Abstract
Forty-one E. bosistoana families were evaluated for the production of heartwood quantity and quality in two sites. High estimated heritabilities of heartwood diameter (HWD) were found in both sites (0.66 and 0.71). The estimated heritabilities of extractives content (EC) were lower with 0.16 and 0.25. Weak genetic correlations between HWD and EC were found in one site, but highly negative (− 0.86) genetic correlations were observed in the other. The G × E interaction had no significant influence on growth traits but a small-level influence on the EC. Five families were selected for tree breeding as they produced both large HWD and high EC in both sites. It was suggested that genetic breeding selection could improve the heartwood quantity and quality of E. bosistoana plantations.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Most tree breeding programs for Eucalyptus species target improvements in tree growth and health, basic density and pulp yield, as plantations were primarily intended for pulp and paper products (Greaves and Borralho 1996; Schimleck et al. 2004; Stackpole et al. 2010). Less work has been done on improving Eucalyptus solid-wood properties; in particular, work on heartwood properties is very scarce. As plantations and their breeding programs have evolved, for some species the range of traits assessed has increased to include natural durability (Bush et al. 2011).
Natural durability describes the resistance of wood to biological decay by fungi, insects or marine borers without wood preservation treatment. The presence of wood extractives is the most important factor explaining the natural durability of wood (Hawley et al. 1924; Rudman 1964). Extractives are deposited when the inner part of the sapwood is transformed into heartwood (Hillis 1987; Taylor et al. 2002).
New Zealand vineyards preponderantly use softwood posts to support the vines. These posts are treated with copper/chrome/arsenate (CCA), as untreated radiata pine is not sufficiently durable to meet commercial standards (Bush et al. 2011). However, these wood preservation treatments result in costly and potentially harmful environmental problems (Townsend and Solo-Gabriele 2006). These concerns have resulted in naturally durable wood being increasingly regarded as a desirable material for posts.
There is significant within-species variability on natural resistance to biodeterioration, which is partly controlled by genetics and possibly amenable to improvement through breeding (Hillis 1987). Genetic variation in natural durability has been reported for various tree species, including Larix sp. (Gierlinger et al. 2004), Thuja plicata, Chamaecyparis nootkatensis (Taylor et al. 2006) or Picea glauca (Yu et al. 2003) as well as some eucalyptus species. Significant variation in fungal decay resistance has been reported in Eucalyptus marginata (Perry et al. 1985), E. cladocalyx (Bush 2011) and E. grandis as well as E. camaldulensis × botryoides clones (Palanti et al. 2010). Bush (2011) also found that methanol extractive content in E. cladocalyx could be improved by breeding, a finding mirrored in Pinus sylvestris with reported heritability ranging from 0.5 to 0.7 for quantities of different extractives (Fries et al. 2000).
Apart from heartwood of good quality, it is also important that the trees produce large quantities of heartwood. Heartwood quantity has also been shown to be under genetic control, for example in E. cladocalyx (Bush 2011) or P. sylvestris (Ericsson and Fries 1999).
For genetic variability, the effect of environmental variables should also be considered. Lima et al. (2000) found significant clone × site interaction for wood density of 26 Eucalyptus clones in four sites at 8 years old. There is also evidence of plantation site influencing heartwood decay resistance in Tectona grandis (Moya and Berrocal 2010). Gierlinger et al. (2004) reported that the heartwood/sapwood ratio and the axial development of heartwood within E. globulus shows high levels of genotype × environment (G × E) interaction. Moraes et al. (2002) suggested that site differences may indirectly influence the amount of heartwood extractives linked to termite resistance in Eucalyptus spp.
Eucalyptus bosistoana can grow in warm temperate climates up to 30–40 m in height and usually has excellent stiffness, high density and hardness (Bootle 2005; Poynton 1979). In addition, E. bosistoana is classified as class 1 durable (Australian Standard 2003), having a life expectancy of more than 25 years in-ground. The New Zealand Dryland Forests Initiative (NZDFI) identified E. bosistoana as the main species to establish an alternative durable wood plantation in New Zealand to meet the insufficient supply of ground-durable timber (Altaner et al. 2017). There are very few studies on the performance variability of E. bosistoana, particularly of its durability as a plantation species (Apiolaza et al. 2011; Davies et al. 2017).
