic COVID-19 caused by the delta variant. In addition, at least one week after the second dose, we found a similar protection rate against COVID-19 infection to the reported VE by Sharif of 73% (69–77%) for adenovirus vector vaccine and 85% (82–88%) for the mRNA vaccine (Sharif et al. 2021). Despite this similarity, our study has the advantage of having taken into account the dynamic nature of this vaccine effectiveness, which indicates a rapid increased protection rate after the second dose for mRNA vaccine compared to the DNA and Inactivated vaccines.
Although mRNA vaccines seem to display very similar results, DNA vaccines appear to be more heterogeneous. Taken as a group, DNA vaccines are the most efficient in the first 3 weeks after vaccination, but that is mostly due to the good results of Sputnik-V and ChAdOx1 nCov-19 during this time-period, while Ad26.COV2.S displays the lowest efficacy and performs significantly worse than three other vaccines. This is particularly intriguing given the sponsor strategy of recommending a single injection in the primary vaccination, while all other vaccines offer a 2-injection primary vaccination. Further comparison would require more prolonged data with one-injection regimens for the other vaccines, which is not currently available. Nevertheless, the results of this analysis do not support a one-shot primary vaccination schedule for Ad26.COV2.S.
The strength of this study includes reconstructed IPD to allow vaccines efficacies comparison at different time-points, thereby reducing differences due to the definition of population for analyses and statistical methods, and the accounting for difference in study design and background risk of COVID-19 during the study. The extracted data exactly matches those reported by authors, suggesting the robustness of our results. However, the small number of randomized studies can be a limitation. This lack of sufficient data on mixed comparison between vaccines makes it challenging to assess a possible incoherence between direct and indirect comparisons, which is the statistical manifestation of intransitivity. Nevertheless, transitivity assumption is also addressed by indirectness that refers to the relevance of the included studies to the research question, which was well considered in our study. Therefore, our findings have a great confidence after considering the within-trials bias, reporting bias, indirectness, and imprecision domains of the Confidence in Network Meta-Analysis (CINeMA) approach (Nikolakopoulou et al. 2020). Additionally, our findings should be interpreted with caution because vaccines are compared using the currently available trial interim data with disparate study population, duration of exposure, type of control, definition and assessment of the primary endpoints, and the high trials risk of bias due to per protocol analysis.
The reduction in vaccine effectiveness, combined with the gap between mass vaccination and pandemic progression, raises questions about herd immunity and reinforces vaccine hesitancy. Therefore, the readjustment of vaccination strategies and policies, especially the possibility to administer booster doses of vaccine, and to develop variant-targeted vaccines are urgently needed to overcome this pandemic.
Conclusion
Among the current COVID-19 vaccines, mRNA-1273 provides a higher protection against COVID-19. Adherence to public health guidelines and long-term surveillance of vaccine efficacy and safety are necessary, especially in the context of circulation of variants of concern.
Availability of data and materials
The data are available upon request (alhassane.diallo@chu-montpellier.fr).
