Abstract
A modified quick, easy, cheap, effective, rugged, and safe (QuEChERS) sample preparation method coupled with gas chromatography–tandem mass spectrometry has been developed for the simultaneous analysis of amide/dinitroaniline/substituted urea herbicides in bivalve shellfish samples. A molecularly imprinted polymer (MIP) synthesized through bulk polymerization exhibited strong group–selective interactions with pigments, including β-carotene, chlorophyll A, and fucoxanthin, in bivalve shellfish extracts. Afterward, a modified QuEChERS method based on MIPs and primary and secondary amines was established to effectively remove matrix components in bivalve shellfish samples. Under the optimal conditions, good linearities were obtained in all of the analytes with R2 larger than 0.9995. Limits of quantification were in the range of 0.03–8.88 μg kg−1, respectively. The recoveries of all of the herbicides spiked at three concentrations of 10, 25, and 50 μg kg−1 in blank bivalve shellfish samples ranged from 81 to 109% with intra-day and inter-day relative standard deviations less than 8%, respectively. Results demonstrated that the proposed QuEChERS method coupled with GC–MS/MS was applied successfully to simultaneously determine 26 amide/dinitroaniline/substituted urea herbicides in bivalve shellfish.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Herbicides, one of the most frequently used group of pesticides, are widely used as weed control in global modern agriculture, and some herbicides are also used to control algae in aquaculture [1,2,3]. However, their residues had been detected in soil, crops, water and sediment due to the continued and indiscriminate use, which resulted in the potential threat on the ecosystem environment [4]. Meanwhile, a large number of researchers had confirmed that the herbicides were toxic and harmful [5, 6]. Their ecotoxicological hazardous effects have attracted increasing attentions. Especially, bivalves, which are the most widely cultured shellfish in aquaculture, are distributed along coasts, where shellfish are easily contaminated by herbicide residues in the aquaculture environment [7]. Therefore, the residue conditions of herbicides in shellfish by potential bioaccumulation should be investigated, and effective analytical techniques should be developed.
The majority of current quantitative analytical methods, such as liquid chromatography–tandem mass spectrometry, sensor detection, gas chromatography–tandem mass spectrometry (GC–MS/MS), and biosensor method, have been used to detect herbicides in different matrices [8,9,10,11,12]. However, shellfish samples usually contain various pigments, lipids, proteins, and other interference matrices, which can affect the separation and ionization properties of analytes [13]. Meanwhile, multiclass herbicides are usually difficult to extract from biological samples because of their different polarities. Therefore, a rapid and effective analytical approach is necessary to obtain satisfactory sensitivity, eliminate interference matrix, and improve determination efficiency.
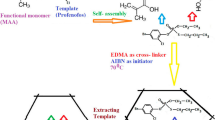
The quick, easy, cheap, effective, rugged, and safe (QuEChERS) sample preparation approach, which is based on simple solvent extraction and dispersive solid-phase extraction (d-SPE) cleanup, was introduced by Anastassiades et al. [14] and has been applied successfully to analyze herbicide residues in various fruits and vegetables. Clean-up was usually carried out using d-SPE by primary secondary amine (PSA), octadecyl (C-18) and graphitized carbon black (GCB) materials. For samples with high pigment contents, which can lead to serious signal interferences, GCB is usually employed as an adsorption sorbent of QuEChERS to remove pigments from a sample matrix [15]. However, GCB not only removes pigment but also adsorbs herbicides with planar and p-benzene ring structures, resulting in poor accuracy and precision [16]. To address this problem, researchers should develop a highly selective adsorption material with the properties of adsorption interfering pigments rather than target analytes. Molecularly imprinted polymers (MIPs) have been widely applied as solid-phase extraction (SPE) sorbents with an excellent molecular recognition capability to targeted compounds [17, 18], thereby providing high extraction selectivity and sample purification efficiency. Few studies have reported MIPs as QuEChERS sorbents to remove interferences of various pigments [16].
This study aimed to develop MIPs with excellent group selectivity and good adsorption capacity to pigments of the bivalve shellfish extracts. The fabricated MIPs were used as sorbents of QuEChERS method for pigment removal in sample pretreatment. A novel determination strategy coupled with a modified QuEChERS method based on MIPs and PSA with GC–MS/MS was successfully developed and validated for the simultaneous qualitative and quantitative analyses of amide/dinitroaniline/substituted urea herbicides in the bivalve shellfish samples.