In this study, we examine the influence of genetic and environmental effects on the variability of properties associated to durability of E. bosistoana, using samples taken from two first-generation progeny trials. Heartwood diameter (HWD), sapwood diameter (SWD) sapwood area (SWA), heartwood extractive content (EC) and growth traits were examined. Near infrared reflectance (NIR) spectroscopy was evaluated as a rapid and cost-effective method to screen samples for EC.
Materials and methods
Materials
The New Zealand Drylands Forest Initiative (NZDFI) planted two E. bosistoana progeny trials in 2010. The trials represented 41 open-pollinated families, established in Martin (Canterbury) and Craven’s Road (Marlborough) on New Zealand’s South Island. Table 1 summarises the characteristics of the two sites.
All trees were planted in an alpha lattice incomplete-block, single-tree-plot design. Incomplete blocks were \(12\,{\text{m}} \times 10.8\,{\text{m}}\), with 30 trees per plot using a 2.4 m × 1.8 m spacing. Each tree represented one family, and no family was repeated within a block. Trials were thinned and pruned in Craven’s Road at age six. There were one family at Martin and five families at Craven’s Road with no surviving trees.
Measurements
Stem diameter at breast height (DBH) was measured in 2017. Samples were collected in May and June 2017, coring all living trees in the trials with a battery-powered, 14-mm inner-diameter increment corer. Bark-to-bark stem cores through the pith were taken ~ 50 cm above the ground. In total, 1115 trees were cored at Martin and 650 trees at Craven’s Road (Table 1). Each core was collected into a cooling box and measured for core length under bark and HWD with a ruler in the green state at the day of collection. The heartwood was highlighted on the cores using a pH indicator (methyl orange). Heartwood changed colour to pink, while the colour of sapwood remained unchanged. SWD was calculated as the difference between the core length and HWD. SWA was calculated assuming circular a cross section.
NIR spectroscopy and model calibration
Measurements of EC and decay resistance in laboratory tests or field trials are time- and cost-consuming. Near infrared spectroscopy was used for rapidly and effectively measuring the EC in thousands of tree cores. Each core was placed into an air-controlled room for a month (25 °C, 60% RH) to obtain a stable moisture content (~ 9%).
NIR spectra were collected from each core sample at the radial–tangential plane, after it had been sanded using P 100 sand paper, with a fibre optics probe (Model N-500, Bruker Optics, Germany) every 5 mm along the heartwood. Spectra were acquired from 9000 to 4000 cm−1 at 4 cm−1 intervals. Thirty-two scans were averaged for each spectrum. EC was predicted for each spectra using a PLS calibration developed by Li and Altaner (2018). The mean EC in heartwood was calculated as an area-weighted average.
Statistical analysis
Estimates of genetic parameters were obtained by fitting bivariate linear mixed models with restricted maximum likelihood (REML) analysis. Considering a single-trait observation yi for a tree, it can be represented with the following model equation:
where xi is a vector linking the fixed effects m to the observation, and bi, fi and ei are the random block, family and residual effects. Expanding the notation to a bivariate case, for each individual we have a vector of two observations yi (phenotypes for trait 1 and 2), and random vectors bi, fi and ei for blocks, families and residuals. Stacking those vectors for all trees produces the model equation:
where y is a vector of phenotypic observations, m is the vector of fixed effects (overall mean), b, f and e are vectors of bivariate random effects for block, family and residual effects. X, Z1 and Z2 are incidence matrices linking observations to the appropriate effects. The vector of expected values (E) and dispersion matrices (Var) were defined as:
where \(\otimes\) and \(\oplus\) are the direct product and direct sum operations, respectively, and
where \(\sigma_{{b_{i} }}^{2}\), \(\sigma_{{f_{i} }}^{2}\) and \(\sigma_{{e_{i} }}^{2}\) represent the block, family and residual variances for trait \(i\), and \(\sigma_{{b_{i} b_{j} }}\), \(\sigma_{{f_{i} f_{j} }}\) and \(\sigma_{{e_{i} e_{j} }}\) are the block, family and residual covariances between traits \(i\) and trait \(j\). The block and residual covariances were 0 when analysing the same trait across sites, as there was no information to estimate them.
