Abstract
The polyphagous pest Tetranychus urticae feeds on over 1100 plant species including highly valued economic crops such as hops (Humulus lupulus). In the key hop production region of the Pacific Northwest of the USA, T. urticae is one of the major arthropod pests. Over the years, T. urticae control has been dominated by the application of various acaricides. However, T. urticae quickly adapts to these acaricides by developing resistance. Here, we determined resistance ratios of T. urticae populations in hops to three acaricides: etoxazole, fenpyroximate, and spirodiclofen. The mechanisms underlying resistance to these and three other acaricides were investigated in 37 field-collected T. urticae populations using a comprehensive diagnostic approach. Our data showed that T. urticae populations exhibited complex adaptation patterns to acaricides. Resistance to abamectin, fenpyroximate, and spirodiclofen by enhanced target metabolic detoxification gene(s) was identified in 100%, 50%, and 20% of populations tested, respectively. Resistance to bifenthrin, bifenazate, and etoxazole by target site insensitivity was pervasive among tested populations. Our study provides new information in understanding the complexity of T. urticae adaptation to multiple acaricides, which will help in designing sustainable pest control strategies for T. urticae on hops and other economically valuable crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Key message
-
1.
IPM-compatible acaricides are mainstays for sustainable Tetranychus urticae management in crop systems, while resistance to these acaricides threatens effective control practices.
-
2.
We developed a comprehensive strategy to examine phenotypic status and underlying mechanisms of adaptation to acaricides with molecular markers.
-
3.
Our study revealed many T. urticae populations collected from hopyards exhibited evidence of resistance to multiple acaricides.
-
4.
This robust proactive diagnostic tactic can help prevent the loss of effectiveness of acaricides in mite control.
Introduction
Phytophagous mites pose a threat to global agricultural production through direct feeding or transmission of pathogens and viruses (Van Leeuwen et al. 2015). The two-spotted spider mite, Tetranychus urticae Koch is a ubiquitous agricultural pest of many major crops, including cotton, soybean, maize, vegetables, fruits, and ornamentals as well as numerous specialty crops such as hops (Grbic et al. 2011; Piraneo et al. 2015; Van Leeuwen et al. 2015). T. urticae has been reported to be one of the major arthropod pests in Pacific Northwest (PNW) hop fields, where it represents above 99% of the US hop acreage (O’Neal et al. 2015; Piraneo et al. 2015). The US hop industry is increasing in value annually, and it was estimated to be worth over $500 million in 2016 (USDA NASS). Although various strategies such as biocontrol using natural enemies, cultural control and population monitoring have been adopted for T. urticae management (McMurtry and Croft 1997; O’Neal et al. 2015), the use of acaricides remains essential to prevent mite populations from reaching economic thresholds (O’Neal et al. 2015). Unfortunately, T. urticae has been documented to quickly develop resistance to all acaricides used for its control, irrespective of mode of action (Van Leeuwen et al. 2013). Development of acaricide resistance will lead to an even greater increase of acaricide application, raising the risk of chemical exposure to non-target beneficial arthropods and cause the complete failure of pest management strategies. Thus, there is a critical need to reveal the mechanisms underlying adaptation to acaricides, and to apply this knowledge for pest management practices in the field (Van Leeuwen et al. 2010; Ilias et al. 2014; Piraneo et al. 2015; Adesanya et al. 2018).
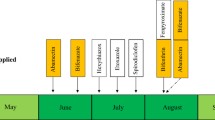
In urban and agro-ecosystems, rotation of multiple pesticides with diverse modes of action is a typical approach to reduce the development of pesticide resistance (Goodhue et al. 2011; Piraneo et al. 2015; Zhu et al. 2016a). According to spray records from PNW hopyards, the acaricides regularly used by growers to suppress T. urticae include abamectin (IRAC group 6), bifenthrin (IRAC group 3A), bifenazate (IRAC group 20D), hexythiazox and etoxazole (IRAC group 10A and 10B), fenpyroximate (IRAC group 21A), and the recently registered acaricides spirodiclofen (IRAC group 23), and acequinocyl (IRAC group 20B) (Piraneo et al. 2015) (Fig. S1). Possessing different modes of action, these chemicals are applied in rotation or combination during the hop growing season from May to August (Piraneo et al. 2015) (Fig. S1a). Our previous study investigated the resistance ratios (RRs) and mechanisms of resistance to abamectin, bifenazate, and bifenthrin in T. urticae populations collected from PNW hopyards (Piraneo et al. 2015). However, the phenotypic and genotypic resistance status of T. urticae populations from hops to etoxazole, fenpyroximate, and spirodiclofen remains unknown.
Due to specific activities against spider mites compared to predatory mites and other non-target beneficial arthropods, etoxazole, fenpyroximate, and spirodiclofen have been used as valuable IPM tools for T. urticae management on hops and other crops (Kim et al. 2005). Etoxazole belongs to a class of acaricides known as mite growth inhibitors (MGIs), which inhibit chitin deposition during embryonic and immature stages of mites (Van Leeuwen et al. 2012; Demaeght et al. 2014). MGIs are highly specific against spider mites and show low or no toxicity to beneficial arthropods and vertebrates (Adesanya et al. 2018). The primary target site of MGIs is the chitin synthase 1 (CHS 1) (Van Leeuwen et al. 2012). Fenpyroximate is a mitochondrial electron transport inhibitor (METI), with the mode of action in inhibition of complex I of the respiratory chain at the ubiquinone site of NADH oxidoreductase (referred as METI-I) (Bajda et al. 2017). Fenpyroximate has a broad-spectrum activity against all life stages of many mite species but is relatively safe to beneficial insects and predatory mites (Motoba et al. 1992; Van Pottelberge et al. 2009). Spirodiclofen belongs to the group of spirocyclic tetronic acid derivatives (keto-enols) and targets on the acetyl-coenzyme A carboxylase (ACCase) in the lipid biosynthesis pathway (referred as LPI) (Van Leeuwen et al. 2009). Spirodiclofen is active against all developmental stages, but particularly against eggs, and shows high activity against female mites through decreasing their fecundity and fertility after tarsal contact (Nauen 2005).
