Abstract
Low phosphorus (P) availability is a major problem for cotton production. Identification of low P tolerant and efficient genotypes would best address these problems. In this regard, thirty cotton genotypes were categorized for dry matter production and P use efficiency (PUE) under low and normal P conditions. The results showed that cotton genotypes displayed considerable variations in root morphology, dry matter production, photosynthesis, PUE, and antioxidant system in response to low and normal P conditions. Genotypes Jimian169, Zhongzhimian2, and Lumianyan16 produced more dry matter have high PUE and were considered strong low P tolerant and efficient, whereas genotypes DES926, TM‑1, and CCRI10 have low dry matter and PUE and were proved to be weak low P tolerant and inefficient. Based on various classification methods, genotypes Jimian169 and DES926 were considered strong and weak low P tolerant cotton genotypes, respectively. For most of the traits, the heritability was high, suggesting that selection under low P is more reliable than under the normal P condition. The use of the identified cotton genotypes and traits could improve cotton breeding activities and help to improve sustainable cotton production.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Cotton is growing mainly as a fiber crop, providing raw materials to the textile industry (Chen et al. 2019; Iqbal et al. 2022a). China is one of the leading cotton producers, consumers, and importers in the world (Asif et al. 2021; Niu et al. 2020). In China, most of the cotton is shifted from the Yellow River and Yangtze River valley to Xinjiang province (Iqbal et al. 2022b). However, Xinjiang is an arid region having low precipitation and high surface evaporation and are therefore facing the issues like scarcity of water resources (Li et al. 2015) and low nutrient availability, especially phosphorus (P) (Iqbal et al. 2022c). Studies have found that the availability of P is very poor in Xinjiang soil due to its calcareous nature, which causes slow shoot development, dark green leaves, flower bud necrosis, yellowing of older leaves, and finally affects cotton yield and quality (Zhang et al. 2020). As a result, the growers applied a huge amount of P fertilizers, however, the solubility and availability of P are low and it is easily fixed by metal ions (Amanullah et al. 2019) and therefore the P use efficiency (PUE) in a growing season is only 10–20% (Iqbal et al. 2019a). This poor availability of P not only inhibits plant growth but also increases the cost of production and environmental pollution (Iqbal et al. 2020c). In addition, low P activities various physiological responses, such as increased root surface area and density that can assist more P uptake (George et al. 2006). The response variation among different cultivars suggests that the genotype which performed better under low P is a better choice rather than increasing P fertilizers (Zhang et al. 2012). There is a huge gap between the crop potential and the actual yield under low P condition, indicating that identification and development of low P tolerant and efficient genotypes are needed to address these issues (Cattivelli et al. 2008).
P plays a vital role in plant growth and development as it is an important component of ATP, phospholipids, and nucleic acid (Wang and Liao 2010). Usually, plants suffer from P deficiency because root hairs can only uptake the available P, however, the solubility, mobility, and availability of P are very low (Wang et al. 2021). In addition, 80% of the applied P fertilizer is not available for the plants (Iqbal et al. 2019a) due to its fixation with iron and aluminum in acidic soils and calcium and magnesium in alkaline soils (Wang et al. 2015). As a result, the use of chemical fertilizer is increased in the last few decades and it may increase by a further 2% if current yields are to be maintained (Niu et al. 2013). Moreover, the increase in P fertilization will increase the cost of production and environmental pollution (Ghaffar et al. 2017). In addition, the production of phosphate fertilizer consumes about 85% of the mined rock phosphate every year. According to the statistics of the International Fertilizer Development Center (IFDC), in 2010, the resources of China’s phosphate rock accounted for only 5.8% of the world’s, and the supply was insufficient, and a large amount of it was de-pendent on imports. However, the world’s proven rock phosphate resources can only be mined at the current rate for 50–70 years (Sandaña and Pinochet 2016). Therefore, identification and development of genotypes with high PUE are the crucial need for high crop production (Chowdhury and Zhang 2021).
Tolerance to low P requires maintenance of plant growth and biomass with limited available P and is reported to occur by two distinct routes namely P uptake efficiency (PUpE) and P utilization efficiency (PUtE) (Iqbal et al. 2019b). PUpE is the plant’s ability to extract P from the soil and is expected to be related to root system traits that increase root surface area or facilitate P acquisition (Rausch and Bucher 2002). PUtE is a function of plant growth, remobilization, and physiological traits that translate P acquired by the roots into yield. Therefore, P efficiency is defined as the ability of plants to produce higher biomass or yield, and/or take up more P under inadequate P conditions (Sandaña and Pinochet 2016). Therefore, an increase in P absorption by a good root system is the best approach for low-input agriculture (Mori et al. 2016). Plants evolved a number of adaptation measures to encounter low P through morphological and metabolic modification, especially in roots that play a great role in the absorption of a poor mobile soil P (Bello 2021). The PUpE mechanism consists of elongation in the root system, reduction in primary root growth, and increased lateral root surface area, root thickness, root hair length, root shoot ratio, and shallow axial root growth (Ruiz et al. 2020). In addition, root size, root dry matter (Srinivasarao et al. 2006), root vigor, and photosynthesis (Ali et al. 2002) are also important for P absorption (Huang et al. 2017). P‑efficient genotypes have a shallow-rooted system that improves P absorption in response to surface P fertilization (Yang et al. 2019). The increase in root morphology at the seedling stage is more important than the reproductive stage because grain yield is ensured by P remobilization from the senescing tissues (Lynch 2019). Although under low P, scientists have focused on root morphology (Sandaña 2016), and suggested that extensive root systems may increase the ability of plants to improve the uptake of P (Bilal et al. 2018). Several researchers have studied that an extensive root system under low P increases the ability of plants to improve root volume for the uptake of more P (Srinivasarao et al. 2006).
PUtE is the ability of plants to convert the uptake P into dry matter or yield and therefore, utilization of the uptake P is very important, especially under low P condition (Sandaña and Pinochet 2016). Since P deficiency restricts plant growth by decreasing leaf photosynthesis and sink capacity, low P affects the shoot more than the root (Iqbal et al. 2019a). P regulates ribulose‑1,5‑biphosphate regeneration, carboxylation, energy supply, stomatal size, and conductance, therefore, low P inhibits photosynthesis and carbon metabolism followed by a poor biomass accumulation (Taliman et al. 2019), due to non-stomatal limitations (Carstensen et al. 2018). Under low P, 60–90% of root P is remobilized from senescing tissues to the developing seeds (Masoni et al. 2007), however, the P remobilization is low under normal P condition (Mazlouzi et al. 2020). Although, the non-stomatal inhibition of photosynthesis is known (Taliman et al. 2019), but the importance of P remobilization for increasing PUtE is still to be elucidated.
Previously, the importance of genetic variation in low P tolerance and PUE has been identified in maize (Bayuelo-Jiménez and Ochoa-Cadavid 2014), potato (Sandaña 2016), wheat (Bilal et al. 2018), lentil (Aski et al. 2022), pearl millet (Gemenet et al. 2015), rice (Inthapanya et al. 2000), brassica (Irfan et al. 2020), mung bean (Reddy et al. 2021), and sorghum (Manske et al. 2000). However, the contribution of PUpE and PUtE greatly varies among various crops, soil P availability, and the environment. It was found in the previous studies that PUpE was more critical than PUtE in common bean (Beebe et al. 2006), wheat (Osborne and Rengel 2002), and maize (Parentoni and Souza Júnior 2008), while PUtE contributed more to PUE in maize (Corrales et al. 2007) and potato (Balemi and Schenk 2009a). Under low P, PUpE is more important for variation in yield and PUE, while PUtE is more important under normal P condition (McDonald et al. 2015). The variation among genotypes for PUE in cotton greatly depends on traits and methods of screening. Consequently, various methods and criteria have been proposed by different researchers to screen genotypes under low P such as total P uptake (Sandaña 2016), dry matter production per unit P (Rahim et al. 2010), and the ratio of physiologically active P to total P (Aziz et al. 2014). The complexity of PUpE and PUtE showed that an increase in PUE might be obtained through various contributing traits instead of a single trait. Therefore, it is important to know the key traits contributing to PUE that could help in the classification and identification of contrasting low P tolerant and efficient cotton genotypes. In this regard, the current study was aimed to (1) evaluate the variation in cotton genotypes for low P tolerance and (2) identify the key traits contributing to PUE that could facilitate the selection of P‑efficient cotton genotypes.