The narrow sense heritability \((h^{2} )\) for trait i was estimated by using variance components from model 2
where \(\sigma_{{f_{i} }}^{2}\),\(\sigma_{{b_{i} }}^{2}\) and \(\sigma_{{e_{i} }}^{2}\) are, respectively, the family, block and residual variance for trait \(i\). The genetic correlations \(( r_{{g_{ij} }} )\) and phenotypic correlation \(( r_{{p_{ij} }} )\) between trait i and trait j were calculated as
where \(\sigma_{{f_{ij} }}^{ }\) is the estimated family covariance between trait \(i\) and trait \(j\), \(\sigma_{{f_{i} }}^{2}\) is the estimated family variance for trait \(i\), and \(\sigma_{{f_{j} }}^{2}\) is the estimated family variance for trait \(j\). The realised genetic gain (\(\Delta G_{R}\)) was computed by subtracting the mean breeding values of selected top ratio wood traits from the total mean of the wood trait. The difference was subsequently calculated in each site.
All models were fit using the asreml-R package (Gilmour et al. 2009) in the R statistical system (R Core Team 2017).
Results and discussion
Differences between family and site in growth and heartwood properties
Table 2 presents summary statistics for the six traits at Martin and Craven’s Road. In both sites, the coefficients of phenotypic variation (CV) were highest for HWD followed by SWA, EC, DBH and SWD. There were site-level differences for the wood traits, with Craven’s Road showing the higher mean for DBH, SWD, SWA and HWD but the lower mean for EC. In detail, DBH and HWD for trees at Craven’s Road were with 101.8 and 42.1 mm significantly larger than those at Martin where DBH averaged 64.2 mm and HWD 31.7 mm. The mean annual increment in DBH was 14.5 mm/year at Craven’s Road and 9.2 mm/year at Martin. The mean SWA were 84.5 and 55.6 cm2 in Craven’s Road and Martin, respectively. However, these larger trees at Craven’s Road had a lower mean EC (7.7%) than those at Martin (9.6%). The EC means were lower than the 12% methanol-soluble extractives reported for E. cladocalyx at 8 years of age (Bush et al. (2011). However, apart from difference between species, older trees are known to produce more extractives than younger trees (Rudman 1964) and methanol extracts slightly different compound mixtures than ethanol which was predicted in this study.
The data showed that trees at Craven’s Road produced more heartwood and more sapwood. As trees at Craven’s Road produced less heartwood extractives during growth than those at Martin suggested that fast growing trees usually produce less EC.
Heritability estimates used a coefficient of relationship of 1/2.5 in our study, which differed from (Apiolaza et al. 2011), who used ¼ for half-sibling E. bosistoana families. The genetic structure of eucalypt populations is known to be complex due to inbreeding and hybridisation (Elliott and Byrne 2003; Hunde et al. 2007; McDonald et al. 2003). However, there is no published information on both the reproductive biology and population structure of E. bosistoana to make informed assumptions. Lower relatedness coefficients pushed heritability estimates outside the parameter space (\(h^{2} > \, 1\)), suggesting deviations from the assembling of half-siblings and the presence of inbreeding effects.
Moderate individual heritability estimates were found for most wood traits (Table 2). The highest heritability estimates were for DBH at Martin \((h^{2} = \, 1.11)\). The estimated heritability for HWD in both sites was similar with 0.66 and 0.71. These values were higher than the findings of Bush et al. (2011), who estimated a heritability of 0.3–0.38 for heartwood proportion in E. cladocalyx and Santos et al. (2004) who presented a heritability of 0.39 for the sapwood/heartwood ratio in E. grandis. EC was less heritable (\(h^{2} = \, 0.16\) in Martin and 0.25 in Craven’s Road). This was lower in one of the sites than the results reported by Wu et al. (2017) for EC who reported a heritability of 0.26 for E. grandis and E. urophylla hybrids and by Bush et al. (2011) who reported a heritability of 0.25 for E. cladocalyx. The estimates of heritability for DBH at age 7 were 0.69 and 1.11 for the two sites. These were higher than the reported h2 of 0.13 estimated at age 7 for E. globulus (Costa e Silva et al. 2009). The high and moderate heritabilities for HWD and EC (Table 2), respectively, together with the substantial variability, suggested potential for improving heartwood quantity and quality via selection for E. bosistoana.