Target site insensitivity is one of the most important mechanisms of acaricide resistance by T. urticae (Van Leeuwen et al. 2010; Van Leeuwen and Dermauw 2016). A non-synonymous mutation I1017F was identified in the non-catalytic domain of the CHS 1 gene that is correlated with resistance to MGIs in T. urticae (Van Leeuwen et al. 2012; Demaeght et al. 2014). A recent study showed that the fenpyroximate resistance in T. urticae has been associated with a H92R mutation in the PSST homologue of complex I (Bajda et al. 2017). The cyclic keto-enol insecticide spirotetramat inhibits insect and spider mite ACCase by interfering with the carboxyltransferase partial reaction (Lümmen et al. 2014). However, so far there is no report of target site insensitivity associated with cyclic keto-enol insecticides resistance in T. urticae. In the greenhouse whitefly Trialeurodes vaporariorum (Westwood), a glutamic acid substitution with lysine in position 645 (E645K) of the ACCase gene was linked to spiromesifen resistance (Karatolos et al. 2012). Two mutations, G323D and G326E were identified in the glutamate-gated chloride channel (GluCl) genes associated with abamectin resistance (Kwon et al. 2010a, b; Dermauw et al. 2012). Studies reported that resistance to bifenazate was tightly linked with mutations or mutation combinations in the mitochondrial cytochrome b (cytb) gene (Van Leeuwen et al. 2008; Van Nieuwenhuyse et al. 2009). Several amino acid substitutions in the voltage-gated sodium channel (VGSC) gene have been associated with pyrethroid resistance in T. urticae (Tsagkarakou et al. 2009; Kwon et al. 2010a, b). Besides target site insensitivity, enhanced metabolic detoxification by cytochrome P450 s, glutathione S-transferases (GSTs), and esterases is another important mechanism responsible for resistance to many acaricides and insecticides (Feyereisen 1995; Stumpf and Nauen 2002; Liu et al. 2006; Demaeght et al. 2013; Zhu et al. 2013; Van Leeuwen and Dermauw 2016). For example, recent studies revealed that the P450 CYP392A16, and the GSTs TuGSTd10, TuGSTd14, and TuGSTm09 are associated with high levels of abamectin resistance in T. urticae (Riga et al. 2014; Pavlidi et al. 2015). Another P450, CYP392A11 was shown to be able to hydroxylate fenpyroximate (Riga et al. 2015). Functional characterization of CYP392E10 confirmed that it can metabolize spirodiclofen and spirotetramat (Demaeght et al. 2013). The high level of spirodiclofen resistance in T. urticae was also potentially linked with enhanced hydrolysis activity of the carboxylesterase CCE04 (Demaeght 2015). The mutations associated with acaricide resistance and enhanced expression of detoxification genes with known functions in acaricide metabolism potentially can be used as molecular markers for resistance monitoring in the field (Zhu et al. 2016a).
The current study examined the phenotypic resistance status within 37 field-collected T. urticae populations to three acaricides: etoxazole, fenpyroximate, and spirodiclofen. Then, we extensively searched for any mutations responsible for resistance to abamectin, bifenthrin, bifenazate, etoxazole, fenpyroximate, and spirodiclofen among these populations. Lastly, we investigated the relative expression of several detoxification genes, including cytochrome P450s, GSTs and a carboxylesterase, that are associated with resistance to abamectin, fenpyroximate, and spirodiclofen.
Materials and methods
Mites
The susceptible T. urticae population was initially collected from weeds in Montana in 1995 and has never been exposed to any pesticides (Piraneo et al. 2015; Morales et al. 2016; Adesanya et al. 2018). Thirty-seven T. urticae populations were collected from seven major locations of commercial hopyards in the Yakima Valley of Washington State from June to September, 2016 (Table S1). One population was collected from Grandview (46°15′13″N 119°54′36″W), nine populations from Harrah (46°24′15″N 120°32′35″W), two populations each from Mabton (46°12′42″N 119°59′47″W) and Moxee (46°33′23″N 120°23′14″W), nine populations from Prosser (46°12′25″N 119°45′56″W), ten populations from Toppenish (46°22′44″N 120°18′43″W), and four populations from White_swan (46°23′40″N 120°42′11″W) (Table S1). Hop’s leaves from each hopyard were randomly sampled covering all the areas of the farm in an X-shaped pattern. Leaves were collected from the upper, middle, and lower parts of hop plants to ensure that each sample was representative of the actual mite population occurring in the hopyard. The leaves were kept in Ziploc® plastic bags in ice coolers and transported immediately to the laboratory, where the number of mites per leaf was counted under a dissecting light microscope. Mites were reared on lima bean plants (Phaseolus lunatus L.) for one generation under laboratory conditions (28 ± 2 °C, 70 ± 5 RH) and then used for subsequent experiments.