Experimental
Chemicals and Reagents
Herbicides (Table S1) (purity > 97.0%) were purchased from Dr. Ehrenstorfer (Augsburg, Germany). Chromatographic-grade n-hexane was procured from Tedia (Fairfield, USA). Octadecylsilane (C18) (50 μm, 60 Å pore size), GCB, and PSA (40–60 μm) were obtained from ANPEL Laboratory Technologies (Shanghai, China). Analytical-grade acetonitrile, acetone, sodium chloride (NaCl), anhydrous magnesium sulfate (MgSO4), anhydrous sodium sulfate (Na2SO4), and sodium acetate (CH3COONa) were provided by Sinopharm Chemical Reagent Corporation (Shanghai, China). β-carotene (purity ≥ 97%) and chlorophyll A (purity ≥ 96%) were bought from Hefei Bomei Biotechnology (Hefei, China). Fucoxanthin (purity ≥ 98%) was purchased from Shanghai Yuanye Bio-Technology. MgSO4 and Na2SO4 were heated in a muffle furnace at 400 °C for 6 h and stored in a desiccator until use. Azobisisobutyronitrile (AIBN) was obtained from Tianjin Guangfu Fine Chemical Research Institute (Tianjin, China). Methacrylic acid (MAA) and ethylene glycol dimethacrylate (EGDMA) were procured from Sigma-Aldrich (Steinheim, Germany).
Stock solutions of individual herbicide standards (1000 mg L−1) were prepared by dissolving 10 mg of each standard into 10 mL of acetone and kept at − 20 °C. A mixed stock solution (1.0 mg L−1) of 26 amide/dinitroaniline/substituted urea herbicides was prepared by diluting the individual stock solutions with n-hexane. All of the solutions were stored at 4 °C in darkness.
Synthesis of MIPs and Non-imprinted Polymers (NIPs)
The MIPs were synthesized by bulk polymerization as previously reported in the literature [19]. Typically, 2.5 mg of β-carotene and 4 mmol MAA were dissolved in 5 mL of toluene: acetonitrile (v/v, 3:7) and ultrasonically agitated for 10 min under nitrogen atmosphere. After the solution was allowed to stand for 30 min, 25 mmol EGDMA and 2 mmol AIBN were added and sonicated for 5 min under nitrogen atmosphere. The solution was purged with dry nitrogen for 5 min and immersed in a water bath at 60 °C for 24 h in a sealed environment. Finally, the bulk polymers were crushed, ground, sieved (40–63 μm), and washed with methanol: acetic acid (v/v, 9:1) by a Soxhlet apparatus to remove the template molecule. The corresponding NIPs were prepared using the same protocol but without a template.
Binding Property Assay
The binding properties of MIPs and NIPs were studied using equilibrium batch rebinding experiments. Typically, 20 mg of polymers was added to 2 mL of toluene: acetonitrile (v/v, 3:7) containing different analyte concentrations (10–500 mg L−1) in a glass vial, respectively. The vials were sealed and underwent shaking at 180 rpm for 2 h at room temperature. The analyte concentrations on the supernatants were analyzed by a UV spectrophotometer at 450 nm. The amounts of polymer bound to the analyte were calculated by subtracting the free analyte from the initial concentration in the vial. All of the experiments were processed in triplicate.
The specific adsorption properties and Scatchard analysis of MIPs were carried out as previously reported [20].
where C0 (mg L−1) is the initial concentration of β-carotene; C (mg L−1) is the concentration of β-carotene after adsorption; V is the volume of the solution; W (mg) is the mass of the polymer; Q and Qmax (μg mg−1) are the amounts of β-carotene adsorbed at equilibrium and saturation, respectively. The definite imprinting factor (IF), that is, the selective alpha, was determined by the following formula to further compare the imprinting effect: IF = QMlP/QNlP, α = (QMlP − QNlP)/QNlP, where QMlP is the amount of the imprinted polymer to adsorb β-carotene of the template molecule, and QNlP is the adsorption capacity of the NIP to the template molecule β-carotene.