Materials and Methods
Plant Materials
Based on the multi-year P fertilizer experiments in the experimental farm of the Cotton Research Institute of the Chinese Academy of Agricultural Sciences (CRI, CAAS), we have found that the amount of available P (3 ± 0.5 mg kg−1) in the soil is low and the cotton genotypes respond differently during the cotton growth period. Therefore, the current study is very important to understand the genotypic difference under low and normal P conditions. Moreover, the selection of the best contrasting P‑efficient cotton genotypes is the basis for the current study. Previously, we studied the agronomic performance and P use efficiency (PUE) of 384 cotton genotypes under low (0.01 mM KH2PO4) and normal (1 mM KH2PO4) P conditions, and based on dry biomass and PUE, 30 genotypes were selected for further screening. The present study was designed to use the selected 30 cotton genotypes to identify the best contrasting P‑efficient cotton genotypes under low and normal P conditions in hydroponic and pot cultures.
Hydroponic Experiment
Thirty cotton genotypes (Table S1) were categorized for various morphophysiological traits under low (0.01 mM KH2PO4) and normal (1 mM KH2PO4) P conditions in hydroponic culture. The experiment was conducted in the greenhouse at CRI, CAAS, Anyang, China (36°06′ N, 114°21′ E). The healthy seeds of each genotype were grown in 50% sand and vermiculite for one week. After germination, uniform seedlings were transplanted into a plastic container (7 L) in the condition of 16/8 h light/dark cycle, 28 °C temperature, and 60% relative humidity. During the first week, a half-strength Hoagland solution was applied followed by a full-strength solution as mentioned in our previous study (Iqbal et al. 2019a). Further, seedlings with two true leaves were exposed to low and normal P conditions. The seedlings were aerated with an electric pump and the solutions were changed every week. After obvious morphological variation, the four-week-old seedlings were harvested and various morphophysiological traits were measured.
Pot Experiment
A pot experiment was conducted in the greenhouse at the CRI, CAAS, Anyang, Henan province, China (36°06′ N, 114°21′ E). The test soil of 0–20 cm low-fertile arable soil was collected from the cotton field and sterilized before the experiment. The thirty cotton genotypes were grown in a black plastic bucket (diameter: 12 cm, height: 10 cm) having soil collected from the cotton field under low (0.01 mM KH2PO4) and normal P (1 mM KH2PO4) conditions. The other nutrients were used at the recommended levels. Except for P treatments, other cultivation and management measures will remain the same for each pot.
Morphological Measurements
The shoot length (cm) of six randomly selected plants from each replication was measured with the help of a ruler and the average has been worked out as the mean shoot length (Iqbal et al. 2020a). After harvesting, the plants were divided into roots and shoots and subsequently dried at 105 °C for 1 h followed by 80 °C for 48 h. After complete drying, the shoot, root, and total dry matter were determined using an electric balance. At the same time, the roots of half of the plants from each genotype were scanned and analyzed through WinRhizo root analyzer system (Iqbal et al. 2020b).
Determination of Leaf Photosynthetic and Chlorophyll Traits
The photosynthetic traits were measured from the third fully expanded leaf by using a portable photosynthesis system (Li-Cor 6800; Li-Cor, Inc., Lincoln, NE, USA) from 9:00 to 11:00 a. m. (Iqbal et al. 2020a). About 50 mg of fresh leaf sample was used to measure chlorophyll and carotenoid contents. The collected samples were cut into small pieces and incubated overnight in acetone: ethanol (1:1) solution for 48 h at 25 °C. Further, the absorbance for chlorophyll and carotenoid was measured according to the standard protocols (Iqbal et al. 2020a).
Phosphorus Concentration and Use Efficiency
P concentration in root and shoot tissues were measured according to Kjeldahl method (Iqbal et al. 2022b; Iqbal et al. 2020d). The grounded sample of 0.2 g from each tissue was digested with H2SO4-H2O2, and the final P concentration was analyzed using the Bran + Luebbe Continuous-Flow AutoAnalyzer III (AA3-Australia). The various PUE-related definitions were measured according to our previous study (Iqbal et al. 2019a).
Determination of Malonaldehyde Contents and Antioxidant Enzymatic Activities
The malonaldehyde (MDA) content in root and shoot was measured by the thiobarbituric acid (TBA) reactions according to the standard protocol (Li et al. 2022). The samples (0.2 g) were extracted in 2 ml of 0.25% TBA prepared in 10% TCA. The extract was heated at 95 °C for 30 min, and then, quickly cooled on ice. The collected extract was centrifuged at 10,000 g for 10 min and absorbance was measured at 532 nm.
For measuring the enzymatic activities, about 0.5 g of root and shoot samples were crushed in liquid nitrogen and 10 ml of 50 mM sodium phosphate buffer consisting of 1% polyvinyl pyrrolidine, 0.2 mmol · L−1 ethylenediamine tetra acetic acid, and 10 mmol · L−1 magnesium chloride was added. The solution was then centrifuged at 12,000 g for 12 min at 4 °C. Finally, the collected supernatant was stored at 4 °C and the POD activity was measured according to the protocol mentioned in the earlier study (Iqbal et al. 2020a).
SOD activity was assayed using the photochemical NBT method. The assay mix (1 ml) contained 50 mM phosphate buffer (pH 7.8), 9.9 mM methionine, 57 mM NBT, 0.025% Triton X‑100, and 0.0044% riboflavin. The photoreduction of NBT was measured at 560 nm. One unit of SOD was defined as the volume of extract that causes inhibition of the photoreduction of NBT by 50%.
Catalase activity was determined in the homogenates by measuring the decrease in absorption at 240 nm as H2O2 and enzyme activity expressed as µmol H2O2 oxidized min−1 g−1 protein. In this case, 50 µl enzyme extract was added to a mixture that contained 50 mM sodium phosphate buffer (pH 7.0) and 10 mM H2O2 to make the volume to 3 ml. Catalase activity was calculated by using an extinction coefficient of 39.4 mM−1 cm−1.
Finally, the collected supernatant was stored at 4 °C, and the SOD, POD, and CAT activities were measured according to the protocol mentioned in earlier studies (Wang et al. 2022).
Methods for Classification of Cotton Genotypes
Method 1
In this method, the genotypes were divided into three groups such as efficient (E), medium (M), and inefficient (I) based on values assigned to each genotype using mean (m) and standard deviation (SD) of each trait under low and normal P in hydroponic and pot culture (Osborne and Rengel 2002) and (Aziz et al. 2011), the mung bean genotypes were classified into. The genotype was considered efficient, if the mean value is more than m + SD, medium if the value is ranging between m = SD and m‑SD, and inefficient when the mean value is less than m‑SD. The score was assigned as 3 to efficient, 2 to medium, and 1 to inefficient for each trait and finally the score of all traits were sum up for each genotype.
Method 2
According to this method, the classification of cotton genotypes was performed using shoot dry matter and PUtE under low and normal P conditions (Fageria and Baligar 1993). The genotypes were classified based on efficiency and responsiveness to normal and low P conditions. The genotype having high dry matter than the mean dry matter of all genotypes was considered efficient, while the genotype having high PUtE than the mean PUtE of all the genotypes was considered responsive. Thus the genotypes were divided into four groups such as (i) efficient and responsive (ii) efficient and non responsive (iii) inefficient and responsive and (iv) inefficient and nonresponsive (Kosar et al. 2003).