Phenotypic and genetic correlations between traits
Table 3 shows the estimated genetic and phenotypic correlations between different traits at Martin and Craven’s Road. Genetic correlations were high between HWD and DBH (\(r_{g} = \, 0.89\) in Martin and \(r_{g} = \, 0.98\) in Craven’s Road). Martin had the smaller DBH and smaller heartwood diameter distribution, which suggested that tree growth constrained heartwood diameter growth. In summary, big trees yield larger heartwood diameter than small trees. Similar positive correlations have been reported among DBH and heartwood diameter in P. pinaster (Pinto et al. 2004), P. radiata (Wilkes 1991), and E. globulus (Miranda et al. 2009).
No genetic correlation (0.13) and weak phenotypic correlation (0.37) was found between HWD and EC at Martin, while at Craven’s Road significant negative genetic correlations between EC and other traits, including HWD (− 0.86), SWD (− 0.89) and DBH (− 0.86) were detected. In 2015 this site was pruned and small trees thinned, which may have contributed to the differences between sites. Negative correlations between growth traits and EC were also reported by Wu et al. (2017) for Eucalyptus hybrid clones. Therefore, growing large-diameter trees on fast growing sites will not necessarily result in the most valuable plantations if durable wood is the target product.
Site influence on wood traits
Genotype by environment (G × E) interaction is a common preoccupation in breeding for growth traits (Alía et al. 1997; Costa e Silva et al. 2006; Ivković et al. 2015). In multivariate genetic evaluation, genetic correlations between the same traits in different sites can be interpreted as a measure of G × E interaction. Consistent rankings across sites (low G × E interaction) are reflected as high genetic correlations.
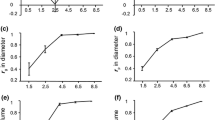
Figure 1 shows the genetic correlations across sites (and their standard errors) for all traits, while Fig. 2 displays the across-site stability for the predicted family values. Rankings were consistent across the two sites for HWD, SWA, SWD, DBH indicated by the high genetic correlations of 0.98, 0.96, 0.93 and 0.95, respectively. Even the lowest genetic correlation (0.6 for EC) suggested only moderate changes in the rankings between sites. Nevertheless, it is important to remember that this is a sample of only two sites, and there is evidence of more variable performance between sites for E. bosistoana in terms of tree height (e.g. Apiolaza et al. 2011).
Changes of family rankings across sites for heartwood diameter (HWD), DBH, extractives content (EC) and sapwood area (SWA) for E. bosistoana at age 7 between two sites. Family values are expressed as deviation from the site mean. BV breeding values, DBH diameter at breast height, EC heartwood extractive content, HWD heartwood diameter, SWA sapwood area
Family selection
Quantity and quality of heartwood are the main timber traits in the NZDFI breeding programme. Families that have both large HWD and high EC should be preferred for breeding. Figure 3 presents the mean of HWD and EC relationship in both sites. It is clear that some families have large HWD in both sides, including families 133, 134, 135, 138, 139, but have also lower EC. These families should not be selected as low EC is likely to result in low natural durability. Similarly families, which produced high EC but small HWD in both sites, for example 118, 121 or 128, should not be selected. In contrast, despite the negative correlation between EC and HWD (Table 3), some families (including 101, 112, 117, 120, 126) produced above average HWD and EC and should be selected. Families, which performed above average for both, HWD and EC, in only one of the sites (e.g. 132 and 115 at Craven’s Road and 107 and 111 at Martin) have potential when matched to site conditions.
Realised genetic gains
The realised genetic gains when selecting the top 10, 20 and 30% of the families for individual traits are presented in Table 4. Genetic gains increased with stronger selection rates for all traits. Estimated genetic gains for DBH in both sites ranged from 17.8 to 34.7 mm. This compared to gains of 11.6–24 mm per year reported for E. globulus when selecting the top 0.6% of individuals (Jarvis et al. 1995). However, since for E. bosistoana the target trait is heartwood, selection for DBH is not appropriate. Instead, selection should consider HWD, which gave estimated gains from 21.9 to 14 mm for Craven’s Road and from 16.7 to 10.5 mm for Martin. Larger genetic gain for HWD, SWD, DBH and SWA can be expected for sites comparable to Craven’s Road. Growing conditions similar to those at Martins only slightly favoured realised genetic gain for EC.
Conclusions
We observed high variation and relatively high heritabilities of HWD (h2 = 0.66–0.71) and DBH (h2 = 0.69–1.11) for 7-year-old E. bosistoana at all sites. It was possible to identify families with superior HWD and EC despite their negative genetic correlation.