Bioassays
Mites were subjected to leaf disk bioassays with three acaricides: etoxazole (Zeal®, 72 WP), fenpyroximate (Fujimite®, 5% SC), and spirodiclofen (Envidor®, 240 g/L SC). Direct spray of chemicals on freshly laid eggs of T. urticae was found to be the most efficient bioassay method for assaying mite growth inhibitors (Adesanya et al. 2018) and was therefore used to calculate the toxicity of the MGI etoxazole. Bioassay arenas were created by placing a fresh lima bean leaf disk (2 cm in diameter) on water-saturated cotton in a clean petri dish. Six to eight gravid adult female mites were allowed to oviposit for 24 h. After oviposition, the adult female mites were removed. The fresh laid eggs were sprayed using a Potter precision spray tower (Burkard Manufacturing, Rickmansworth, Herts, UK) with 2 ml of etoxazole solution with varying concentrations, ranging from 0 (control with Millipore-filtered water only) to the field-recommended dose on hops, 300 ppm a.i. The Potter spray tower was calibrated to deliver 2.0 ± 0.1 mg/cm2 of liquid under 1.1 kg/cm2 of pressure. Each dose was replicated 3–5 times. After spraying, the total number of eggs laid was recorded using a light microscope. After approximately 5 days, the number of eggs that successfully hatched into larvae was counted. Female adults were used for the bioassay of fenpyroximate, as described by Piraneo et al. (2015). Larvae were used for the bioassay of spirodiclofen, following the procedure similar as Van Pottelberge et al. (2009). Briefly, gravid adult female mites were allowed to lay eggs on fresh lima bean leaf disks (2 cm in diameter). The period of oviposition was less than 24 h to ensure synchronous hatching of eggs into larvae. The larvae (48 h after hatching) were subjected to varying doses of spirodiclofen using the precision Potter spray system. After 48 h, the mortality was scored for the sprayed adults or larvae. Mites were counted as dead if they did not respond to a gentle tap by a fine camel-hair brush. The dose–mortality response was adjusted to the control treatment using Abbot’s formula (Abbott 1925). Probit analysis was used to estimate LC50 values, slopes and 95% confidence intervals (POLO Probit 2014). The statistical comparison of LC50 values was performed according to non-overlapping of 95% CIs (Liu and Yue 2000).
Detection of resistance-associated point mutations
gDNA was extracted from 100 to 150 individuals of susceptible and 36 field-collected T. urticae populations using the DNeasy Blood & Tissue kit (QIAGEN®). PCR were carried out in a ProFlex PCR system (Thermo Fisher Scientific, USA). Each PCR contained 1 µL gDNA (100 ng/µL), 4 µL PCR buffer (5 ×), 0.8 µL dNTP mix (10 mM), 0.6 µL forward and reverse primers (Table S5) for each target gene (stock 10 µM), 0.8 µL Phusion High-Fidelity DNA Polymerase (Thermo Scientific, Pittsburgh, PA), and 12.8 µL ddH2O. PCR was conducted under the following cycling parameters: 94 °C for 3 min 50 s, 35 cycles of 94 °C for 30 s, 50–58 °C for 30 s, and 72 °C for 1 min, with a final extension for 10 min at 72 °C. After the PCR, 2 µl of the PCR product was run on agarose gels for evaluation of PCR product integrity. The raw PCR products were submitted to Functional Biosciences (Madison, WI) for purification and sequencing. The sequences obtained were analyzed using BioEdit 7.01 software (Ibis Biosciences, Carlsbad, CA). The presence or absence of mutations was determined by inspection of sequencing chromatographs. The frequency of resistance alleles was estimated by comparing the height of the wild-type and resistance allele peaks in chromatographs based on the methods in Bajda et al. (2017).
RNA extraction, cDNA synthesis, and qRT-PCR
Total RNA was extracted from 250 female T. urticae adults with TRIZOL reagent (Invitrogen®) following the manufacturer’s protocol. Nanodrop spectrophotometer ND-2000 (Thermo Scientific, Pittsburgh, PA) was used to quantify and qualify the total RNA. The total RNA was treated with DNase I (Ambion Inc. Austin, TX) to remove the contaminating genomic DNA (gDNA). cDNA synthesis was then performed with M-MLV reverse transcriptase (Promega, Madison, WI) and 2 µg of DNase I-treated total RNA as template. qRT-PCR was performed on a CFX96™ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA). Each reaction contained 5 µL iQ™ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA), 0.4 μL forward and reverse gene specific primers (stock 10 µM) (Table S5), 1.0 μL cDNA, and 3.6 μL ddH2O. The optimized program used included an initial incubation of 95 °C for 3 min, 40 cycles of denaturation at 95 °C for 10 s, and annealing/extension at 60 °C for 30 s, followed by a final melting curve cycle of 95 °C for 10 s, 65 °C for 5 s, and 95 °C for 10 s. The reactions were set up in a 96-well full-skirted PCR plate (USA Scientific, Ocala, FL) with three technical replicates and at least three biological replicates. Three negative (no-template) controls were included in each plate. Two previously identified most stable housekeeping genes, CycA and Rp49, were used as the reference genes (Morales et al. 2016). Both PCR efficiency and correlation coefficient (R2) values were taken into consideration in evaluating the primers. Primer pairs with PCR efficiency higher than 94% or lower than 110% and R2 values > 0.99 were considered as qualified for further analysis. The 2−ΔΔCT method was used to calculate the relative gene expression (Huggett et al. 2005). Student’s t test was used to test for significant difference in gene expression between susceptible and field-collected T. urticae populations.
Results
Phenotypic resistance levels of T. urticae populations on PNW hops
The toxicities of 3 registered acaricides, etoxazole, fenpyroximate, and spirodiclofen, were assessed on 31, 28, and 21 of total 37 field-collected T. urticae populations, respectively. The LC50 of a susceptible T. urticae population to etoxazole was 0.65 ppm a.i. (Table 1). The LC50s of field T. urticae populations ranged from 16.0 to 217.4 ppm a.i., which are below the field dose of etoxazole (300 ppm a.i.). The RRs of the field populations to etoxazole ranged from 24.72 to 336.59 (Table 1). Based on RRs, 52% and 48% of the tested populations had moderate (100 > RR ≥ 10) and high (RR ≥ 100) resistance to etoxazole, respectively (Fig. 1). Three hopyards, Harrah_1, Toppenish_6, and Toppenish_7, were sampled twice during the hops growing season (Table S1). The RR of Harrah_1 increased slightly from 120.71 to 122.23 (Table 1). The RR of Toppenish_6 decreased slightly from 120.09 to 112.69, while that of Toppenish_7 increased 1.7-fold in the 2 months (Table 1; Table S1). In the etoxazole bioassay, the mortality at field dose ranged from 63.2% to 86.5%. The highest resistance level to etoxazole was recorded in the sample collected from Prosser_1 in 2016 (RR = 336.59) (Table 1). However, no Prosser population collected from 2015 showed high levels of etoxazole resistance (Adesanya et al. 2018).