Method 3
In this method, the genotypes were classified on the basis of efficiency and responsiveness (Gerhardt et al. 2017). Based on the deviation of mean shoot dry matter of each genotype in relation to mean shoot dry matter under both low and normal P conditions. The data were plotted in a scatter plot, showing deviation under normal P (responsiveness) on the x‑axis and deviation under low P (efficiency) on the y‑axis. This method also classified the genotypes into four groups such as (i) efficient and responsive (ii) efficient and non-responsive (iii) inefficient and responsive and (iv) inefficient and non-responsive.
Method 4
This method is based on the stress tolerance score calculated from the total dry matter of each genotype as shown in the following equations (Negarestani et al. 2019) and (Grzesiak et al. 2019).
Where, C and T represent the total dry matter (TDM) of genotypes under normal and low P conditions, respectively. xC and xT showed mean total dry matter of genotypes under low and normal P conditions, respectively. Finally, the values of all indices were summed up to get the value of the stress tolerance score as shown in the following equation (Negarestani et al. 2019).
Method 5
The cluster ranking groups were obtained based on cluster analysis (Ravikiran et al. 2018) using various studied morphophysiological traits subjected to Hierarchical cluster analysis in Origin Pro 2018 (Iqbal et al. 2019a).
Statistical Analysis
Genetic traits were measured from analysis of variance for all the studied traits in thirty cotton genotypes under low and normal P conditions in both hydroponic and pot cultures. All the traits were measured from the mean square between cotton genotypes and P conditions such as the phenotypic variance component, with the estimator σ2P = MSg / r (MSg stands for the mean square of the genotypes and r for replications); genotypic variance component, with the estimator σ2G = (MSg − MSe) / r (MSe is the error mean square); coefficient of experimental variation, with the estimator CVe% = 100.(MSe)1/2 / µ (where µ is the average of treatments); coefficient of genotypic variation, with the estimator CVg% = 100.(σ2G)1/2 / µ; variation index, with the estimator VI = CVg / CVe; and heritability, with the estimator H2 = σ2G / σ2P (Almasy and Blangero 2010).
The experiment was laid out on a randomized complete block design having three replications and the obtained data were analyzed using Statistix 8.1. The means were separated by the least significant value at a 5% level of significance. Figures were drawn in Origin Pro 2018 (Origin Lab Corporation, Northampton, MA, USA).
Results
Descriptive Statistics and Analysis of Variance of Measured Traits
The descriptive statistics of measured traits as explanatory variables of PUE among 30 cotton genotypes grown in low and normal P conditions are presented in Tables S2 and S3. A high range of differences among the studied traits was recorded, demonstrating that P affects the agronomic, physiological, and biochemical traits in cotton genotypes (Tables S2 and S3). In hydroponic culture, the highest percentage reduction was noticed for net photosynthetic rate (−38.76) followed by shoot P accumulation (−33.52) and shoot PUpE (−33.52) under low P condition compared to normal P condition. Whereas the highest gain was noticed for root volume (179.65) followed by root surface area (62.09), and root shoot ratio (43.45) under low P condition (Table S2). Among the 30 genotypes, the total PUpE ranged from 8.96 to 10.34 and 6.21 to 7.62 under normal and low P conditions, respectively, whereas PUtE ranged from 197.9 to 244.9 and 165.4 to 204.3 under normal and low P conditions, respectively. In pot culture, the highest percentage reduction was noticed for chlorophyll b (−53.68) followed by shoot P accumulation (−41.70) and shoot PUpE (−41.70) under low P compared to that of normal P conditions. Whereas the highest gain was noticed for root volume (213.15) followed by root surface area (78.34), and root length (60.24) under low P condition (Table S3). Among the 30 genotypes, the total PUpE ranged from 3.67 to 4.43 and 2.17 to 2.84 under normal and low P conditions, respectively, whereas PUtE ranged from 163 to 209 and 133 to 201 under normal and low P conditions, respectively.
Classification of Cotton Genotypes for Phosphorus Use Efficiency
Method 1
Among various studied traits, shoot dry matter, root dry matter, root shoot ratio, net photosynthetic rate, PUpE, and PUtE were selected for genotype classification. Cotton genotypes showed significant differences for the selected traits considered for scoring under both low and normal P conditions. In hydroponic culture, the genotypes Jimian169, Zhongzhimian2, and Lumianyan16 recorded the highest score (17 out of 18), while the lowest score was recorded from DES926 and TM‑1 (7 out of 18) under low P condition. Whereas under normal P condition, Jimian169, Zhongzhimian2, and Lumianyan16 recorded the highest score (17 out of 18), while the lowest score was recorded from DES926 and TM‑1 (7 out of 18) followed by Dalingmian69 (8 out of 18), and CCRI10 and Nongdamian7 (9 out of 18) (Table S4). Finally, the points score of cotton genotypes under both low and normal P was summed up to know the overall genotypic performance (Table S4). For overall performance among the genotypes, Jimian169, Zhongzhimian2, and Lumianyan16 recorded the highest score (34 out of 36), while the lowest score was recorded from DES926 and TM‑1 (14 out of 18) followed by Dalingmian69 (16 out of 36) and CCRI10 (18 out of 36) (Table S4).
In pot culture, the highest score was recorded from genotypes Jimian169 and Zhongzhimian2 (17 out of 18), while the lowest was from DES926, TM‑1, CCRI10, and Dalingmian69 (7 out of 18) under low P condition. Under normal P condition, Jimian169, Zhongzhimian2, and Lumianyan16 recorded the highest score (17 out of 18), while the lowest score was recorded from DES926, TM‑1, and CCRI10 (7 out of 18) (Table S5). The sum of the score under low P and normal P conditions showed that genotypes Jimian169 and Zhongzhimian2 have the highest score (34 out of 36), while DES926, TM‑1, and CCRI10 have the lowest score (14 out of 18) followed by Dalingmian69 (16 out of 36) (Table S5).
Method 2
In this method, the 30 cotton genotypes were categorized for P efficiency and responsiveness into four groups based on shoot dry matter and PUtE under both low P and normal P. In hydroponic culture, the genotypes Jimian169, Zhongzhimian2, Lumianyan16, Deltapine15, CCRI60, CCRI41, Lumianyan28, Lumian1138, CCRI16, and CCRI35 under low P condition and genotypes Jimian169, Zhongzhimian2, Lumianyan16, M‑8124-1159, CCRI17, Zhongyuan9114, CCRI60, CCRI19, CCRI41, Jia-1‑7, ZhongzhimianBD13, and Lumianyan28 under NP condition were classified in ER group. Moreover, the genotypes DES926, TM‑1, CCRI10, Nongdamian7, Dalingmian69, CCRI12, Zhongzhimian86‑1, Xinluzao49, Zhongyuan9114, Yuekangmian10, Jimian11, Xinluzhong5, and CCRI50 were grouped in the INR under low P condition (Fig. 1). However, under NP conditions genotypes DES926, TM‑1, CCRI10, Dalingmian69, Nongdamian7, CCRI35, CCRI12, Xinluzhong5, CCRI16, Zhongzhimian86‑1, Lumian1138, Yuekangmian10, CCRI49, Jimian11, Xinluzao49, and Deltapine15 were grouped into INR (Fig. 1b). Similarly, genotypes Jimian169, Zhongzhimian2, Lumianyan16, and CCRI60 were grouped in ER under both low P and normal P conditions. While genotypes DES926, TM‑1, CCRI10, Nongda-mian7, Dalingmian69, CCRI12, Zhongzhimian86‑1, Xinluzao49, Yuekangmian10, Jim-ian11, and Xinluzhong5 were categorized in INR under both P conditions (Fig. 1).