There was no practically significant G × E interaction for growth traits, while there was a small level of G × E interaction for EC. We observed both phenotypic and genetic variation for HWD and EC between the families, indicating that there is potential to improve heartwood quantity and quality in E. bosistoana by selection to ensure trees with abundant heartwood of good quality.
References
Alía R, Moro J, Denis JB (1997) Performance of Pinus pinaster provenances in Spain: interpretation of the genotype by environment interaction. Can J For Res 27:1548–1559
Altaner C, Murray TJ, Morgenroth J (eds) (2017) Durable eucalypts on drylands: protecting and enhancing value. New Zealand School of Forestry, Christchurch
Apiolaza LA, Mcconnochie R, Millen P (2011) Introducing durable species to New Zealand drylands: genetics of early adaptation of Eucalyptus bosistoana. In: Walker J (ed) Developing a eucalypt resource: learning from Australia and elsewhere, vol 137. Wood Technology Research Centre, Canterbury
Australian Standard (2003) Timber-natural durability ratings. AS 5604e2003. Standards Australia, Homebush
Bootle KR (2005) Wood in Australia. Types, properties, and uses, 2nd edn. McGraw-Hill, North Ryde
Bush D (2011) Selecting and breeding eucalypts for natural durability. In: Walker JCF (ed) Developing a eucalypt resource: learning from Australia and elsewhere. Wood Technology Research Centre, Blenheim, pp 125–136
Bush D, McCarthy K, Meder R (2011) Genetic variation of natural durability traits in Eucalyptus cladocalyx (sugar gum). Ann For Sci 68:1057
Costa e Silva J, Potts BM, Dutkowski GW (2006) Genotype by environment interaction for growth of Eucalyptus globulus in Australia. Tree Genet Genomes 2:61–75. https://doi.org/10.1007/s11295-005-0025-x
Costa e Silva J, Borralho NM, Araújo JA, Vaillancourt RE, Potts BM (2009) Genetic parameters for growth, wood density and pulp yield in Eucalyptus globulus. Tree Genet Genomes 5:291–305
Davies NT, Apiolaza LA, Sharma M (2017) Heritability of growth strain in Eucalyptus bosistoana: a Bayesian approach with left-censored data. N Z J For Sci 47:5
Elliott C, Byrne M (2003) Genetic diversity within and between natural populations of Eucalyptus occidentalis (Myrtaceae). Silvae Genet 52:169–173
Ericsson T, Fries A (1999) High heritability for heartwood in north Swedish Scots pine. Theor Appl Genet 98:732–735. https://doi.org/10.1007/s001220051128
Fries A, Ericsson T, Gref R (2000) High heritability of wood extractives in Pinus sylvestris progeny tests. Can J For Res 30:1707–1713
Gierlinger N, Jacques D, Grabner M, Wimmer R, Schwanninger M, Rozenberg P, Pâques LE (2004) Colour of larch heartwood and relationships to extractives and brown-rot decay resistance. Trees 18:102–108. https://doi.org/10.1007/s00468-003-0290-y
Gilmour AR, Gogel BJ, Cullis BR, Thompson R (2009) ASReml User Guide Release 3.0. VSN International Ltd, Hemel Hempstead, HP1 1ES, UK. http://www.vsni.co.uk
Greaves B, Borralho N (1996) The influence of basic density and pulp yield on the cost of eucalypt kraft pulping: a theoretical model for tree breeding. Appita J 49:90–95
Hawley LF, Fleck LC, Richards CA (1924) The relation between durability and chemical composition in wood. Ind Eng Chem Res 16:699–700
Hillis W (1987) Heartwood and tree exudates. Springer, Berlin
Hunde T, Gizachew B, Harwood C (2007) Genetic variation in survival and growth of Eucalyptus globulus ssp. globulus in Ethiopia. Aust For 70:48–52
Ivković M, Gapare W, Yang H, Dutkowski G, Buxton P, Wu H (2015) Pattern of genotype by environment interaction for radiata pine in southern Australia. Ann For Sci 72:391–401
Jarvis SF, Borralho N, Potts B (1995) Implementation of a multivariate BLUP model for genetic evaluation of Eucalyptus globulus in Australia. Paper presented at the eucalypt plantations: improving fibre yield and quality, Hobart, Australia