The LC50 of the susceptible T. urticae population to fenpyroximate was 9.5 ppm a.i. (Table 2). The LC50s of field T. urticae populations to fenpyroximate ranged from 25.0 to 228.5 ppm a.i. LC50s in two populations, Toppenish_2 and Toppenish_3, are above the field dose of fenpyroximate (180 ppm a.i.), suggesting the acaricide tested tends to lack general efficiency in these populations. Mortality at the field dose of fenpyroximate varied from 52% to 100%, though 100% mortality of mites was only observed in the Prosser_7 population (Table 2). The RRs of field populations ranged from 2.63 to 24.05 (Table 2). Based on RRs, 75% and 25% of the populations had low and moderate resistance, respectively (Fig. 1). The highest resistance level to fenpyroximate was recorded in the samples collected from Toppenish_3 (RR = 24.05) (Table 2).
The LC50 of the susceptible T. urticae population to spirodiclofen was 1.9 ppm a.i. (Table 3). The LC50s of field T. urticae populations ranged from 6.0 to 83.8 ppm a.i., which are below the field dose of spirodiclofen (300 ppm a.i). Mortality at field dose ranged from 71% to 100%. One hundred percent mortality of mites at field dose of spirodiclofen was observed in the Mabton_1 and Prosser_7 populations. The RRs of field T. urticae populations to spirodiclofen varied from 3.16 to 44.11 (Table 3). Based on RRs, low and moderate levels of resistance were observed in 25.7% and 74.3% of tested populations, respectively (Fig. 1). The highest resistance level to spirodiclofen was recorded in the samples collected from Harrah_2 (RR = 44.11) (Table 3).
Distribution pattern of resistance-associated mutations in T. urticae populations from hops
The presence of 19 resistance-associated mutations in six acaricide target genes, GluCl1 and GluCl3 (targets of abamectin), voltage-gated sodium channel (VGSC, target of bifenthrin), cytochrome b (cytb, target of bifenazate), CHS 1 (target of etoxazole), PSST (target of fenpyroximate), and ACCase (target of spirodiclofen) in 36 T. urticae populations was evaluated by diagnostic PCR and sequencing analysis (Table S2). Populations were genotypically scored as wild-type, resistant, or mixed at each gene locus based on the sequencing chromatographs. The allele frequency of each resistant allele was also estimated by inspection of chromatographs (Tables S3 and S4). In total, five mutations were detected: M918L, F1534S, and F1538I in VGSC; G126S in cytb; and I1017F in CHS 1 (Fig. 2). There were no mutations found in GluCl1 and GluCl3, PSST, and ACCase (Table S4), indicating that target site insensitivity may not be important in T. urticae resistance to abamectin, fenpyroximate, or spirodiclofen in hopyards.
Multiple resistance-associated mutations on target site genes identified from T. urticae populations collected from hopyards. Pink fill indicates susceptible allele (S), gray fill indicates the resistant allele (R), and blue fill indicates both alleles were detected in the population (S/R). No fill indicates no data available. (Color figure online)
We sequenced three VGSC fragments containing regions of domain II, domain II-III interlinker, and domain III, and evaluated nine candidate pyrethroid resistance-associated loci (Table S2). Three mutations, M918L, F1534S, and F1538I were detected in 25% (9 populations), 28% (10 populations) and 83% (30 populations) of the 36 T. urticae populations collected from hops, respectively (Fig. 2; Table S3).
We also looked for 5 bifenazate resistance-associated mutations (G126S, I136T, S141F, D161G, and P262T) in the mitochondrial Cyt b gene by performing PCR and sequencing an 828 bp fragment of the T. urticae cyt b gene (Table S2). The G126S mutation was detected in 72% (26) of 36 populations, which all were a mix of the resistant and susceptible alleles (Fig. 2; Table S4). The G126S mutation alone only results in low to moderate bifenazate resistance and does not contribute to cross-resistance between bifenazate and acequinocyl (Van Leeuwen et al. 2008; Van Nieuwenhuyse et al. 2009). Other cytb mutations or mutation combinations, such as P262T, G126S + I136T, and G126S + S141F, that have been reported to confer high levels of bifenazate resistance in T. urticae (Van Leeuwen et al. 2008; Van Nieuwenhuyse et al. 2009) were not observed in any of T. urticae populations that we tested.
Resistance to MGIs (etoxazole, hexythiazox, and clofentezine) has been previously linked to a single non-synonymous mutation (I1017F) in the CHS 1 gene (Van Leeuwen et al. 2012; Demaeght et al. 2014). Our initial survey of four T. urticae populations collected from hops in 2015 found that this mutation was detected in one of these populations (Adesanya et al. 2018). Here, we found the I1017F mutation in 83% (30) of the 36 T. urticae populations we examined (Fig. 2).