Abbreviations
- RCT:
-
randomized controlled trials
- NR:
-
not reported
- IRR:
-
incidence risk ratio
- COVID-19:
-
coronavirus disease 2019
- US:
-
United States
- UK:
-
United Kingdom
- HIV:
-
human immunodeficiency virus
- HBV:
-
hepatitis B virus
- HCV:
-
hepatitis C virus
- RT-PCR:
-
reverse transcriptase-polymerase chain reaction
- LD:
-
low dose
- SD:
-
standard dose
- VP:
-
viral particle
- NMA:
-
random-effects network meta-analysis
- VE:
-
vaccine efficacy
References
Al Kaabi N, Zhang Y, Xia S, Yang Y, Al Qahtani MM, Abdulrazzaq N et al (2021) Effect of 2 inactivated SARS-CoV-2 vaccines on symptomatic COVID-19 infection in adults: a randomized clinical trial. JAMA 6 326(1):35
Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R et al (2021) Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 384(5):403–416
Dagan N, Barda N, Kepten E, Miron O, Perchik S, Katz MA et al (2021) BNT162b2 mRNA COVID-19 vaccine in a nationwide mass vaccination setting. N Engl J Med 384(15):1412–1423
Guyot P, Ades A, Ouwens MJ, Welton NJ (2012) Enhanced secondary analysis of survival data: reconstructing the data from published Kaplan-Meier survival curves. BMC Med Res Methodol 12(1):9
Harder T, Kulper-Shiek W, Reda S et al (2021) Effectiveness of COVID-19 vaccines against SARS-CoV-2 infection with the Delta (B.1.617.2) variant: second interim results of a living systematic review and meta-analysis, 1 January to 25 August 2021. Euro Surveill 26(41)
Heath PT, Galiza EP, Baxter DN, Boffito M, Browne D, Burns F, et al (2021) Safety and efficacy of NVX-CoV2373 COVID-19 vacchttps://doi.org/10.1056/NEJMoa2107659
Jara A, Undurraga EA, González C, Paredes F, Fontecilla T, Jara G, et al (2021) Effectiveness of an Inactivated SARS-CoV-2 Vaccine in Chile. N Engl J Med. https://doi.org/10.1056/NEJMoa2107715
Kissling E, Hooiveld M, Sandonis Martín V, Martínez-Baz I, William N, Vilcu A-M, et al (2021) Vaccine effectiveness against symptomatic SARS-CoV-2 infection in adults aged 65 years and older in primary care: I-MOVE-COVID-19 project, Europe, December 2020 to May 2021. Eurosurveillance 26(29). Disponible sur: https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2021.26.29.2100670
Logunov DY, Dolzhikova IV, Shcheblyakov DV, Tukhvatulin AI, Zubkova OV, Dzharullaeva AS et al (2021) Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet 397(10275):671–681
Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S et al (2021) Effectiveness of COVID-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med 385(7):585–594
Nikolakopoulou A, Higgins JPT, Papakonstantinou T, Chamani A, Del Giovane C, Egger M et al (2020) CINeMA: an approach for assessing confidence in the results of a network meta-analysis. PLoS Med 17(4):e1003082
Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S et al (2020) Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N Engl J Med. 383(27):2603–2615
Puranik A, Lenehan PJ, Silvert E, Niesen MJM, Corchado-Garcia J, O’Horo JC, et al. (2021) Comparison of two highly-effective mRNA vaccines for COVID-19 during periods of Alpha and Delta variant prevalence. Public and Global Health. Disponible sur: http://medrxiv.org/lookup/doi/10.1101/2021.08.06.21261707
Rohatgi A (2021) WebPlotDigitize. Pacifica: ankitrohatgi@hotmail.com.
Sadoff J, Gray G, Vandebosch A, Cárdenas V, Shukarev G, Grinsztejn B et al (2021) Safety and efficacy of single-dose Ad26.COV2.S vaccine against COVID-19. N Engl J Med 384(23):2187–2201
Schwarzer G, Carpenter JR, Rücker G (2015) Meta-Analysis with R. Springer International Publishing, Cham
Sharif N, Alzahrani KJ, Ahmed SN, Day SK (2021) Efficacy, immunogenicity and safety of COVID-19 vaccines: a systematic review and meta-analysis. Front Immunol 12:714170
Tanriover MD, Doğanay HL, Akova M, Güner HR, Azap A, Akhan S et al (2021) Efficacy and safety of an inactivated whole-virion SARS-CoV-2 vaccine (CoronaVac): interim results of a double-blind, randomised, placebo-controlled, phase 3 trial in Turkey. Lancet 398(10296):213–222
Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK et al (2021) Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. The Lancet 397(10269):99–111
Author information
Authors and Affiliations
Contributions
AD conceived the study design, analyzed the data, and drafted the manuscript, MCB supervised data collection and critical revision of the manuscript, MHD collected and analyzed the data, and AM and FG interpreted and substantially revised the manuscript. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
No applicable.
Consent for publication
Not applicable
Competing interests
We declare no competing interests in relation to this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(PDF 539 kb)
Rights and permissions
About this article
Cite this article
Diallo, A., Carlos-Bolumbu, M., Diallo, M.H. et al. Efficacy of approved vaccines to prevent COVID-19: a systematic review and network meta-analysis of reconstructed individual patient data from randomized trials. J Public Health (Berl.) 31, 1463–1472 (2023). https://doi.org/10.1007/s10389-022-01707-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10389-022-01707-1