Classification of cotton genotypes based on P utilization efficiency and shoot dry matter under low (a) and normal (b) P condition in hydroponic culture according to Fageria and Baligar (1993). This classification divides genotypes into four groups i.e., ER efficient and responsive, IR in-efficient but responsive, ENR efficient but non-responsive, INR in-efficient and nonresponsive. Numbers from P1 to P30 represent cotton genotypes mentioned in Table S1
In pot culture, the genotypes Jimian169, Zhongzhimian2, Lumianyan16, Lumianyan28, CCRI41, Xinluzao49, CCRI19, Lumian1138, CCRI60, Zhongyuan9114, Yuekangmian10, and Deltapine15 were grouped in ER under low P condition (Fig. 2). Similarly, genotypes Jimian169, Zhongzhimian2, Lumianyan16, CCRI19, Xinluzao49, CCRI17, CCRI60, CCRI41, M‑8124-1159, Lumianyan28, Handan284, Jia-1‑7, CCRI50, ZhongzhimianBD13, and Yuekangmian10 were categorized in ER under normal P condition (Fig. 2). The genotypes DES926, TM‑1, CCRI10, Zhongzhimian86‑1, CCRI12, CCRI50, Nongdamian7, Dalingmian69, CCRI17, Jimian11, CCRI49, Xinluzhong5, and CCRI35 under low P condition and genotypes DES926, TM‑1, CCRI10, Dalingmian69, Nongdamian7, CCRI16, Xinluzhong5, CCRI12, CCRI35, Jimian11, and Zhongyuan9114 while under normal P condition were grouped in INR. Except, Lumian1138, Zhongyuan9114, and Deltapine15, the genotypes placed in ER under low P condition were similar to that under normal P condition (Fig. 2). Moreover, the genotypes categorized in INR under low P condition were similar to that of normal P except Zhongzhimian86‑1, CCRI50, CCRI17, and CCRI49.
Classification of cotton genotypes based on P utilization efficiency and shoot dry matter under low (a) and normal (b) P condition in pot culture according to Fageria and Baligar (1993). This classification divides genotypes into four groups i.e., ER efficient and responsive, IR in-efficient but responsive, ENR efficient but non-responsive, INR in-efficient and nonresponsive. Numbers from P1 to P30 represent cotton genotypes mentioned in Table S1
Method 3
In this method, cotton genotypes were classified for P efficiency and responsiveness based on the deviation of the mean PUtE of each genotype in relation to mean PUtE under both low and normal P conditions (Figs. S1a and S1b). In hydroponic culture, the graph showed that genotypes Jimian169, Zhongzhimian2, Lumianyan16, Lumianyan28, CCRI60, CCRI41, Jia-1‑7, ZhongzhimianBD13, CCRI19, and CCRI17 were categorized in ER suggesting that the mean PUtE is higher than the mean under both low P and normal P conditions. Moreover, genotypes DES926, TM‑1, CCRI10, Xinluzao49, Nongdamian7, Yuekangmian10, Dalingmian69, Xinluzhong5, CCRI12, Jimian11, Zhongzhimian86‑1, and Handan284 were classified as INR indicating their mean PUtE is lower than the combined mean under both low and normal P conditions (Fig. S1a). Likewise, in pot culture, the graph showed that genotypes Jimian169, Zhongzhimian2, Lumianyan16, CCRI19, Lumianyan28, CCRI60, CCRI41, Xinluzao49, Handan284, ZhongzhimianBD13, Yuekangmian10, CCRI49, and Deltapine15 were placed in ER, while DES926, TM‑1, CCRI10, Dalingmian69, Nongdamian7, Xinluzhong5, CCRI12, and Jimian11 were categorized as INR (Fig. S1b).
Method 4
This method is based on the stress tolerance score (STS) calculated from total dry matter with seven stress tolerance indices. In hydroponic culture, the highest STS score was recorded by Jimian169 (74.22) followed by Zhongzhimian2 (65.68) and Lumianyan16 (65.45). While the lowest was recorded by DES926 (21.96) followed by TM‑1 (23.75), Dalingmian69 (25.54), and Nongdamian7 (25.77) (Table 1). Similarly, in pot culture, the highest STS score was obtained by Jimian169 (42.71), Zhongzhimian2 (65.68), and Lumianyan16 (37.35). However, genotypes DES926 (9.31), TM‑1 (9.67), and CCRI10 (9.73) have the lowest STS values (Table 2).
Method 5
The hierarchical cluster analysis was performed to classify cotton genotypes based on various morphophysiological traits contributing to PUE (Fig. 3). As shown in the figure, thirty cotton genotypes were classified into four main groups. The hierarchical cluster classification showed that genotypes Jimian169, Zhongzhimian2, and Lumianyan16 were grouped in cluster 1, while DES926, TM‑1, and CCRI10 were grouped in cluster 4 indicating their contrasting behavior under both hydroponic and pot experiments (Fig. 3).
Genetic Parameters and Multivariate Analysis for Key Traits Mining
In the hydroponic experiment, the coefficient of genetic variation (CVg) varies between 3.14 and 41.52%. The highest genetic variation was observed in root diameter (41.52%), root P accumulation (41.18%), total P accumulation (38.53%), and shoot P accumulation (38.39%), while the lowest in intercellular CO2 concentration (3.14%). In case of heritability (H2), the highest H2 was recovered for root length (1.00%), root surface area (0.99%), shoot P accumulation (0.97%), and total P accumulation (0.97%), while the lowest for carotenoid contents (0.42%), transpiration rate (0.43%), and root volume (0.48%). Generally, the CVg, VI, and H2 values were high for most of the studied traits, except for intercellular CO2 concentration, transpiration rate, carotenoid contents, and stomatal conductance (Table 3). Similarly, in the pot experiment, the CVg range from 3.01 to 55.94%. The highest genetic variation was observed in root P accumulation (55.94%) shoot P accumulation (53.84%), and total P accumulation (53.29%), and the lowest was in intercellular CO2 concentration (3.01%). Following the results of the hydroponic experiment, the highest H2 was observed for root length (1.00%) and root surface area (0.99%), and the lowest for root shoot ratio (0.05%) (Table 4).
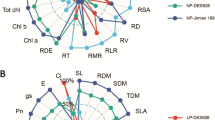
Principal component analysis was performed using different studied traits to identify the key traits. In hydroponic culture, the PC1 contributed 59.5% and was associated with P conditions, while cotton genotypes were associated with PC2 and shared 18.5% of the total variation. Shoot dry matter, shoot P accumulation, and total P accumulation were the key traits contributing to PC1, and root morphological traits like root surface area, root dry matter, root length, root volume, and root diameter mainly contributed to PC2 (Fig. 4a and Table S6). In pot culture, the loading plot of PC1 and PC2 contributed 59.5 and 20.1%, respectively. The traits like shoot P accumulation, root P accumulation, shoot dry matter, total plant dry matter, transpiration rate, root, shoot, and total P concentration contributed to PC1. However, shoot MDA, root MDA, root surface area, root length, root diameter, and root volume contributed to the PC2 (Fig. 4b and Table S6). The greater distance between the low and normal P conditions showed that cotton genotypes are highly responsive to P, especially in low P conditions.
Principal component analysis (PCA) biplot of morphophysiological traits of 30 cotton genotypes grown under low and normal P conditions in hydroponic (a) and pot (b) cultures. The eigenvectors are shown in Table S1. (SL Shoot length, SDM shoot dry matter, RDM root dry matter, TDM total dry matter, RSR root shoot ratio, RL root length, RSA root surface area, RV root volume, RD root diameter, Chl a chlorophyll content, Chl b chlorophyll b contents, Car carotenoid contents, Pn photosynthetic rate, gs stomatal conductance, E transpiration rate, Ci intercellular CO2 concentration, RP root P concentration, SP shoot P concentration, TP total P concentration, RPA root P accumulation, SPA shoot P accumulation, TPA total P accumulation, PUpE P uptake efficiency, PUtE P utilization efficiency, RMDA root malondialdehyde content, SMDA shoot malondialdehyde content, RSOD root superoxide dismutase activity, SSOD shoot superoxide dismutase activity, RPOD root peroxidase activity, SPOD shoot peroxidase activity, RCAT root catalase activity, SCAT shoot catalase activity)
Further correlation analysis was performed to reveal the relationship between various morphophysiological and biochemical traits with PUE (Fig. 5). In the hydroponic experiment, PUE traits had a strong positive correlation with maximum traits except for root shoot ratio, transpiration rate, and intercellular CO2 concentration. PUpE had a strong positive correlation (r > 0.56) with shoot dry matter, total plant dry matter, root surface area, root diameter, stomatal conductance, shoot P accumulation and total P accumulation. Similarly, PUtE had a strong positive correlation (r > 0.70) with shoot dry matter, total plant dry matter, root diameter, shoot P concentration, shoot P accumulation, total P accumulation, and PUpE (Fig. 5a). In the pot experiment, except root shoot ratio, transpiration rate, and intercellular CO2 concentration, all other studied traits have a positive relationship with PUE traits. PUpE had a strong positive correlation (r > 0.65) with shoot dry matter, total plant dry matter, root diameter, shoot P concentration, shoot P accumulation, and total P accumulation. Similarly, PUtE had a strong positive correlation (r > 0.80) with shoot dry matter, total plant dry matter, root diameter, shoot P accumulation, and total P accumulation (Fig. 5b).
