Abstract
Objective
This work presents a highly-accelerated, self-gated, free-breathing 3D cardiac cine MRI method for cardiac function assessment.
Materials and methods
A golden-ratio profile based variable-density, pseudo-random, Cartesian undersampling scheme was implemented for continuous 3D data acquisition. Respiratory self-gating was achieved by deriving motion signal from the acquired MRI data. A multi-coil compressed sensing technique was employed to reconstruct 4D images (3D+time). 3D cardiac cine imaging with self-gating was compared to bellows gating and the clinical standard breath-held 2D cine imaging for evaluation of self-gating accuracy, image quality, and cardiac function in eight volunteers. Reproducibility of 3D imaging was assessed.
Results
Self-gated 3D imaging provided an image quality score of 3.4 ± 0.7 vs 4.0 ± 0 with the 2D method (p = 0.06). It determined left ventricular end-systolic volume as 42.4 ± 11.5 mL, end-diastolic volume as 111.1 ± 24.7 mL, and ejection fraction as 62.0 ± 3.1%, which were comparable to the 2D method, with bias ± 1.96 × SD of −0.8 ± 7.5 mL (p = 0.90), 2.6 ± 3.3 mL (p = 0.84) and 1.4 ± 6.4% (p = 0.45), respectively.
Conclusion
The proposed 3D cardiac cine imaging method enables reliable respiratory self-gating performance with good reproducibility, and provides comparable image quality and functional measurements to 2D imaging, suggesting that self-gated, free-breathing 3D cardiac cine MRI framework is promising for improved patient comfort and cardiac MRI scan efficiency.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
2D cardiac cine MRI is considered the clinical standard for evaluating global cardiac function. Data acquisition is usually performed with a series of breath-holds to cover the entire left ventricle. This results in patient discomfort and can be particularly challenging for patients who have impaired breath-hold capability, or patients who have difficulty in cooperating with breath holding instructions. Furthermore, breath-hold positions may change from slice to slice, leading to slice misregistration errors, and thus reducing the accuracy of cardiac function quantification. Free-breathing 3D cardiac cine imaging techniques can overcome these limitations by improving patient comfort, avoiding the slice misregistration errors, providing contiguous volumetric coverage with thinner slice thickness that enables improved accuracy in measurements of cardiac function, and providing increased signal-to-noise ratio (SNR) and allowing visualization of reconstructed images in any desired orientation which also greatly simplifies specification of the imaging volume.
Free-breathing 3D cardiac cine MRI, however, has several challenges that impede its acceptance for wide clinical use. First, it requires a reliable respiratory motion compensation strategy. Gating is usually applied based on a respiratory motion signal, so that only data with minimal motion blurring (such as at end-expiration) are used for image reconstruction [1–7]. Other approaches have been developed to correct for respiratory motion by registering MRI data or images at different motion states and then combining all of them [8–10]. For both motion compensation strategies, a respiratory motion signal is required, which can be obtained from either external devices or an intrinsically acquired self-navigator. Respiratory bellows has been shown to be a reliable device for obtaining respiratory motion signal [11], however, its setup requires additional preparation and post-processing (especially the synchronization of the bellows signal with acquired k-space data). Self-navigation is an attractive alternative for free-breathing cardiac cine imaging [1, 10, 12–14]. It enables a simplified scan set-up with improved scan efficiency, and has also been demonstrated to have similar imaging performance as respiratory bellows in free-breathing cardiac cine imaging [3, 4].
Acceleration of 3D cardiac cine MRI is usually needed and this has been achieved using different fast imaging techniques, such as non-Cartesian acquisition and parallel imaging [1, 3, 10, 15–17]. K–t parallel imaging techniques, such as k–t SENSE and k–t GRAPPA, have also been used to exploit spatial–temporal redundancy in the dynamic image series, thus enabling improved imaging performance with higher acceleration rates [6, 18–20]. Compressed sensing (CS) is another powerful approach that can be used to accelerate 3D cardiac cine imaging [21, 22]. CS circumvents previous limits on imaging speed and efficiency, allowing efficient reconstruction of images from incoherently undersampled data by exploiting image sparsity. It can be combined with parallel imaging to exploit joint sparsity from multiple coils for further improved imaging performance [23–30]. Successful implementation of CS requires specific undersampling schemes, such as random undersampling that generate incoherent aliasing artifacts that have the appearance of added noise, and a non-linear reconstruction algorithm to remove incoherent artifacts by enforcing image sparsity. Since its introduction to MRI, a variety of CS techniques have been developed with encouraging imaging performance for a variety of clinical applications.