Expression pattern of resistance-associated metabolic genes
Recent functional studies have revealed that several P450s, GSTs and one esterase contribute resistance to abamectin (Riga et al. 2014; Pavlidi et al. 2015), fenpyroximate (Riga et al. 2015), and spirodiclofen (Demaeght et al. 2013) in T. urticae (Table 4). These genes can serve as molecular markers to evaluate the acaricide resistance in T. urticae populations on hops. Thus, we randomly chose one to two population(s) from each of the hop production districts used in this study [Harrah_3, Harrah_8, Mabton_2, Moxee_1, Prosser_1, Prosser_9, Toppenish_7, Toppenish_7_1, White_swan_1, and White_swan_4 (Table S1)] to examine transcription levels of these genes as possible mediators of acaricide resistance.
Overexpression of CYP392A16, two TuGSTs of the delta class (TuGSTd10 and TuGSTd14), and one of the mu class (TuGSTm09) have been highly associated with abamectin or multiple acaricide resistance (Dermauw et al. 2013). Figure 3 displays the relative expression of these four abamectin-associated genes in 10 field-collected T. urticae populations compared to the susceptible one. In general, all of these four genes show significantly higher expression (≥ 2-fold, p value ≤ 0.05) than the susceptible strain in at least one of the field-collected T. urticae populations (Fig. 3), suggesting P450- or GST-associated abamectin resistance is common in T. urticae populations on hops. CYP392A16 and TuGSTd14 were significantly overexpressed (≥ 2-fold, p value ≤ 0.05) in seven out of the 10 tested T. urticae populations (Figs. 3a, c). TuGSTd10 and TuGSTm09 showed overexpression (≥ 2-fold, p value ≤ 0.05) in five and three of the T. urticae field populations, respectively (Fig. 3b, d).
A recent study revealed that a cytochrome P450, CYP392A11 plays a role in fenpyroximate resistance in T. urticae (Riga et al. 2015). Therefore, we used CYP392A11 as a molecular marker to evaluate the fenpyroximate resistance in T. urticae populations from hops. As shown in Fig. 4a, CYP392A11 was significantly overexpressed (≥ 2-fold, p value ≤ 0.05) in five out of 10 tested T. urticae populations. The highest overexpression levels of CYP392A11 occurred in the Toppenish_7 and Toppenish_7_1 populations, with about 16-fold overexpression relative to the susceptible strain (Fig. 4a), which is consistent with the moderate level of fenpyroximate resistance in these populations (Table 2).
Molecular analysis of spirodiclofen resistance in T. urticae suggested a P450 (CYP392E10) and an alternative allele of carboxyl/choline esterase-4 (CCE04) were strongly associated with high levels of spirodiclofen resistance (Demaeght et al. 2013; Demaeght 2015). These two genes can be used as molecular markers to evaluate P450 or esterase-associated spirodiclofen resistance in the field. CYP392E10 was only overexpressed (≥ 2-fold, p value ≤ 0.05) in the Prosser_9 and Toppenish_7 populations (Fig. 4b), both of which exhibited moderate levels of spirodiclofen resistance (Table 3). CCE04 was only overexpressed in the Toppenish_7 population (Fig. 4c). This is despite our finding that 74.3% of 21 T. urticae populations from hops exhibited moderate levels of resistance to spirodiclofen (Table 3), suggesting other mechanisms besides CYP392E10- and CCE04-mediated detoxification are responsible.
Multiple acaricide resistance in T. urticae populations
With the availability of molecular markers, we summarized the genotypic resistance status in nine randomly selected field-collected T. urticae populations to six commonly used acaricides in hopyards. As shown in Table 4, abamectin resistance by enhanced target P450 and/or GST gene(s)’s expression was observed in all nine populations tested. In addition, target site insensitivity-mediated resistance to bifenthrin, bifenazate, and etoxazole was identified in all nine populations tested as well. However, metabolic detoxification-associated resistance to fenpyroximate and spirodiclofen only occurred in four and two out of nine populations, respectively (Fig. 4; Table 4). Among these nine populations, one population (Toppenish_7) contains resistance-associated genotype to all six acaricides evaluated (Table 4). Taken together, all nine T. urticae populations simultaneously harbor resistance-associated genotypes to multiple acaricides (average = 4.7), raising the specter of “super mites” that possess resistance to multiple acaricides.
Discussion
Narrow-spectrum and reduced-risk acaricides are mainstays for sustainable T. urticae management in crop systems. To retain the utility of these IPM-compatible acaricides, it is necessary to develop a comprehensive strategy to frequently examine the phenotypic status and underlying mechanisms of adaptation to acaricides in field populations of mites. Our study found that T. urticae populations in hopyards exhibit a moderate to high levels of resistance to etoxazole, but resistance to fenpyroximate and spirodiclofen remains at low to moderate levels. Our data revealed a complex genetic acaricide adaptation pattern toward six commonly used acaricides in PNW hopyards. Resistance to abamectin, fenpyroximate, and spirodiclofen by enhanced metabolic detoxification-associated gene(s) was detected. P450 and/or esterase gene(s)-mediated resistance to fenpyroximate and spirodiclofen only occurred in a few populations. However, target P450 and/or GST genes-associated resistance to abamectin was ubiquitous. In addition, target site insensitivity-mediated resistance to bifenthrin, bifenazate, and etoxazole was identified in a wide range of T. urticae populations from hops. Many of these T. urticae populations collected from PNW hopyards exhibited evidence of resistance to multiple acaricides, indicating efforts need to be made to slow down the development of such resistance.