MIPs Characterization
The size and morphological characteristics of MIPs and NIPs were investigated using an S-4800 cold field-emission scanning electron microscope (Hitachi, Japan). The pore volume and surface area of MIPs and NIPs were measured using an ASAP 2020 accelerated surface area and porosimetry analyzer (Micromeritics Instrument Corporation, Norcross, GA), and 50 mg of the dried polymer was used and degassed at 200 °C for 24 h under nitrogen flow prior to measurement. The chemical structure and bonding of MIPs were confirmed using a Fourier transform infrared spectroscope (FTIR, Nicolet, Thermo, USA).
Sample Preparation by QuEChERS with d-SPE Cleanup Using the MIP
Bivalve shellfish samples were purchased from a local aquatic market in Ningbo. In general, 2 g of bivalve shellfish samples were weighed into a 50 mL polypropylene centrifuge tube and homogenized at 8000 rpm for 10 min. Then, 5 mL of 1% acidic acetonitrile and 2 g of NaCl were added. After vortexing for 2 min, 0.3 g of Na2SO4 and 1.7 g of CH3COONa were added. The mixture was shaken for 2 min and centrifuged at 5000 rpm for 5 min. The samples were extracted repeatedly. The supernatants were combined and evaporated to 1.0 mL under nitrogen at 30 °C. The solution was transferred into a 2 mL centrifuge tube containing 150 mg of Na2SO4, 50 mg of MIPs, and 25 mg of PSA. After the solution was shaken for 15 min, the tube was centrifuged at 8000 rpm for 1 min. Afterward, 450 μL of the extract (the upper layer) was decanted into an autosampler vial, and 50 μL of internal standard was added for GC–MS/MS analysis.
Conventional Solid-Phase Extraction
Briefly, the 2.00 g of bivalve shellfish samples were weighed into a 15 mL polypropylene tube and 5 mL acetonitrile were added. Afterwards, the mixture was homogenized for 2 min in a blender at 9.0 × 103 rpm (IKA, Staufen, Germany). Subsequently, the blender was rinsed with 5 mL acetonitrile and pooled in the polypropylene tube. Furthermore, 5 g NaCl and 0.3 g Na2SO4 were added and vortexed for 30 s. The mixture was centrifuged at 5.0 × 103g for 5 min, and the supernatant was collected and concentrated to 2 mL for SPE procedure. The cartridges GCB/NH2 (500 mg/6 mL, ANPLE, China) was selected and conditioned with 4 mL acetonitrile/toluene (3:1, v/v). The supernatant were directly percolated through the cartridges. Afterwards, the SPE cartridges were eluted with 10 mL acetonitrile/toluene (3:1, v/v), and the eluent was evaporated to dryness under nitrogen atmosphere at 40 °C. The samples were re-suspended in 0.5 mL of n-hexane (4 g equivalent sample) and passed through a 0.22 μm filter membrane for GC–MS/MS analysis.
GC–MS/MS
The samples were analyzed via an Agilent 7890B GC system coupled to an Agilent 7000D triple-quadrupole mass spectrometer (Agilent Technologies, USA) with electron ionization in a multiple reaction monitoring mode. The GC system was equipped with an DB-5 MS UI fused silica capillary column (30 m × 0.25 mm, i.d., 0.25 μm, Agilent). The samples (1.0 μL) were injected using a splitless injection mode. The GC–MS/MS conditions were as follows: helium was used as a carrier gas at a constant flow of 1.0 mL min−1, and injector temperature was set at 280 °C. The GC oven temperature profile was started at 80 °C, maintained for 1 min, increased to 170 °C at a rate of 40 °C min−1, increased to 200 °C at a rate of 5 °C min−1, and ramped to 210 °C at 2 °C min−1, then increased to 310 °C at 10 °C min−1. The final temperature was kept for 5 min, and the total run time was 29.25 min. The transfer line and ion source temperatures were both 280 °C. The collision energies of the parent ions and the quantitative daughter ions of the herbicides are summarized in Table S1, and GC–MS/MS chromatograms of herbicides at 25.0 μg kg−1 are illustrated in Fig. S1.
Results and Discussion
MIP Preparation and Characterization
The molar ratios of the template to the functional monomer play an important role in enhancing the specific affinity of MIPs. The low proportion of the template to the functional monomer usually produces a low affinity capacity, whereas its high proportion results in relatively high nonspecific binding sites [21]. In Table S2, when the molar ratio of the template molecule to the functional monomer was 1:4, the MIPs had the highest adsorption capacity and imprinting effect with an IF of 2.2. Therefore, the MIPs were synthesized with a template-to-functional monomer ratio of 1:4.