Relationships between morphophysiological traits of 30 cotton genotypes grown under low and normal P conditions in hydroponic (a) and pot (b) culture. Where red color shows a positive correlation and blue color shows a negative correlation. * shows significant differences at p < 0.05. All the traits have been defined in Fig. 4
Discussion
The morphophysiological variation at the seedling stage in the controlled condition is a reliable approach to screen genotypes for stress tolerance and future breeding program (Duangpan et al. 2018). Low P stress affects plant growth and productivity (Zhang et al. 2014). Previously, it was observed that low P response varied among species as well as in different genotypes of the same specie (Bilal et al. 2018). Currently, genotypic variation in thirty cotton genotypes was analyzed using various morphophysiological traits to identify the P‑efficient and low P‑tolerant genotypes. In both hydroponic and pot cultures, cotton genotypes showed a considerable variation in various morphophysiological traits under low and normal P conditions, which are in line with the results obtained from the previous study (Neji et al. 2015). The variation among the traits was high under low P which is consistent with the variation in agronomic traits among sugarcane genotypes (da Silveira et al. 2014). Thus, abiotic stress, especially low P stress resulted in a great genotypic variation, suggesting that each genotype shows a different stress response. However, the variation among genotypes under both low and normal P conditions was more as compared to the earlier studies (Neji et al. 2015).
Different criteria have been proposed to evaluate PUE, such as total plant P (Sandaña and Pinochet 2016), dry matter production per unit P application (Rahim et al. 2010), and the ratio of active P to P uptake by the plant (Aziz et al. 2014). In chickpea, great variation was noted in tissue dry matter, tissue P concentration, PUpE, and PUtE under low P condition (Pang et al. 2018). In the current study, we used root dry matter, shoot dry matter, net photosynthetic rate, root shoot ratio, PUpE, and PUtE to identify the contrasting P‑efficient cotton genotypes. The main reason is relying on one or two traits is not sufficient and the aim must be clear before starting the classification of genotypes for PUE or breeding for P‑efficient genotypes (Aziz et al. 2011). On the basis of the selected traits, cotton genotypes were classified into three groups such as P‑efficient, P‑inefficient, and medium P‑efficient as suggested by earlier studies (Manske et al. 2000). However, this classification is not perfect and the genotype may be efficient under low P but could not produce sufficient dry matter under normal P condition. Therefore, genotype classification was performed on both low and normal P conditions and genotypes performed better under both conditions is desired hence the point scores were summed for classification (Tables S4 and S5). Finally, genotypes Jimian169, Zhongzhimian2, and Lumianyan16 recorded the highest score and were considered P‑efficient, while the lowest score was recorded from DES926, TM‑1, and CCRI10 and was classified as P‑inefficient, the remaining genotypes were considered as medium P‑efficient (Tables S4 and S5).
Similarly, in another method, genotypes were categorized into four groups based on P efficiency and responsiveness such as genotype with a high dry matter under low P and positive response to P application was considered P‑efficient and responsive (ER), and genotypes with low dry matter and positive response to P was classified as P‑inefficient and responsive (IR), genotypes with the high dry matter but no response to P application were grouped into P‑efficient and nonresponsive (ENR), and genotypes with low dry matter and no response to P application were presented as inefficient and nonresponsive (INR) (Neto et al. 2016). Among these groups, ER is the most desirable for breeding, followed by ENR, while INR genotypes were considered undesirable (Akhtar et al. 2008). Based on this classification, cotton genotypes Jimian169, Zhongzhimian2, and Lumianyan16 were mainly classified in ER, while that DES926, TM‑1, and CCRI10 as in the INR group in both hydroponic and pot culture. The reason behind this might be that genotypes DES926 and TM‑1 are American genotypes, while Jimian169, Zhongzhimian2, and Lumianyan16 are developed in China. Thus, it was suggested that the performance of cotton genotypes in low P is linked to the environmental variables, climatic conditions, and soil physicochemical properties where the genotypes have been developed. Importantly, according to this classification, our results indicated that the American genotypes DES926 and TM‑1 performed poor than Chinese origin genotypes Jimian169, Zhongzhimian2, and Lumianyan16 and therefore these genotypes might be used as model genotypes for breeding P‑efficient and low P tolerant genotypes.
Moreover, some scientists have classified genotypes based on the stress tolerance index using total plant dry matter as each genotype under low and normal P conditions (Grzesiak et al. 2019). This includes SSI, TI, and SI as susceptibility indices and presents a negative relationship with dry matter and thus dividing the genotypes into tolerant and susceptible (Sareen et al. 2012). However, MPI, GMPI, and STI are the tolerance indices indicating positive relation with dry matter and classify the genotypes with high dry matter and stress tolerance (Khodarahmpour et al. 2011). In the present study, it was clear that two indices MPI and SSI were able to explain the most percent of the variation among the studied indices. However, GMPI, MPI, and STI are the suggested indices to classify tolerant genotypes (Mohammadi et al. 2011). Conversely, tolerance and susceptible indices separately cannot identify tolerant genotypes with high dry matter (Khayatnezhad et al. 2010), therefore, a combination of both will be used to identify tolerant genotypes. In the current study, the highest stress tolerance score was noted for Jimian169 and was considered a low P tolerant genotype, while the lowest score was recorded for DES926 and was considered a low P susceptible genotype (Tables 1 and 2). Previously, STI was used to screen genotypes for drought tolerance in wheat, millet, and sorghum (Grzesiak et al. 2019; Negarestani et al. 2019) and for PUE in mung bean. This method classifies genotypes with high dry matter production and resilience under stress conditions. The current study provides new insights into genotypes screening and identification of the genotypic response of cotton genotypes under low and normal P conditions.
PCA and hierarchical cluster analysis has also been used for the identification of P‑efficient genotypes based on various morphophysiological traits (Pan et al. 2008). In addition, various studies have investigated that the PUE of a genotype is contributed by various morphophysiological traits that help in increasing the P uptake (PUpE) and its utilization to maintain growth and produce more dry matter under low P (PUtE) (Balemi and Schenk 2009b). In the current study, the PCA and hierarchical cluster analysis, with a strong relationship between dry matter and PUE, divided the genotypes into P‑efficient and P‑inefficient characterized by a difference in various morphophysiological traits in response to low and normal P conditions under hydroponic and pot culture. Moreover, the rest of the genotypes that showed distinct responses to low P, such as a decrease in dry matter and an increased shoot to root ratio (Frydenvang et al. 2015), were classified as medium P‑efficient genotypes. Interestingly, P‑efficient cotton genotypes established a good root system and high antioxidant enzymatic activities than P‑inefficient cotton genotypes, especially under low P. The increase in root proliferation and development of a better root system is an adaptive strategy of the plants against low P to increase P uptake (Iqbal et al. 2019a). In addition, comparatively high antioxidant enzymatic activities of P‑efficient genotypes under low P play a key role in low P tolerance. Thus, it was considered that the dry matter and antioxidant enzymatic activities could be used for classification of tolerant, efficient, and responsive genotypes under low P condition (Akhtar et al. 2008).