Because of their relative safety and unique mode of action, MGIs are widely used in field and greenhouses for T. urticae management (Zhu et al. 2016b). Etoxazole and hexythiazox have been used on hops since they were registered in the 1980s and 2000s, respectively (Adesanya et al. 2018). Based on spray records we examined, hop farmers applied etoxazole or hexythiazox two to four times each year (Fig. S1a) (Piraneo et al. 2015). In 2016, approximately 60% of the hop farms in Washington were treated with etoxazole or hexythiazox (Fig. S1b). Moreover, cross-resistance between etoxazole and hexythiazox and from MGIs to bifenazate and bifenthrin had been reported (Adesanya et al. 2018). Frequent usage of MGIs and cross-resistance are likely responsible for the moderate to high levels of etoxazole resistance observed in the T. urticae populations in this study (Figs. 1, 2). Hence, farmers should limit the use of etoxazole and other MGIs to avoid control failure (Demaeght et al. 2014; Adesanya et al. 2018).
According to the spray records we obtained, Class I METI acaricides, which include fenpyroximate and pyridaben, were only applied to 16.5% of all hop farms in 2016 (Fig. S1b). Fenpyroximate was only sprayed once in the growing season (Fig. S1a) (Piraneo et al. 2015). It is possible that the reduced application of these acaricides is responsible for the lack of resistance we observed. LPIs including spirodiclofen and spirotetramat are recently registered acaricides on hops. However, in 2016 approximately 83.8% of the hop farms in Washington were treated with LPIs (Fig. S1b). Therefore, monitoring of resistance plays an essential role in preventing the loss of effectiveness of LPIs in mite control.
Pyrethroids have been registered for use in hopyards for more than 20 years. Our previous study reported that one mutation, F1538I, was observed in 66.7% of tested T. urticae populations collected from hopyards in 2013 (Piraneo et al. 2015). Here, the mutation F1538I alone and the presence of two or multiple mutations of M918L + F1538I, F1534S + F1538I, and M918L + F1534S + F1538I were detected in all 30 tested populations with complete genotypic data (Fig. 2). This is the first study to report the presence of the M918L and F1534S mutations in T. urticae populations. The M918L mutation, which is a variant of the “super-kdr” (M918T) mutation, was originally identified from cypermethrin-resistant Aphis gossypii populations (Carletto et al. 2010), and then in Hyalella azteca populations (Weston et al. 2013). The “super-kdr” M918T was reported in the tomato red spider mite, Tetranychus evansi, in the absence of L1014F (kdr), and was associated with the pyrethroid resistance (Van Leeuwen et al. 2008). In the present study, the mutation M918L was observed with F1534S (in 2 out of 9 populations) and F1538I (in 8 out of 9 populations) (Fig. 2), suggesting a probably high level of pyrethroid resistance in these T. urticae populations. The mutation F1534C was first identified in pyrethroid resistant mosquitos Aedes aegypti and A. albopictus (Kawada et al. 2009; Kasai et al. 2011). The functional properties of this mutation have been evaluated through in vitro expression in Xenopus oocytes. It was found that the phenylalanine (F) to cysteine (C) change reduced sodium channel sensitivity dramatically to type I pyrethroids (e.g., bifenthrin) but not type II pyrethroids (e.g., deltamethrin) (Hu et al. 2011). Therefore, the presence of F1534S in 10 tested T. urticae populations from hops is likely to confer resistance to bifenthrin. Interestingly, the F1534S mutation was always found together with the F1538I mutation (Fig. 2). F1538I occurs in domain IIIS6 of the VGSC and was previously identified as one of the primary pyrethroid resistance mutations linked to high level resistance in multiple arthropod species, including T. urticae (Soderlund and Knipple 2003; Van Leeuwen et al. 2010). We detected F1538I in almost all field-collected T. urticae populations (30), except for six populations for which we lack complete diagnostic data. In fact, this mutation has been fixed in 15 of our collected T. urticae populations (Fig. 2). These results indicate that pyrethroid resistance of T. urticae populations in hopyards is increasing, which reflects the prolonged selection from the consistent usage of pyrethroids over the years. The use of pyrethroids, especially the type I pyrethroids, in hopyards should be reduced in order to avoid control failure and maintain the usefulness of these IPM-compatible acaricides.
Acaricides are one of the cornerstones of effective mite management efforts in agriculture worldwide. However, extensive usage of a limited number of acaricides for a prolonged period could cause the escalated accumulation of resistance-associated genes in populations. Therefore, it is not surprising that there are so many field populations exhibiting resistance genotypes to multiple acaricides. Compared with phenotypic bioassays alone, the comprehensive diagnostic approach we used here with both phenotypic bioassays and molecular markers provides a more accurate and proactive tactic for evaluating acaricide resistance and allowing efficient acaricide rotation, which can help prevent the loss of effectiveness of acaricides in mite control.
Author contributions
DBW and FZ conceived and designed the experiments; MW, AWA, and MAM performed the experiments; MW, AWA, and FZ analyzed the data; DBW, LCL, MDL, and FZ contributed new reagents/analytic tools; MW, AWA, and FZ wrote the original draft; all authors contributed in revising and preparing the paper for submission.