The surface morphology and size distribution of the fabricated MIPs were observed through SEM. In Fig. 1, the average size of the resulting MIPs and NIPs was approximately 40–60 μm, and the pores were embedded in a network of MIPs and NIPs. No distinct difference was observed in size distributions and morphological structures between MIPs and NIPs, but the surface area of MIPs (MIPs, 259 m2 g−1; NIPs, 235 m2 g−1) was higher than that of NIPs, thereby providing more recognition sites with specific adsorption to target molecules.
The FTIR spectra of MIPs and NIPs (Fig. S2) further confirmed the successful imprinting sites onto MIPs. The O–H stretching vibration at 3590 cm−1 indicated the successful polymerization of MAA and crosslinker. The symmetrical and asymmetrical stretching vibrations of C–H in methyl were 2987 and 2954 cm−1, respectively. The stretching vibration of C=O in the polymer appeared at 1715 cm−1. The symmetrical and asymmetrical stretching vibration of C–H in methylene displayed at 1456 and 1388 cm−1, respectively. The symmetrical and asymmetrical stretching vibration of C–O–C were 1261 and 1161 cm−1, respectively, confirming the successful polymerization of EGDMA. C–H surface bending vibration and surface rocking vibration in vinyl were 960 and 881 cm−1, respectively. The results demonstrated that MIPs and NIPs exhibited similar primary compositions and major backbones.
Binding Property Assay
MIPs had specific and group-specific binding capability, and the binding properties were the most important constants during sample purification. Therefore, the binding properties of MIPs were evaluated via an adsorption isotherm experiment and through Scatchard analysis. In Fig. 2, the adsorption capacity of MIPs was higher than that of NIPs over the tested concentrations range of 10–50 μg mL−1. Scatchard analysis indicated that the binding sites of MIPs were heterogeneous and classified into two distinct groups. Kd and Qmax were calculated in accordance with the slopes and intercepts of the two linear regression equations. The linear regression equation of MIPs of the upper line was Q/Cf = − 0.2066Q + 1.0248 (R2 = 0.9734). The linear regression equation of the lower line was Q/Cf = − 0.0267Q + 0.3691 (R2 = 0.9137). Kd and Qmax of the high-affinity binding sites were 1.02 mg L−1 and 4.96 mg g−1, respectively. Kd and Qmax of the low-affinity binding sites were 0.37 mg L−1 and 13.82 mg g−1, respectively. These results demonstrated that recognition sites were created onto the MIPs by molecular imprinting. The results of the static adsorption of MIPs indicated that 20 mg of MIPs could selectively adsorb above 90% β-carotene at 10 mg L−1. The high binding affinity and capacity of MIPs to β-carotene enabled its selective removal from complex shellfish matrices.
Chlorophyll A and fucoxanthin, which have partially similar structures and functional groups, were chosen to gain insights into recognition capability. In Fig. 3, the MIPs exhibited the strongest specific affinity and binding amounts to β-carotene with an IF of 2.4. The binding amounts of MIPs to those of β-carotene and its structurally related compounds, chlorophyll A and fucoxanthin, than the NIPs, and the structural effects of the analytes on the binding properties of MIPs were observed during the binding procedure. The results indicated that the MIPs had high group-specific binding for β-carotene and its structurally related compounds, such as chlorophyll A and fucoxanthin, in appropriate solvents because of the imprinting effect. β-Carotene, chlorophyll A, and fucoxanthin are the most common pigments in aquaculture [22]. Therefore, the MIPs with group selectivity could be applied as QuEChERS sorbents in pigment removal in shellfish extracts.
The kinetic adsorption of β-carotene onto the MIPs was further investigated (Fig. S3). The adsorption rate of β-carotene by MIPs was very fast, almost reaching equilibrium at 15 min. This finding indicated that the MIPs had relatively fast adsorption kinetics.
Optimization of Sample Pretreatment
QuEChERS involve extraction partitioned from the matrix by salt adding. To obtain the satisfactory results, the QuEChERS procedure was optimized. As shown in Fig. S4. The recoveries of herbicides under the anhydrous Na2SO4 were higher compared with those obtained under the anhydrous MgSO4. Especially, the recoveries of ethalfluralin and trifluralin were improved from 49% (anhydrous MgSO4) to 100% (anhydrous Na2SO4) and from 45% (anhydrous MgSO4) to 92% (anhydrous Na2SO4), respectively. The satisfactory recoveries of herbicides ranged from 81 to 121% were acquired. The above results demonstrated that the anhydrous Na2SO4 is effective to elicit phase separation compared with anhydrous MgSO4 [23].