Some sampling patterns have also been proposed in recent years for continuous data acquisition on a Cartesian grid with golden-angle rotation. These include Radial view ordering (VDRad) [31], Golden angle Cartesian acquisition with Spiral Profile ordering (G-CASPR) [32], GOlden-angle CArtesian Randomized Time-resolved (GOCART) sampling [33], spiral phyllotaxis sampling on a Cartesian grid [34], and comprEssed Sensing PaRtial SubSampling (ESPReSSo) incorporating variable-density Poisson Disc and partial Fourier acquisition [35]. VDRad was recently adopted for pediatric cardiac MRI (ROCK) [22], in which an intravascular contrast agent was used for enhancing blood signal.
Recently, we proposed a novel pseudo-random, variable-density, undersampling strategy called CIRcular Cartesian UnderSampling (CIRCUS) [36], which integrates the desirable features of variable-density randomization and flexible interleaving trajectories on a 3D Cartesian grid and is suitable for CS techniques. The trajectory can be segmented into multiple interleaves, by applying a golden-ratio profile for continuous k-space coverage. Thus, it enables retrospective data sorting without the need to pre-define temporal frames for dynamic imaging applications. Additionally, oversampling of the center of k-space in CIRCUS helps to reduce sensitivity to motion, and also offers the possibility to extract a respiratory motion signal from the acquired data for retrospective respiratory gating. The performance of CIRCUS has previously been demonstrated in both phantom imaging and in vivo studies, including brain imaging, dynamic contrast-enhanced (DCE) wrist MRI, and intracranial 4D flow imaging [36–38]. A high acceleration fator of R = 20 was achieved by combining CIRCUS with a multicoil dynamic compressed sensing reconstruction framework (k–t SPARSE-SENSE) [39, 40] for DCE wrist MRI [37].
In this study, we extend the application of CIRCUS to free-breathing 3D cardiac cine imaging. The method combines the CIRCUS acquisition, retrospective cardiac gating and respiratory self-gating, and k–t SPARSE-SENSE for highly-accelerated self-gated free-breathing 3D cardiac cine imaging. The performance of the proposed technique is compared to conventional breath-hold multi-slice 2D cine imaging for evaluation of image quality and cardiac function quantification.
Materials and methods
CIRCUS Sampling
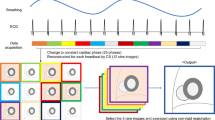
CIRCUS is implemented in the k y –k z plane of a 3D Cartesian acquisition grid [36]. Here, k x is referred to as the frequency encoding axis and k y –k z are referred to as the two phase-encoding dimensions. This sampling strategy features a radial- or spiral-like interleaving scheme, in which sampling points on Cartesian coordinates are selected based on the golden-ratio profile [36, 41]. Such a sampling scheme enables continuous data acquisition for dynamic imaging with flexible image reconstruction of an image series with arbitrary temporal resolution that can be chosen retrospectively. It also allows arbitrary sorting of the acquired data for self-navigation and naturally provides a variable-density random acquisition for the application of compressed sensing methods. The oversampled k-space centers enable extraction of physiological motion signals (e.g., cardiac and respiration) retrospectively from acquired k-space data. As shown in [36], a variety of sampling patterns can be generated with CIRCUS for different applications. In this study, a 3D balanced steady-state free precession (bSSFP) sequence was modified to implement a spiral-like CIRCUS pattern with a smooth transition of readout lines in order to minimize eddy current effects [36]. In addition, partial Fourier was applied in both the readout dimension and phase-encoding dimensions, resulting in 75% partial acquisition along all three encoding directions.
Data acquisition
This study was approved by the Institutional Review Board at our institution and written informed consent was obtained from all subjects before MRI scans. Cardiac imaging was conducted in eight healthy volunteers (4 females, age = 29.9 ± 5.7 years, heart rate = 62.0 ± 8.5 bpm) on a 3.0T MR scanner (750 Wide Bore, GE Medical Systems, Milwaukee, WI, USA) equipped with an 8-channel cardiac coil. For each subject, both conventional breath-hold multi-slice 2D cardiac cine bSSFP imaging and the proposed free-breathing 3D cine imaging with CIRCUS were performed in a short axis orientation in a random order.