References
Abbott WS (1925) A method of computing the effectiveness of an insecticide. J Econ Entomol 18:265–267
Adesanya AW, Morales MA, Walsh DB, Lavine LC, Lavine MD, Zhu F (2018) Mechanisms of resistance to three mite growth inhibitors of Tetranychus urticae in hops. Bull Entomol Res 108:23–34
Bajda S, Dermauw W, Panteleri R, Sugimoto N, Douris V, Tirry L, Osakabe M, Vontas J, Van Leeuwen T (2017) A mutation in the PSST homologue of complex I (NADH:ubiquinone oxidoreductase) from Tetranychus urticae is associated with resistance to METI acaricides. Insect Biochem Mol Biol 80:79–90
Carletto J, Martin T, Vanlerberghe-Masutti F, Brevault T (2010) Insecticide resistance traits differ among and within host races in Aphis gossypii. Pest Manag Sci 66:301–307
Demaeght PA (2015). A genomic approach to investigate resistance mechanisms in the two-spotted spider mite Tetranychus urticae. Ph.D., University of Amsterdam
Demaeght P, Dermauw W, Tsakireli D, Khajehali J, Nauen R, Tirry L, Vontas J, Lummen P, Van Leeuwen T (2013) Molecular analysis of resistance to acaricidal spirocyclic tetronic acids in Tetranychus urticae: CYP392E10 metabolizes spirodiclofen, but not its corresponding enol. Insect Biochem Mol Biol 43:544–554
Demaeght P, Osborne EJ, Odman-Naresh J, Grbic M, Nauen R, Merzendorfer H, Clark RM, Van Leeuwen T (2014) High resolution genetic mapping uncovers chitin synthase-1 as the target-site of the structurally diverse mite growth inhibitors clofentezine, hexythiazox and etoxazole in Tetranychus urticae. Insect Biochem Mol Biol 51:52–61
Dermauw W, Ilias A, Riga M, Tsagkarakou A, Grbic M, Tirry C, Van Leeuwen T, Vontas J (2012) The cys-loop ligand-gated ion channel gene family of Tetranychus urticae: implications for acaricide toxicology and a novel mutation associated with abamectin resistance. Insect Biochem Mol Biol 42:455–465
Dermauw W, Wybouw N, Rombauts S, Menten B, Vontas J, Grbic M, Clark RM, Feyereisen R, Van Leeuwen T (2013) A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proc Natl Acad Sci USA 110:E113–E122
Feyereisen R (1995) Molecular biology of insecticide resistance. Toxicol Lett 82–83:83–90
Goodhue RE, Bolda M, Farnsworth D, Williams JC, Zalom FG (2011) Spotted wing drosophila infestation of California strawberries and raspberries: economic analysis of potential revenue losses and control costs. Pest Manag Sci 67:1396–1402
Grbic M, Van Leeuwen T, Clark RM, Rombauts S, Rouze P, Grbic V et al (2011) The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479:487–492
Hu Z, Du Y, Nomura Y, Dong K (2011) A sodium channel mutation identified in Aedes aegypti selectively reduces cockroach sodium channel sensitivity to type I, but not type II pyrethroids. Insect Biochem Mol Biol 41:9–13
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6:279–284
Ilias A, Vontas J, Tsagkarakou A (2014) Global distribution and origin of target site insecticide resistance mutations in Tetranychus urticae. Insect Biochem Mol Biol 48:17–28
Karatolos N, Williamson MS, Denholm I, Gorman K, Ffrench-Constant R, Nauen R (2012) Resistance to spiromesifen in Trialeurodes vaporariorum is associated with a single amino acid replacement in its target enzyme acetyl-coenzyme A carboxylase. Insect Mol Biol 21:327–334
Kasai S, Ng LC, Lam-Phua SG, Tang CS, Itokawa K, Komagata O, Kobayashi M, Tomita T (2011) First detection of a putative knockdown resistance gene in major mosquito vector, Aedes albopictus. Jpn J Infect Dis 64:217–221
Kawada H, Higa Y, Komagata O, Kasai S, Tomita T, Thi Yen N, Loan LL, Sanchez RA, Takagi M (2009) Widespread distribution of a newly found point mutation in voltage-gated sodium channel in pyrethroid-resistant Aedes aegypti populations in Vietnam. PLoS Negl Trop Dis 3:e527
Kim SS, Seo SG, Park JD, Kim SG, Kim DI (2005) Effects of selected pesticides on the predatory mite, Amblyseius cucumeris (Acari: Phytoseiidae). J Entomol Sci 40:107–114
Kwon DH, Clark JM, Lee SH (2010a) Cloning of a sodium channel gene and identification of mutations putatively associated with fenpropathrin resistance in Tetranychus urticae. Pest Biochem Physiol 97:93–100
Kwon DH, Yoon KS, Clark JM, Lee SH (2010b) A point mutation in a Glutamate-gated chloride channel confers abamectin resistance in the two-spotted spider mite, Tetranychus urticae Koch. Insect Mol Biol 19:583–591
Liu N, Yue X (2000) Insecticide resistance and cross-resistance in the house fly (Diptera: Muscidae). J Econ Entomol 93:1269–1275
Liu N, Zhu F, Xu Q, Pridgeon JW, Gao XW (2006) Behavioral change, physiological modification, and metabolic detoxification: mechanisms of insecticide resistance. Acta Entomol Sin 49:671–679
Lümmen P, Khajehali J, Luther K, Van Leeuwen T (2014) The cyclic keto-enol insecticide spirotetramat inhibits insect and spider mite acetyl-CoA carboxylases by interfering with the carboxyltransferase partial reaction. Insect Biochem Mol Biol 55:1–8
Mcmurtry JA, Croft BA (1997) Life-styles of Phytoseiid mites and their roles in biological control. Annu Rev Entomol 42:291–321
Morales MA, Mendoza BM, Lavine LC, Lavine MD, Walsh DB, Zhu F (2016) Selection of reference genes for expression studies of xenobiotic adaptation in Tetranychus urticae. Int J Biol Sci 12:1129–1139
Motoba K, Suzuki T, Uchida M (1992) Effect of a new acaricide, fenpyroximate, on energy metabolism and mitochondrial morphology in adult female Tetranychus urticae (two-spotted spider mite). Pest Biochem Physiol 43:37–44