The chemical composition of shellfish extracts was incompatible with the GC–MS/MS system, resulting in serious negative effects on signal response. Therefore, the rapid and effective purification procedures were required to obtain satisfactory accuracy and precision. In this study, the developed MIPs exhibited high adsorption capacity and effective removal efficiency in pigment composition. Other matrices included organic acids, fatty acids and sugars in shellfish extracts except the pigments. Therefore, a combination of 25 mg of PSA, which can retain many polar compounds of organic acids, fatty acids, and sugars [14], but it gives not satisfying results in the case of samples with high contents of pigment [24], and an appropriate amount of MIPs was applied to subsequent experiments. First, the effect of the amounts of MIPs on the matrix removal of bivalve shellfish extracts was determined. Figure S5 shows that when the amounts of MIPs reached 50 mg, the recoveries of the amide/dinitroaniline/substituted urea herbicides ranged from 82 to 120% with a relative standard deviation below 7%. However, the recoveries of some amide herbicides decreased as the amounts of MIPs further increased possibly because nonspecific interactions occurred between the amide group of herbicides and the hydroxyl group of MIPs, resulting in their low recoveries. Simultaneously, the recoveries of some herbicides were below 80% when the amounts of MIPs used was less than 30 mg possibly because low amounts of MIPs favored the insufficient removal of matrix components, resulting in matrix suppression. Therefore, 50 mg of MIPs was used as QuEChERS sorbents in subsequent experiments.
Method Validation
The method was validated under the optimized experimental conditions by determining linearity, limits of detection (LOD), and quantification (LOQ), matrix effects, accuracy, and precision.
The matrix effects (ME, %), leading to the poor accuracy and precision of experimental results and probably originating from the signal interferences between the analyte and the coeluting substance [25]. During the QuEChERS procedure, the MIPs as the sorbent exhibited a high pigment removal capability, which was evidenced by the change in the color of bivalve shellfish extracts to colorless after purification was completed. However, the ME evaluation was significant because of the presence of other compounds except the pigments. The matrix effect was examined by comparing the slopes of the calibration curve obtained using the solvent and extracted matrix, and the slope ratio values that ranged from 0.8 to 1.2 could be considered insignificant [26]. The ME results were grouped into 3 classes: a high ME (less than − 50% or higher than + 50%), a medium ME (between − 50% and − 20% or + 20% and + 50%) and a low ME (between + 20% and − 20%). Figure S6 shows the ME of each herbicides, 16% showed low ME, 46% showed medium ME and 38% showed high ME. To avoid the ME, matrix matched calibration standards were used for quantification to compensate for the ME. Therefore, the matrix-matched calibrations were selected to compensate matrix effects. The linearity was evaluated by studying six-level calibration curves constructed from a set of herbicide standards prepared in the solvent and in matrix-matched shellfish extracts within a concentration range of 0.005–0.25 μg kg−1, and each concentration was analyzed in triplicate. All analyses yielded high correlation coefficients (R2) above 0.9995.
The LOQs of each herbicides were determined by calculating a signal-to-noise ratio of 10, respectively. The LOQs were in the range of 0.03–8.88 μg kg−1, respectively (Table 1). The LOQ of ethalfluralin (0.03 μg kg−1) was below its MRL in bivalve shellfish established by EU regulation (0.1 μg kg−1), indicating that the sensitivity of the optimized method can fully meet the needs of the proposed application. The recoveries and precisions were determined by measuring six samples at three spiking levels of 10, 25, and 50 μg kg−1, the recoveries of the amide/dinitroaniline/substituted urea herbicides ranged from 84 to 106% with relative standard deviation (RSD %) below 6% at the three spiking levels (Table 2), which were within the acceptable range. The method precision was assessed by determining repeatability (inter-day) RSD range of 0.6–8% and reproducibility (intra-day) RSD range of 0.9–8% reproducibility was determined by analyzing spiked from 3 different days. These results demonstrated that the proposed QuEChERS based on MIPs showed excellent accuracy and precision for the simultaneous extraction and purification of 26 amide/dinitroaniline/substituted urea herbicides in bivalve shellfish samples.