Heritability is an important breeding component that shows the relative contribution of genetic and environmental factors in the genetic variation of a trait and its transmission into further generations (Mazid et al. 2013). The results of the current study showed that the heritability of most of the studied traits was high in both low and normal P conditions (Tables 3 and 4), indicating that the variation among genotypes for the traits was under strong genetic control. Among the studied traits, a high heritability was recorded for root morphological and PUE traits, suggesting that the selection of these traits directly under low P would be more effective than the indirect selection under normal P (Gemenet et al. 2016). In line with our result, high heritability for growth and PUE traits were recorded in many crops (Ao et al. 2010). Conversely, a low heritability was recorded for photosynthetic traits (Tables 3 and 4), suggesting that variation in these traits was mainly controlled by the environment and cannot be used for breeding programs. This is supported by the contrasting performance of the genotypes as American genotypes DES926 and TM‑1 performed poorly than that of Chinese origin genotypes Jimian169, Zhongzhimian2, and Lumianyan16. These results were in line with the results obtained from B. hybridum germplasms, where low heritability was found for photosynthetic traits and suggested that the lines were developed in a different bioclimatic conditions as photosynthesis is mainly controlled by environmental factors (Gururani et al. 2015). Thus, we hypothesized that the photosynthesis of each genotype fit to the environment where it has been developed.
A strong positive relationship has been observed between dry matter and PUE traits that could be very useful for breeders to select genotypes with high dry matter and PUE. Similarly, a previous study has reported a negative relationship between dry matter and P concentration, suggesting that selection under such conditions may provide undesirable low-yield genotypes (Missaoui and Young 2016). Moreover, root morphological traits have a poor relationship with PUE traits which is supported by the results of an earlier study (Ao et al. 2010). Despite the significant increase in root length under low P, there was an obvious difference in root dry matter between low and normal P. This pattern suggested a contrasting dry matter partitioning between the shoot and root and a preferential translocation of P to the shoot as an adaptive mechanism under low P (Ao et al. 2010).
Conclusions
Classification of cotton germplasms is critical to understand P‑efficient and low P‑tolerant genotypes under low and normal P conditions is important for breeding PUE. A significant genotypic variation among the genotypes was found for root morphology, dry matter production, photosynthetic traits, PUE traits, and antioxidant enzymatic activities under low and normal P conditions. The P efficiency and tolerance of cotton genotypes to low P vary with traits and classification methods. The classification of cotton genotypes into different groups by using the described methods is important to know the minor discrepancy in the genotypes for low P tolerance and efficiency. Among these methods, the stress tolerance score is the method that visualizes the contrasting behaviors among the genotypes for dry matter production and resilience to low P. Based on various classification methods, genotype Jimian169 was found strong low P tolerant and efficient, while DES926 as weak low P tolerant and inefficient genotypes. In addition, traits like root dry matter, shoot dry matter, root to shoot ratio, net photosynthetic rate, P uptake efficiency, and P utilization efficiency are important in the categorization of cotton genotypes for PUE. The current study has laid the foundation for breeding a P-efficient cotton genotype that could grow well under low P fertilization and hence reduce the cost of production and environmental pollution.
References
Akhtar MS, Oki Y, Adachi T (2008) Intraspecific variations of phosphorus absorption and Remobilization, P forms, and their internal buffering in brassica Cultivars exposed to a P-stressed environment. J Integr Plant Biol 50(6):703–716
Ali Y, Krishnamurthy L, Saxena N, Rupela O, Kumar J, Johansen C (2002) Scope for genetic manipulation of mineral acquisition in chickpea. In: Food security in nutrient-stressed environments: Exploiting plants’ genetic capabilities. Springer, Dordrecht, pp 165–176
Almasy L, Blangero J (2010) Variance component methods for analysis of complex phenotypes. Cold Spring Harb Protoc. https://doi.org/10.1101/pdb.top77
Amanullah, Iqbal A, Khan A, Khalid S, Shah A, Parmar B, Muhammad A (2019) Integrated management of phosphorus, organic sources, and beneficial microbes improve dry matter partitioning of maize. Commun Soil Sci Plant Anal 50(20):2544–2569
Ao J, Fu J, Tian J, Yan X, Liao H (2010) Genetic variability for root morph-architecture traits and root growth dynamics as related to phosphorus efficiency in soybean. Funct Plant Biol 37(4):304–312
Asif I, Dong Q, Wang X, Gui H, Zhang H, Pang N, Song M (2021) Genotypic variation in root morphology, cotton subtending leaf physiology and fiber quality against nitrogen. J Cotton Res 4(1):1–14
Aski M, Mehra R, Mishra GP, Singh D, Yadav P, Rai N, Singh MP (2022) Genotypic variation in root architectural traits under contrasting phosphorus levels in Mediterranean and Indian origin lentil genotypes. PeerJ 10:e12766
Aziz T, Rahmatullah, Maqsood MA, Sabir M, Kanwal S (2011) Categorization of Brassica cultivars for phosphorus acquisition from phosphate rock on basis of growth and ionic parameters. J Plant Nutr 34(4):522–533
Aziz T, Finnegan PM, Lambers H, Jost R (2014) Organ-specific phosphorus-allocation patterns and transcript profiles linked to phosphorus efficiency in two contrasting wheat genotypes. Plant Cell Environ 37(4):943–960
Balemi T, Schenk MK (2009a) Genotypic difference of potato in carbon budgeting as a mechanism of phosphorus utilization efficiency. Plant Soil 322(1):91–99
Balemi T, Schenk MK (2009b) Genotypic variation of potato for phosphorus efficiency and quantification of phosphorus uptake with respect to root characteristics. J Plant Nutr Soil Sci 172(5):669–677
Bayuelo-Jiménez JS, Ochoa-Cadavid I (2014) Phosphorus acquisition and internal utilization efficiency among maize landraces from the central Mexican highlands. Field Crop Res 156:123–134
Beebe SE, Rojas-Pierce M, Yan X, Blair MW, Pedraza F, Munoz F, Lynch JP (2006) Quantitative trait loci for root architecture traits correlated with phosphorus acquisition in common bean. Crop Sci 46(1):413–423
Bello SK (2021) An overview of the morphological, genetic and metabolic mechanisms regulating phosphorus efficiency via root traits in soybean. J Soil Sci Plant Nutr 21(2):1013–1029
Bilal HM, Aziz T, Maqsood MA, Farooq M, Yan G (2018) Categorization of wheat genotypes for phosphorus efficiency. PLoS ONE 13(10):e205471
Carstensen A, Herdean A, Schmidt SB, Sharma A, Spetea C, Pribil M, Husted S (2018) The impacts of phosphorus deficiency on the photosynthetic electron transport chain. Plant Physiol 177(1):271–284
Cattivelli L, Rizza F, Badeck FW, Mazzucotelli E, Mastrangelo AM, Francia E, Stanca AM (2008) Drought tolerance improvement in crop plants: an integrated view from breeding to genomics. Field Crop Res 105(1–2):1–14