Li Y, Altaner C (2018) Predicting extractives content of Eucalyptus bosistoana F. Muell. heartwood from stem cores by near infrared spectroscopy. Spectrochim Acta A Mol Biomol Spectrosc. 198:78–87. https://doi.org/10.1016/j.saa.2018.02.068
Lima J, Breese M, Cahalan C (2000) Genotype–environment interaction in wood basic density of Eucalyptus clones. Wood Sci Technol 34:197–206
McDonald M, Rawlings M, Butcher P, Bell J (2003) Regional divergence and inbreeding in Eucalyptus cladocalyx (Myrtaceae). Aust J Bot 51:393–403
Miranda I, Gominho J, Pereira H (2009) Variation of heartwood and sapwood in 18-year-old Eucalyptus globulus trees grown with different spacings. Trees 23:367–372
Moraes JC, Zanetti R, Amaral Castro N, Zanuncio J, Andrade H (2002) Effect of Eucalyptus species and soil type on infestation levels of heartwood termites (Insecta: Isoptera) in reforested areas of Brazil. Sociobiology 39:145–153
Moya R, Berrocal A (2010) Wood colour variation in sapwood and heartwood of young trees of Tectona grandis and its relationship with plantation characteristics, site, and decay resistance. Ann For Sci 67:109
Palanti S, Susco D, Feci E (2010) Natural durability of eucalypt from Italian plantations against fungi and cerambicid Trichoferus holosericeus Rossi. Eur J Wood Wood Prod 68:59–62
Perry D, Lenz M, Watson J (1985) Relationships between fire, fungal rots and termite damage in Australian forest trees. Aust For 48:46–53
Pinto I, Pereira H, Usenius A (2004) Heartwood and sapwood development within maritime pine (Pinus pinaster Ait.) stems. Trees 18:284–294
Poynton RJ (ed) (1979) Eucalyptus bosistoana F. Mueller. In: Tree planting in Southern Africa: the eucalypts. Department of Forestry, South Africa
R Core Team (2017) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Rudman P (1964) Durability in the genus Eucalyptus. Aust For 28:242–257
Santos PETd, Geraldi IO, Garcia JN (2004) Estimates of genetic parameters of wood traits for sawn timber production in Eucalyptus grandis. Genet Mol Biol 27:567–573
Schimleck LR, Kube PD, Raymond CA (2004) Genetic improvement of kraft pulp yield in Eucalyptus nitens using cellulose content determined by near infrared spectroscopy. Can J For Res 34:2363–2370
Stackpole DJ, Vaillancourt RE, Downes GM, Harwood CE, Potts BM (2010) Genetic control of kraft pulp yield in Eucalyptus globulus. Can J For Res 40:917–927
Taylor AM, Gartner BL, Morrell JJ (2002) Heartwood formation and natural durability—a review. Wood Fiber Sci 34:587–611
Taylor AM, Gartner BL, Morrell JJ, Tsunoda K (2006) Effects of heartwood extractive fractions of Thuja plicata and Chamaecyparis nootkatensis on wood degradation by termites or fungi. J Wood Sci 52:147–153. https://doi.org/10.1007/s10086-005-0743-6
Townsend TG, Solo-Gabriele H (2006) Environmental impacts of treated wood. CRC Press, Boca Raton
Wilkes J (1991) Heartwood development and its relationship to growth in Pinus radiata. Wood Sci Technol 25:85–90
Wu S, Zhu Y, Xu J, Lu Z, Chen G, Song P, Guo W (2017) Genetic variation and genetic gain for energy production, growth traits and wood properties in Eucalyptus hybrid clones in China. Aust For 80:57–65
Yu Q, Yang D-Q, Zhang S, Beaulieu J, Duchesne I (2003) Genetic variation in decay resistance and its correlation to wood density and growth in white spruce. Can J For Res 33:2177–2183
Acknowledgements
We like to thank NZDFI for access to growth data. This work was supported by funding from the MBIE Partnership for Speciality Wood Products (contract FFRX1501).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
We declare no conflict of interest.
Additional information
Communicated by Martina Meincken.
Rights and permissions
About this article
Cite this article
Li, Y., Apiolaza, L.A. & Altaner, C. Genetic variation in heartwood properties and growth traits of Eucalyptus bosistoana. Eur J Forest Res 137, 565–572 (2018). https://doi.org/10.1007/s10342-018-1125-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10342-018-1125-0