Clean-up sorbents play an important role in achieving satisfactory accuracy and precision. However, the use of inappropriate materials could adsorb target analytes to some degree, resulting in their low accuracy. C18 and GCB were the mostly used sorbents to remove a nonpolar matrix. Therefore, C18, GCB, MIPs, and NIPs were evaluated for their capability to remove interferences. The results are shown in Fig. 4. GCB not only removes pigments but also adsorbs the analytes with planar molecules or molecules containing planar aromatic rings, resulting in poor accuracy and precision such that the recoveries of trifluralin (45%), benfluralin (45%), alachlor (29%), isoproturon (7%), and nitralin (47%) were below 50%. The amide/dinitroaniline/substituted urea herbicides purified by the C18 sorbent had wide recoveries in the range of 28–121%, and the recoveries of ethalfluralin (39%), trifluralin (36%), benfluralin (36%), profluralin (45%), fluchloralin (47%), isoproturon (28%), and nitralin (33%) were below 50%. However, the recoveries of the amide/dinitroaniline/substituted urea herbicides purified by the modified QuEChERS based on MIPs focused in the range of 81–112%. By contrast, the wide recoveries in the range of 12–116% were obtained for the NIPs, and the recoveries of over 60% the amide/dinitroaniline/substituted urea herbicides were below 50%. These results indicated that the developed QuEChERS method based on the MIPs could provide the best stable recoveries for the amide/dinitroaniline/substituted urea herbicides in bivalve shellfish samples, and the MIPs could remove pigments and had no adsorption effect on the amide/dinitroaniline/substituted urea herbicides, which played an important role in pretreatment for the removal of pigments and other interfering matrixes.
The robustness of the established method was evaluated by calculating the recoveries and RSDs at the spiked concentration of 25 μg kg−1 in three different bivalve shellfish samples of razor clam, clam, and mussel. The results are summarized in Table 1. The mean recoveries of razor clam, clam, and mussel ranged from 80 to 120%, from 80 to 117%, and from 80 to 118%, respectively, and their RSDs were below 10%.
Comparison Between QuEChERS and Conventional SPE (C-SPE) Method
The proposed method was compared with other reported methods in the literature for the extraction and determination of the multi-residues herbicides in shellfish from the viewpoints of extraction method, clean-up procedure, matrix, and LOQs to further evaluate the modified QuEChERs with GC–MS/MS method [27,28,29]. The comparison results (Table S3) showed that the current method exhibited comparable LOQs and precision compared with the other reported methods and was more convenient than that obtained with the other reported methods. Especially, the modified QuEChERs method exhibited a lower detection limit compared with those of the SPE method (LOQs: 0.06–12 μg kg−1). The above results indicated that the modified QuEChERs had an excellent adsorption capacity for the pigments and could be applied to enrich the amide/dinitroaniline/substituted urea herbicides in bivalve shellfish samples.
Application to Real Samples
As above reported in this study, C18 and GCB are the pre-existing and commercial adsorbents to remove pigments. However, the recoveries of trifluralin, benfluralin, alachlor, isoproturon and nitralin after the QuEChERS procedure based on the C18 and GCB were all below 50% in the spiked bivalve shellfish samples. Furthermore, we found that the C18 and GCB exhibited lower pigment removal efficiency than that of MIPs, which are apt to contaminate ion source and instrument. Therefore, to further demonstrate the real and practical advantages of the developed QuEChERS method based on the MIPs, the shellfish products were purchased from Ningbo market and analyzed by the developed QuEChERS and C-SPE following GC–MS/MS analysis. As shown in Table S4. Atrazine, trifluralin, dimethenamid, cyanazine and pendimethalin were detected in the samples analyzed. The results obtained by the developed QuEChERs are in good agreement with those obtained by C-SPE. Therefore, the current study indicated that the developed QuEChERS based on the MIPs exhibited excellent potential in real application.