Chen B, Wang Q, Bücking H, Sheng J, Luo J, Chai Z, Feng G (2019) Genotypic differences in phosphorus acquisition efficiency and root performance of cotton (Gossypium hirsutum) under low-phosphorus stress. Crop Pasture Sci 70(4):344–358
Chowdhury RB, Zhang X (2021) Phosphorus use efficiency in agricultural systems: A comprehensive assessment through the review of national scale substance flow analyses. Ecol Indic 121:107172
Corrales I, Amenós M, Poschenrieder C, Barceló J (2007) Phosphorus efficiency and root exudates in two contrasting tropical maize varieties. J Plant Nut 30(6):887–900
Duangpan S, Buapet P, Sujitto S, Eksomtramage T (2018) Early assessment of drought tolerance in oil palm D× P progenies using growth and physiological characters in seedling stage. Plant Genet Resour 16(6):544–554
Fageria N, Baligar V (1993) Screening crop genotypes for mineral stresses. Workshop on adaption of plants to soil stresses. Lincoln. proceedings
Frydenvang J, van Maarschalkerweerd M, Carstensen A, Mundus S, Schmidt SB, Pedas PR, Husted S (2015) Sensitive detection of phosphorus deficiency in plants using chlorophyll a fluorescence. Plant Physiol 169(1):353–361
Gemenet DC, Hash CT, Sanogo MD, Sy O, Zangre RG, Leiser WL, Haussmann BI (2015) Phosphorus uptake and utilization efficiency in West African pearl millet inbred lines. Field Crop Res 171:54–66
Gemenet DC, Leiser WL, Beggi F, Herrmann LH, Vadez V, Rattunde HF, Haussmann BI (2016) Overcoming phosphorus deficiency in West African pearl millet and sorghum production systems: promising options for crop improvement. Front Plant Sci 7:1389
George T, Turner BL, Gregory P, Cade-Menun B, Richardson A (2006) Depletion of organic phosphorus from Oxisols in relation to phosphatase activities in the rhizosphere. Eur J Soil Sci 57:47–57
Gerhardt I, Amaral Júnior A, Guimarães L, Schwantes I, Santos A, Kamphorst S, Myers G (2017) Intraspecific variability of popcorn S7 lines for phosphorus efficiency in the soil. Embrapa Milho e Sorgo-Artigo em periódico indexado (ALICE)
Ghaffar S, Stevenson RJ, Khan Z (2017) Effect of phosphorus stress on Microcystis aeruginosa growth and phosphorus uptake. Plos One 12(3):e0174349
Grzesiak S, Hordyńska N, Szczyrek P, Grzesiak MT, Noga A, Szechyńska-Hebda M (2019) Variation among wheat (Triticum easativum L.) genotypes in response to the drought stress: I–selection approaches. J Plant Interact 14(1):30–44
Gururani MA, Venkatesh J, Tran LSP (2015) Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol Plant 8(9):1304–1320
Huang G, Hayes PE, Ryan MH, Pang J, Lambers H (2017) Peppermint trees shift their phosphorus-acquisition strategy along a strong gradient of plant-available phosphorus by increasing their transpiration at very low phosphorus availability. Oecologia 185(3):387–400
Inthapanya P, Sihavong P, Sihathep V, Chanphengsay M, Fukai S, Basnayake J (2000) Genotype differences in nutrient uptake and utilisation for grain yield production of rainfed lowland rice under fertilised and non-fertilised conditions. Field Crop Res 65(1):57–68
Iqbal A, Gui H, Zhang H, Wang X, Pang N, Dong Q, Song M (2019a) Genotypic variation in cotton genotypes for phosphorus-use efficiency. Agronomy 9(11):689
Iqbal A, Song M, Shah Z, Alamzeb M, Iqbal M (2019b) Integrated use of plant residues, phosphorus and beneficial microbes improve hybrid maize productivity in semiarid climates. Acta Ecol Sinica 39(5):348–355
Iqbal A, Dong Q, Wang X, Gui H, Zhang H, Zhang X, Song M (2020a) High nitrogen enhance drought tolerance in cotton through antioxidant enzymatic activities, nitrogen metabolism and osmotic adjustment. Plants 9(2):178
Iqbal A, Dong Q, Wang X, Gui H, Zhang H, Zhang X, Song M (2020b) Variations in nitrogen metabolism are closely linked with nitrogen uptake and utilization efficiency in cotton genotypes under various nitrogen supplies. Plants 9(2):250
Iqbal A, Dong Q, Wang X, Gui HP, Zhang H, Pang N, Song M (2020c) Nitrogen preference and genetic variation of cotton genotypes for nitrogen use efficiency. J Sci Food Agric 100(6):2761–2773
Iqbal A, Qiang D, Zhun W, Xiangru W, Huiping G, Hengheng Z, Meizhen S (2020d) Growth and nitrogen metabolism are associated with nitrogen-use efficiency in cotton genotypes. Plant Physiol Biochem 149:61–74
Iqbal A, Jing N, Qiang D, Xiangru W, Huiping G, Hengheng Z, Meizhen S (2022a) Physiological characteristics of cotton subtending leaf are associated with yield in contrasting nitrogen-efficient cotton genotypes. Front Plant Sci 13:825116–825116
Iqbal A, Qiang D, Xiangru W, Huiping G, Jing N, Leilei L, Meizhen S (2022b) N‑efficient cotton genotype grown under low nitrogen shows relatively large root system, high biomass accumulation and nitrogen metabolism. Agron J 114(1):582–600
Iqbal A, Qiang D, Xiangru W, Huiping G, Hengheng Z, Xiling Z, Meizhen S (2022c) Low phosphorus tolerance in cotton genotypes is regulated by root morphology and physiology. Growth and nitrogen metabolism are associated with nitro-gen-use efficiency in cotton genotypes. J Plant Growth Regulat. https://doi.org/10.1007/s00344-022-10829-5
Irfan M, Abbas M, Shah JA, Akram MA, Depar N, Memon MY (2020) Categorization and identification of brassica genotypes for phosphorus utilization efficiency. Int J Agric Biol 23(1):227–234
Khayatnezhad M, Zaeifizadeh M, Gholamin R (2010) Investigation and selection index for drought stress. Aust J Basic Appl Sci 4(10):4815–4822
Khodarahmpour Z, Choukan R, Bihamta M, Majidi HE (2011) Determination of the best heat stress tolerance indices in maize (Zea mays L.) inbred lines and hybrids under Khuzestan province conditions
Kosar H, Gill M, Aziz T, Tahir M (2003) Relative phosphorus utilization efficiency of wheat genotypes in hydroponics. Pak J Agric Sci. 40(1-2):28–32
Li H, Liu J, Li G, Shen J, Bergström L, Zhang F (2015) Past, present, and future use of phosphorus in Chinese agriculture and its influence on phosphorus losses. AMBIO 44(2):274–285
Li L, Qian Q, Hengheng Z, Qiang D, Asif I, Huiping G, Mirezhatijiang K, Meizhen S, Xiling Z, Xiangru W (2022) Ameliorative effects of silicon against salt stress in Gossypium hirsutum L. Antioxidants 11:1520
Lynch JP (2019) Root phenotypes for improved nutrient capture: an underexploited opportunity for global agriculture. New Phytol 223(2):548–564
Manske G, Ortiz-Monasterio J, Van Ginkel M, Gonzalez R, Rajaram S, Molina E, Vlek P (2000) Traits associated with improved P‑uptake efficiency in CIMMYT’s semidwarf spring bread wheat grown on an acid Andisol in Mexico. Plant Soil 221(2):189–204
Masoni A, Ercoli L, Mariotti M, Arduini I (2007) Post-anthesis accumulation and remobilization of dry matter, nitrogen and phosphorus in durum wheat as affected by soil type. Eur J Agron 26(3):179–186
Mazid M, Rafii M, Hanafi M, Rahim H, Shabanimofrad M, Latif M (2013) Agro-morphological characterization and assessment of variability, heritability, genetic advance and divergence in bacterial blight resistant rice genotypes. S Afr J Bot 86:15–22
McDonald G, Bovill W, Taylor J, Wheeler R (2015) Responses to phosphorus among wheat genotypes. Crop Past Sci 66(5):430–444
Missaoui AM, Young J (2016) Genetic gain from selection and potential for improving alfalfa phosphorus uptake and removal from soils heavily amended with poultry litter. Euphytica 209(2):495–506
Mohammadi M, Karimizadeh R, Abdipour M (2011) Evaluation of drought tolerance in bread wheat genotypes under dryland and supplemental irrigation conditions. Aust J Crop Sci 5(4):487–493