Each slice of the 2D cine images was acquired during one breath-hold with prospective electrocardiograph (ECG)-gating with the following imaging parameters: FOV = 34.0 × 25.5 cm2, TR/TE = 4.4/2.0 ms, flip angle = 60o, readout bandwidth = ±125 kHz, slice thickness = 8 mm, image matrix = 224 × 144, number of slices = 12–15, views per segment = 16, and the number of reconstructed cardiac phases = 20. The data acquisition time was 2.7 ± 0.5 min, and the actual total scan time (including resting periods between breath holds) was 9.3 ± 2.4 min.
The imaging parameters for 3D imaging were: FOV = 34.0 × 25.5 cm2, TR/TE = 4.1/1.7 ms, flip angle = 60°, readout bandwidth = ±125 kHz, slice thickness = 4 mm, image matrix = 256 × 144, and number of slices = 30–34. Respiratory bellows was used to track respiratory gating, and both ECG and bellows signals were saved for retrospective gating in image reconstruction. The 3D cine imaging sequence was performed for 2.5 ± 0.3 min with an undersampling factor (R) of 3.0 ± 0.5 (with respiratory gating efficiency of 100%). To test the reproducibility of 3D imaging method, we also acquired a second scan (hereafter referred to as scan #2) for 3.0 ± 0.2 min with R = 2.4 ± 0.6 (with respiratory gating efficiency of 100%).
To demonstrate the feasibility of the proposed free-breathing 3D imaging technique for clinical use, we acquired data from two patients; one with atrial fibrillation and the other with an aortic root aneurysm. We imaged the ventricles in the short axis view (similar imaging setting as the volunteer scans), and also imaged the left atrium and the aorta (with higher through plane resolution <3 mm), in a scan time of under 3 min at 3T.
Image reconstruction
Cardiac gating
The ECG signal, which was saved during data acquisition, was used to synchronize the acquired 3D cine data into a composite cardiac cycle. Specifically, given a user defined number of views per segment, the acquired readout lines were assigned to indices of corresponding cardiac phases based on the relevant ECG trigger. With the CIRCUS acquisition, the number of views per segment can be retrospectively selected and it was set at 10 in this study, resulting in a fixed temporal resolution of 41 ms in all datasets. The number of reconstructed cardiac phases N was chosen as the maximal number contained in at least 85% of the cardiac cycles. Cardiac cycles with a number of cardiac phases 30% more or less than N were rejected. N was 24.0 ± 3.5 in our datasets, varying according to the heart rate of each subject.
Respiratory gating
With variable-density CIRCUS acquisition, the k-space central line along k x (k y = k z = 0) was repeatedly acquired and cross-correlation of corresponding signals from all coils was performed to obtain respiratory motion signal [13]. The principle correlation of that data was derived by applying principle component analysis (PCA), which yielded a 1D signal curve containing motion information within the FOV. Band-pass filtering was applied to remove the higher frequency variation resulting from cardiac motion. The derived motion signal was then used for accepting or rejecting data for image reconstruction based on a histogram of the curve and the given gating efficiency [3]. Similar to most of the current respiratory self-gating approaches, our self-gating method was based on relative motion. We used an acceptance rate (%) rather than absolute displacement to evaluate the gating efficiency. With the interleaving features of CIRCUS acquisition, we could retrospectively choose a series of respiratory gating efficiencies, with each of them associated with different acceleration factors. We compared the images using a series of gating efficiencies (25–100% in increments of 25%) to find a range with reasonable properties in terms of image quality, motion blurring and data undersampling. The motion signal obtained from the bellows was used as a reference for the gating method.
Sparse image reconstruction
The undersampled 3D cine datasets with both cardiac and respiratory gating were then reconstructed using k–t SPARSE-SENSE that exploits joint sparsity along the temporal dimension with a total variation constraint. Image reconstruction (e.g., reconstruction algorithm, selection of regularization parameters) was performed as described before in [39, 40] and was implemented in MATLAB (The MathWorks, Natick, MA, USA) on a Macintosh with Intel Core i7 Processor (3.1 GHz) and 16 GB memory. In this work, a range of regularization parameters were tested in two subjects and then the optimal value was selected by visual assessment of image quality and temporal profile of the reconstructed images. Since the imaging protocol was similar in all subjects, the same regularization parameter was applied for all subsequent reconstruction.