Conclusions
A MIP-based modified QuEChERS sample preparation method combined with GC–MS/MS detection was developed to analyze amide/dinitroaniline/substituted urea herbicides in different bivalve shellfish species. MIPs as the sorbent of the modified QuEChERs could be used to purify the samples and remove pigments from bivalve shellfish extracts. The developed method met the requirements for herbicide determination in bivalve shellfish samples. The performance parameters, such as selectivity, precision, and accuracy, of all of the test herbicides were in an acceptable range of 81–109% and had an excellent repeatability RSD below 8%. The developed method was successfully used for the analysis of the amide/dinitroaniline/substituted urea herbicide residues in different bivalve shellfish species and provided a potential strategy for pollutant residue analysis in biological samples.
References
Kumar A, Shalini Sharma G, Naushad M, Kumar A, Kalia S, Guo C, Mola GT (2017) Facile hetero-assembly of superparamagnetic Fe3O4/BiVO4 stacked on biochar for solar photo-degradation of methyl paraben and pesticide removal from soil. J Photochem Photobio A-Chem 337:118–131
Benbrook CM (2016) Trends in glyphosate herbicide use in the United States and globally. Environ Sci Eur 28:3
Li MH, Wang JT, Jiao CN, Wang C, Wu QH, Wang Z (2016) Graphene oxide framework: an adsorbent for solid phase extraction of phenylurea herbicides from water and celery samples. J Chromatogr A 1469:17–24
Marin-Benito JM, Barba V, Ordax JM, Sanchez-Martin MJ, Rodriguez-Cruz MS (2018) Recycling organic residues in soils as amendments: effect on the mobility of two herbicides under different management practices. J Environ Manage 224:172–181
Mela M, Guiloski IC, Doria HB, Randi MAF, Ribeiro CAO, Pereira L, Maraschi AC, Prodocimo V, Freire CA, Assis HCS (2013) Effects of the herbicide atrazine in neotropical catfish (Rhamdia quelen). Ecotoxicol Environ Saf 93:13–21
Villaverde JJ, Sevilla-Moran B, Lopez-Goti C, Calvo L, Alonso-Prados JL, Sandin-Espana P (2018) Photolysis of clethodim herbicide and a formulation in aquatic environments: fate and ecotoxicity assessment of photoproducts by QSAR models. Sci Total Environ 615:643–651
Cruzeiro C, Rodrigues-Oliveira N, Velhote S, Pardal MA, Rocha E, Rocha MJ (2016) Development and application of a QuEChERS-based extraction method for the analysis of 55 pesticides in the bivalve Scrobicularia plana by GC–MS/MS. Anal Bioanal Chem 408:3681–3698
Chang GR, Chen HS, Lin FY (2016) Analysis of banned veterinary drugs and herbicide residues in shellfish by liquid chromatography-tandem mass spectrometry (LC/MS/MS) and gas chromatography-tandem mass spectrometry (GC/MS/MS). Mar Pollut Bull 113:579–584
Kaczynski P (2017) Clean-up and matrix effect in LC-MS/MS analysis of food of plant origin for high polar herbicides. Food Chem 230:524–531
Wang P, Peteinatos G, Li H, Gerhards R (2016) Rapid in-season detection of herbicide resistant Alopecurus myosuroides using a mobile fluorescence imaging sensor. Crop Prot. 89:170–177
Köck M, FarréM Martínez E, Gajda-Schrantz K, Ginebreda A, Navarro A, Alda ML, Barceló D (2010) Integrated ecotoxicological and chemical approach for the assessment of pesticide pollution in the Ebro River delta (Spain). J Hydrol 383:73–82
Swainsbury DJK, Friebe VM, Frese RN, Jones MR (2014) Evaluation of a biohybrid photoelectrochemical cell employing the purple bacterial reaction centre as a biosensor for herbicides. Biosens Bioelectron 58:172–178
Brovedani V, Sosa S, Poli M, Forino M, Varello K, Tubaro A, Pelin M (2016) A revisited hemolytic assay for palytoxin detection: limitations for its quantitation in mussels. Toxicon 119:225–233
Anastassiades M, Lehotay SJ, Stajnbaher D, Schenck FJ (2003) Fast and easy multiresidue method employing acetonitrile extraction/partitioning and “dispersive solid-phase extraction” for the determination of pesticide residues in produce. J AOAC Int 86(2):412–431
Han YT, Zou N, Song L, Li YJ, Qin YH, Lin SW, Li XS, Pan CP (2015) Simultaneous determination of 70 pesticide residues in leek, leaf lettuce and garland chrysanthemum using modified QuEChERS method with multi-walled carbon nanotubes as reversed-dispersive solid-phase extraction materials. J Chromatogr B 1005:56–64