Mori A, Fukuda T, Vejchasarn P, Nestler J, Pariasca-Tanaka J, Wissuwa M (2016) The role of root size versus root efficiency in phosphorus acquisition in rice. J Exp Bot 67(4):1179–1189
Negarestani M, Tohidi-Nejad E, Khajoei-Nejad G, Nakhoda B, Mohammadi-Nejad G (2019) Comparison of different multivariate statistical methods for screening the drought tolerant genotypes of pearl millet (Pennisetum americanum L.) and sorghum (Sorghum bicolor L.). Agronomy 9(10):645
Neji M, Geuna F, Taamalli W, Ibrahim Y, Smida M, Badri M, Gandour M (2015) Morpho-phenological diversity among Tunisian natural populations of Brachypodium hybridum. J Agric Sci 153(6):1006–1016
Neto AP, Favarin JL, Hammond JP, Tezotto T, Couto HT (2016) Analysis of phosphorus use efficiency traits in Coffea genotypes reveals Coffea arabica and Coffea canephora have contrasting phosphorus uptake and utilization efficiencies. Front Plant Sci 7:408
Niu J, Gui H, Iqbal A, Zhang H, Dong Q, Pang N, Yang G (2020) N‑use efficiency and yield of cotton (G. hirsutum L.) are improved through the combination of N‑fertilizer reduction and N‑efficient cultivar. Agronomy 11(1):55
Niu YF, Chai RS, Jin GL, Wang H, Tang CX, Zhang YS (2013) Responses of root architecture development to low phosphorus availability: a review. Ann Bot 112(2):391–408
Osborne L, Rengel Z (2002) Screening cereals for genotypic variation in efficiency of phosphorus uptake and utilisation. Aust J Agric Res 53(3):295–303
Pan XW, Li WB, Zhang QY, Li YH, Liu MS (2008) Assessment on phosphorus efficiency characteristics of soybean genotypes in phosphorus-deficient soils. Agric Sci China 7(8):958–969
Pang J, Zhao H, Bansal R, Bohuon E, Lambers H, Ryan MH, Siddique KH (2018) Leaf transpiration plays a role in phosphorus acquisition among a large set of chickpea genotypes. Plant Cell Environ 41(9):2069–2079
Parentoni SN, Souza Júnior CL (2008) Phosphorus acquisition and internal utilization efficiency in tropical maize genotypes. Pesq agropec bras 43:893–901
Rahim A, Ranjha A, Waraich E (2010) Effect of phosphorus application and irrigation scheduling on wheat yield and phosphorus use efficiency. Soil Environ 29(1):15–22
Rausch C, Bucher M (2002) Molecular mechanisms of phosphate transport in plants. Planta 216(1):23–37
Ravikiran K, Krishnamurthy S, Warraich A, Sharma P (2018) Diversity and haplotypes of rice genotypes for seedling stage salinity tolerance analyzed through morpho-physiological and SSR markers. Field Crop Res 220:10–18
Reddy VRP, Dikshit HK, Mishra GP, Aski M, Singh A, Bansal R, Nair RM (2021) Comparison of different selection traits for identification of phosphorus use efficient lines in mungbean. PeerJ 9:e12156
Ruiz S, Koebernick N, Duncan S, Fletcher DM, Scotson C, Boghi A, Brown L (2020) Significance of root hairs at the field scale–modelling root water and phosphorus uptake under different field conditions. Plant Soil 447(1):281–304
Sandaña P (2016) Phosphorus uptake and utilization efficiency in response to potato genotype and phosphorus availability. Eur J Agron 76:95–106
Sandaña P, Pinochet D (2016) Phosphorus acquisition of wheat, pea and narrow-leafed lupin under different P supplies. J Soil Sci Plant Nutr 16(2):537–549
Sareen S, Tyagi B, Tiwari V, Sharma I (2012) Response estimation of wheat synthetic lines to terminal heat stress using stress indices. J Agric Sci 4(10):97–104
da Silveira G, de Costa PMA, Kist V, de Almeida CF, Baffa DCF, Barbosa MHP (2014) Genetic variation affecting agronomic traits in sugarcane in response to high and low phosphorus availability. Agron J 106(6):2296–2304
Srinivasarao C, Ganeshamurthy A, Ali M, Venkateswarlu B (2006) Phosphorus and micronutrient nutrition of chickpea genotypes in a multi-nutrient-deficient typic ustochrept. J Plant Nutr 29(4):747–763
Taliman NA, Dong Q, Echigo K, Raboy V, Saneoka H (2019) Effect of phosphorus fertilization on the growth, photosynthesis, nitrogen fixation, mineral accumulation, seed yield, and seed quality of a soybean low-phytate line. Plants 8(5):119
Wang X, Shen J, Liao H (2010) Acquisition or utilization, which is more critical for enhancing phosphorus efficiency in modern crops? Plant Sci 179(4):302–306
Wang N, Xiangru W, Qian Q, Asif I, Hengheng Z, Jianbin S, Qiang D, Qinghua X, Xiaohong L, Huiping G, Meizhen S, Xiling Z, Gentu Y (2022) Analysis of the effects of mepiquat chloride priming on the seedling growth-promoting in cotton under salt stress by multi-omics. Ind Crops Prod 186:115296
Wang R, Guo S, Li N, Li R, Zhang Y, Jiang J, Sun Q (2015) Phosphorus accumulation and sorption in calcareous soil under long-term fertilization. PLoS ONE 10(8):e135160
Wang Y, Chen YF, Wu WH (2021) Potassium and phosphorus transport and signaling in plants. J Integ Plant Biol 63(1):34–52
Yang Y, Tong Y, Li X, He Y, Xu R, Liu D, Liao H (2019) Genetic analysis and fine mapping of phosphorus efficiency locus 1 (PE1) in soybean. Theor Appl Genet 132(10):2847–2858
Zhang F, Cui Z, Chen X, Ju X, Shen J, Chen Q, Fan M (2012) Integrated nutrient management for food security and environmental quality in China. Adv Agron 116:1–40
Zhang JY, Cruz De Carvalho MH, Torres-Jerez I, Kang Y, Allen SN, Huhman DV, Udvardi MK (2014) Global reprogramming of transcription and metabolism in Medicago truncatula during progressive drought and after rewatering. Plant Cell Environ 37(11):2553–2576
Zhang Q, Liu J, Liu X, Li S, Sun Y, Lu W, Ma C (2020) Optimizing water and phosphorus management to improve hay yield and water- and phosphorus-use efficiency in alfalfa under drip irrigation. Food Sci Nutr 8(5):2406–2418
Funding
We greatly appreciate the financial support from the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences, Cotton Research Institute, Chinese Academy of Agricultural Sciences, State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences (Grant number; CB2021C10), and Basic Research Service Fund of Central Public Welfare Scientific Institutions (Grant number; 1610162021025).
Author information
Authors and Affiliations
Contributions
Asif Iqbal: Conceptualization, Data curation, Investigation, Methodology, Validation, Visualization, Writing—original draft, Writing—review & editing. Dong Qiang: Conceptualization, Methodology, Project administration, Resources, Software, Supervision, Writing—original draft, Writing—review & editing. Wang Xiangru: Data curation, Resources, Software, Visualization. Gui Huiping: Formal analysis, Methodology. Zhang Hengheng: Data curation, Formal analysis. Zhang Xiling: Funding acquisition, Project administration, Resources, Software, Supervision, Writing—review & editing. Song Meizhen: Conceptualization, Funding acquisition, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing—review & editing.
Ethics declarations
Conflict of interest
A. Iqbal, D. Qiang, W. Xiangru, G. Huiping, Z. Hengheng, Z. Xiling and S. Meizhen declare that they have no competing interests.
Additional information
Asif Iqbal and Dong Qiang contributed equally to this work.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Iqbal, A., Qiang, D., Xiangru, W. et al. Genotypic Variation in Cotton Genotypes for Low Phosphorus Tolerance and Efficiency Under Different Growth Conditions. Gesunde Pflanzen 75, 1975–1993 (2023). https://doi.org/10.1007/s10343-022-00823-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10343-022-00823-y