Nauen R (2005) Spirodiclofen: mode of action and resistance risk assessment in tetranychid pest mites. J Pestic Sci 30:272–274
O’Neal SD, Walsh DB, Gent DH, Barbour JD, Boydston RA, George AE, James DG, Sirrine JR (2015) Field guide for integrated pest management in hops. U.S. Hop Industry Plant Protection Committee, Pullman, WA
Pavlidi N, Tseliou V, Riga M, Nauen R, Van Leeuwen T, Labrou NE, Vontas J (2015) Functional characterization of glutathione S-transferases associated with insecticide resistance in Tetranychus urticae. Pestic Biochem Physiol 121:53–60
Piraneo TG, Bull J, Morales MA, Lavine LC, Walsh DB, Zhu F (2015) Molecular mechanisms of Tetranychus urticae chemical adaptation in hop fields. Sci Rep 5:17090
Riga M, Tsakireli D, Ilias A, Morou E, Myridakis A, Stephanou EG, Nauen R, Dermauw W, Van Leeuwen T, Paine M, Vontas J (2014) Abamectin is metabolized by CYP392A16, a cytochrome P450 associated with high levels of acaricide resistance in Tetranychus urticae. Insect Biochem Mol Biol 46:43–53
Riga M, Myridakis A, Tsakireli D, Morou E, Stephanou EG, Nauen R, Van Leeuwen T, Douris V, Vontas J (2015) Functional characterization of the Tetranychus urticae CYP392A11, a cytochrome P450 that hydroxylates the METI acaricides cyenopyrafen and fenpyroximate. Insect Biochem Mol Biol 65:91–99
Soderlund DM, Knipple DC (2003) The molecular biology of knockdown resistance to pyrethroid insecticides. Insect Biochem Mol Biol 33:563–577
Stumpf N, Nauen R (2002) Biochemical markers linked to abametin resistance in Tetranychus urticae (Acari: Tetranychidae). Pest Biochem Physiol 72:111–121
Tsagkarakou A, Van Leeuwen T, Khajehali J, Ilias A, Grispou M, Williamson MS, Tirry L, Vontas J (2009) Identification of pyrethroid resistance associated mutations in the para sodium channel of the two-spotted spider mite Tetranychus urticae (Acari: Tetranychidae). Insect Mol Biol 18:583–593
Van Leeuwen T, Dermauw W (2016) The molecular evolution of xenobiotic metabolism and resistance in chelicerate mites. Annu Rev Entomol 61:475–498
Van Leeuwen T, Vanholme B, Van Pottelberge S, Van Nieuwenhuyse P, Nauen R, Tirry L, Denholm I (2008) Mitochondrial heteroplasmy and the evolution of insecticide resistance: non-Mendelian inheritance in action. Proc Natl Acad Sci USA 105:5980–5985
Van Leeuwen T, Vontas J, Tsagkarakou A, Tirry L (2009) Mechanisms of acaricide resistance in the two-spotted spider mite Tetranychus urticae. In: Ishaaya I, Horowitz A (eds) Biorational control of arthropod pests. Springer, Berlin
Van Leeuwen T, Vontas J, Tsagkarakou A, Dermauw W, Tirry L (2010) Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: a review. Insect Biochem Mol Biol 40:563–572
Van Leeuwen T, Demaeght P, Osborne EJ, Dermauw W, Gohlke S, Nauen R, Grbic M, Tirry L, Merzendorfer H, Clark RM (2012) Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods. Proc Natl Acad Sci USA 109:4407–4412
Van Leeuwen T, Dermauw W, Grbic M, Tirry L, Feyereisen R (2013) Spider mite control and resistance management: does a genome help? Pest Manag Sci 69:156–159
Van Leeuwen T, Tirry L, Yamamoto A, Nauen R, Dermauw W (2015) The economic importance of acaricides in the control of phytophagous mites and an update on recent acaricide mode of action research. Pestic Biochem Physiol 121:12–21
Van Nieuwenhuyse P, Van Leeuwen T, Khajehali J, Vanholme B, Tirry L (2009) Mutations in the mitochondrial cytochrome b of Tetranychus urticae Koch (Acari: Tetranychidae) confer cross-resistance between bifenazate and acequinocyl. Pest Manag Sci 65:404–412
Van Pottelberge S, Van Leeuwen T, Nauen R, Tirry L (2009) Resistance mechanisms to mitochondrial electron transport inhibitors in a field-collected strain of Tetranychus urticae Koch (Acari: Tetranychidae). Bull Entomol Res 99:23–31
Weston DP, Poynton HC, Wellborn GA, Lydy MJ, Blalock BJ, Sepulveda MS, Colbourne JK (2013) Multiple origins of pyrethroid insecticide resistance across the species complex of a nontarget aquatic crustacean, Hyalella azteca. Proc Natl Acad Sci USA 110:16532–16537
Zhu F, Gujar H, Gordon JR, Haynes KF, Potter MF, Palli SR (2013) Bed bugs evolved unique adaptive strategy to resist pyrethroid insecticides. Sci Rep 3:1456
Zhu F, Lavine L, O’neal S, Lavine M, Foss C, Walsh D (2016a) Insecticide resistance and management strategies in urban ecosystems. Insects. https://doi.org/10.3390/insects7010002
Zhu KY, Merzendorfer H, Zhang W, Zhang J, Muthukrishnan S (2016b) Biosynthesis, turnover, and functions of chitin in insects. Annu Rev Entomol 61:177–196
Acknowledgements
The authors thank Tora Brooks, Dan Groenendale, Peng Wilson, and members of the Agricultural Entomology lab of WSU IAREC at Prosser, WA, for their help with field mite collection. This research was funded by the United States Department of Agriculture National Institute of Food and Agriculture Specialty Crop Research Initiative (SCRI) (Award Number 2014-51181-22381), the Hop Research Council, the Washington Hop Commission, the Washington State Commission on Pesticide Registration. M.W. was supported by the Education Department of Fujian Province of China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Y. Gao.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, M., Adesanya, A.W., Morales, M.A. et al. Multiple acaricide resistance and underlying mechanisms in Tetranychus urticae on hops. J Pest Sci 92, 543–555 (2019). https://doi.org/10.1007/s10340-018-1050-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10340-018-1050-5