Bareki SB, Janes M, Ronald M, Charlotta T, Nelson T (2013) A novel molecularly imprinted polymer for the selective removal of chlorophyll from heavily pigmented green plant extracts prior to instrumental analysis. J Chem. 2013:1–4
Gama MR, Bottoli CBG (2017) Molecularly imprinted polymers for bioanalytical sample preparation. J Chromatogr B 1043:107–121
Sorribes-Soriano A, Esteve-Turrillas FA, Armenta S, Montoya A, Herrero-Martinez JM, Guardia M (2018) Magnetic molecularly imprinted polymers for the selective determination of cocaine by ion mobility spectrometry. J Chromatogr A 1545:22–31
Iturralde I, Paulis M, Leiza JR (2014) The effect of the crosslinking agent on the performance of propranolol imprinted polymers. Euro Pol J 53:282–291
García-Calzón JA, Díaz-García ME (2007) Characterization of binding sites in molecularly imprinted polymers. Sensor Actuat B-Chem 123:1180–1194
Lu HZ, Xu SF (2017) Functional monomer-template-QDs sandwich structure for mesoporous structured bovine hemoglobin imprinted ratiometric luorescence sensor. Talanta 165:482–488
Shahidi F, Metusalach Brown JA (1998) Carotenoid pigments in seafoods and aquaculture. Crit Rev Food Sci Nutr 38:1–67
Hou XL, Wu YL, Chen RX, Zhu Y, Lv Y, Xu XQ (2014) Evaluation of two modified quick, easy, cheap, effective, rugged and safe (QuEChERS) sample preparation methods for the analysis of baclofen and gabapentin in feeds by liquid chromatography tandem mass spectrometry. J Pharm Biomed Anal 88:53–59
Tomasz R, Tomasz T (2015) A review of recent developments and trends in the QuEChERS sample preparation approach. Open Chem 13:980–1010
Dong HF, Tang H, Chen DZ, Xu T, Li L (2014) Analysis of 7 synthetic musks in cream by supported liquid extraction and solid phase extraction followed by GC MS/MS. Talanta 120:248–254
EC-European Commission (2014) Guidance document on analytical quality control and validation procedures for pesticide residues analysis in food and feed, SANCO/12571/2013. EC, Brussels
Bichon E, Dupuis M, Bizec LB, Andre F (2006) LC–ESI-MS/MS determination of phenylurea and triazine herbicides and their dealkylated degradation products in oysters. J Chromatogr B 838:96–106
Rodríguez-González N, González-Castro MJ, Beceiro-González E, Muniategui-Lorenzo S (2015) Development of a Matrix Solid Phase Dispersion methodology for the determination of triazine herbicides in mussels. Food Chem 173:391–396
Carafa R, Wollgast J, Canuti E, Ligthart J, Dueri S, Hanke G, Eisenreich SJ, Viaroli P, Zaldívar JM (2007) Seasonal variations of selected herbicides and related metabolites in water, sediment, seaweed and clams in the Sacca di Goro coastal lagoon (Northern Adriatic). Chemosphere 69:1625–1637
Acknowledgements
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (LR16C190001), the National Natural Science Foundation of China (No. 31772856), Natural Science Foundation of Ningbo (2018A610065), the project by Ningbo Science and Technology Bureau (2016C51002), the Technology Innovation Team of Ningbo city (2015C110018), and the K.C. Wong Magna Fund in Ningbo University.
Funding
This study was funded by the Zhejiang Provincial Natural Science Foundation of China (LR16C190001), the National Natural Science Foundation of China (No. 31772856), Natural Science Foundation of Ningbo (2018A610065), the project by Ningbo Science and Technology Bureau (2016C51002), the Technology Innovation Team of Ningbo city (2015C110018), and the K.C. Wong Magna Fund in Ningbo University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yu, Xr., Zhang, Zm., Li, W. et al. Development and Application of the Dispersive Solid-Phase Extraction Method Based on Molecular Imprinted Polymers for Removal of Matrix Components of Bivalve Shellfish Extracts in the GC–MS/MS Analysis of Amide/Dinitroaniline/Substituted Urea Herbicides. Chromatographia 82, 961–970 (2019). https://doi.org/10.1007/s10337-019-03729-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10337-019-03729-6