Data analysis
The correlation coefficient and linear regression between external bellows gating signal and the derived respiratory self-gating signals were calculated. The images obtained with 2D cine imaging and 3D cine imaging with different reconstruction schemes were blinded and scored by two cardiac experts with over 15-years experience in cardiovascular MRI, using a 5-point scale: 4-excellent, 3-good, 2-fair, 1-poor, 0-non-diagnostic, in terms of perception of sharpness, contrast and artifact level. Image sharpness was measured on the left ventricle (LV) cavity-myocardium interface, averaged over three locations around the LV and 3 cardiac phases including end-systolic, early-diastolic and end-diastolic phases. The measurement of sharpness is defined as the inverse of the distance between 20 and 80% of the intensity of the curve that crosses the myocardium and LV chamber [42]. This was measured using in-house software developed in MATLAB. In addition, the LV functions, including end-systolic volume (ESV), end-diastolic volume (EDV), and ejection fraction (EF, defined as (EDV − ESV)/EDV × 100) were also measured. OsiriX imaging software (OsiriX Foundation, Geneva, Switzerland) was used to segment the LV chamber semi-automatically based on a region-growing algorithm [43], performed twice by two operators.
Statistical analysis
Image scores, sharpness and functional measurements were compared between 3D image sets reconstructed using respiratory bellows-gating and self-gating, respectively. The same comparisons were also performed between 2D and self-gated 3D imaging methods. Wilcoxon signed rank test was used to compare the qualitative ordinal image scores. Linear regression and Bland–Altman plots of the sharpness and functional measurements were used to evaluate the association and difference between the two gating methods with 3D imaging, as well as that between 2D and 3D self-gated methods. The correlation coefficients, linear fitting slope and offsets, mean bias and confidence variations (±1.96 × SD) were calculated between two measurements. p values were obtained using a two-tailed, paired-sample, t test, with a value <0.05 considered to have statistical significance.
Results
Data was successfully acquired and reconstructed for all subjects.
Figure 1 shows representative cardiac cine sampling patterns using CIRCUS. As scan time progressed, the k y –k z plane was filled with pseudo-random variable-density sampling points and the k-space center lines (k y = k z = 0) were repeatedly sampled. For the representative case shown in Fig. 1, within the whole scan time of 140 s, the averaged time interval between adjacent k-space center line acquisitions was 168.0 ± 51.2 ms, which allowed sufficient tracking of respiratory motion (that typically varies over 2–5 s.)
a k y –k z sampling patterns at every other cardiac phase are displayed at three representative scan times of 20, 50 and 140 s. Sampling patterns combined from all the cardiac phases are also shown as the column on the right. b Sampling patterns with respiratory gating (with different gating efficiencies)
With retrospective cardiac gating, k-space sampling patterns at individual cardiac phases were filled with samples as scan time progressed, as shown in Fig. 1a, where k y –k z sampling patterns at every other cardiac phase are displayed at three representative scan times of 20, 50 and 140 s. Sampling patterns combined from all cardiac phases are also shown in the column on the right. Figure 1b shows the sampling patterns during the entire scan (140 s) with retrospective respiratory gating (with four different gating efficiencies).
The respiratory self-gating signals correlated well with the bellows signals (0.87 ± 0.11 correlation coefficient), and the linear regression between the respiration motion curves (after normalization) had a slope of 1.00 ± 0.17 and a zero offset.
The average image reconstruction time was about 1.5 h for each case. The undersampling factors varied according to the chosen gating efficiencies on the data acquired in 2.5 min: R = 3.0 ± 0.5, 4.0 ± 0.6, 6.0 ± 0.9, and 12.1 ± 1.9 for 100, 75, 50 and 25% gating efficiencies, respectively.
Table 1 summarizes the image scores and LV sharpness. Generally, 3D imaging with a lower gating efficiency provides higher sharpness and image scores. There was no significant difference in image quality scoring between 2D and 3D free breathing imaging with either respiratory bellows or self-gating when gating efficiency is 25%, or between the two gating methods in 3D imaging giving the same gating efficiency (p > 0.05). The scores from two observers were correlated (r = 0.65). The difference in sharpness measurement between two gating methods in 3D imaging was not significant (p > 0.05), but it was significant between 2D and 3D imaging (independent of the gating efficiency). Figure 2 shows that the images acquired with 3D imaging are increasingly blurred when an increased amount of data (with more motion corruption) are used for reconstruction, which is also demonstrated on the quantitative sharpness measurements in Table 1. Respiratory gating efficiencies of 25 and 50% consistently provided reasonable image quality throughout all cases for both bellows and self-gating methods.
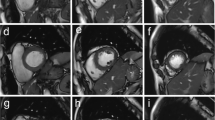
Figure 3 shows the short-axis (SA) images, reformatted in views along the vertical long axis (VLA) and the horizontal long axis (HLA) at peak-systolic and end-diastolic phases, acquired with the conventional 2D breath-hold imaging and the proposed free-breathing 3D imaging methods. The images obtained with 3D imaging methods were comparable to those with 2D imaging without the obvious slice misregistration errors of the 2D data that results from different positions of the multiple breath-holds. The self-gated 3D data provided similar image quality to that obtained with bellows gating. Figure 4 shows reformatted HLA images from four cases representing varying degrees of slice misregistration errors with 2D breath-hold imaging while 3D imaging provides clear alignment along the slice encoding direction. Notice that even for the young healthy subjects in this study, slice misregistration is present.
Images obtained with 2D and 3D imaging methods. Representative end-systolic and -diastolic images of short-axis (SA) (top row), reformatted vertical long axis (VLA) (second row), and horizontal long axis (HLA) (third row) views were obtained with 2D breath-hold (left block), 3D free-breathing with bellows gating (middle block) and 3D free-breathing with self-gating (right block) methods. 3D imaging provides comparable images to those with 2D imaging method, which suffers from slice misregistration errors due to different positions of the multiple breath-holds
Table 2 summarizes the left ventricular functional measurements in eight subjects with 2D and 3D imaging methods (given a respiratory gating efficiency of 25%). The measurements from two observers were highly correlated (r = 0.99). The corresponding Linear Regression and Bland–Altman plots are displayed in Fig. 5, showing that the correlations in LVESV and LVEDV measurements were significant between 2D and 3D self-gating imaging (p < 0.05) and their differences were not significant (p > 0.05). Correlations of the LVEF measurements were not significant likely due to a narrow range of calculated LVEFs (55–68%) in our young healthy subjects, but the differences in the LVEF measurements were not significant (p = 0.45). No significant differences were found for LVESV, LVEDV and LVEF measurements between self-gating and bellows gating, or between two repeated 3D imaging scans.
We successfully applied the developed self-gating free-breathing methods on patients. Figure 6a shows the short-axis view cardiac cine images (as well as reformatted images) from a patient with atrial fibrillation, using our proposed free-breathing 3D imaging acquired in 2.5 min; and Fig. 6b shows images of the left atrium in two orthogonal views, acquired in 2.6 min with a higher through-plane resolution (1.6 × 2.2 × 2.8 mm). In a patient with aortic root aneurysm (Fig. 6c), our proposed free-breathing 3D imaging, which was applied for 3 min with spatial resolution of 1.4 × 1.9 × 2.6 mm, provided a clear delineation of the aortic root (last sub-image) compared to those with the clinically available methods: breath-hold 2D imaging (axial view acquisition with through-plan resolution of 8 mm) and 3D MRA (without ECG gating). Figure 6b, c were acquired about 4–5 min after administration of Gadolinium contrast.
Images acquired on patients using the proposed self-gating free-breathing 3D imaging: a in a short-axis view covering the ventricles in 2.5 min with 1.3 × 1.8 × 5.0 mm; b in a coronal view covering left atrium in 2.6 min with a higher through-plane resolution 1.6 × 2.2 × 2.8 mm; and c in an oblique view covering the aorta in 3 min with 1.4 × 1.9 × 2.6 mm. a, b Were acquired on a patient with atrial fibrillation and c on a patient with aortic root aneurysm. In c the proposed free-breathing 3D imaging provided clear definition of aortic root (last sub-image) compared to those obtained with the clinically available breath-hold 2D imaging (low through-plan resolution) and 3D MRA (no ECG gating) methods
Discussion
In this study, highly-accelerated, self-gated, free-breathing, whole heart 3D cine imaging was successfully achieved using a combination of a CIRCUS acquisition with k–t SPARSE-SENSE. Its performance on healthy volunteers has shown that the proposed method provided comparable results to those acquired with conventional breath-hold 2D cine imaging method in terms of image quality and functional quantification. The proposed 3D imaging approach allowed significantly shorter acquisition times of <3 min (vs ~9.3 min in breath-hold 2D) during free breathing, with higher through-plane spatial resolution (4.0 vs 8.0 mm in 2D) and higher temporal resolution (41 vs 70 ms in 2D).
We imaged the heart in the same short-axis view as in conventional breath-hold 2D cine imaging but in a relatively short scan time of 2–3 min. Recent studies [22, 44, 45] in free-breathing, self-gated 3D cardiac cine imaging based on Cartesian acquisition and compressed sensing reconstruction provided motivation for exploring methods to image the whole heart with isotropic resolution (specifically higher through-plane resolution) and a simpler orientation. That usually requires a longer scan time and could suffer from lower blood signal due to smaller voxel size and reduced inflow effect. Although this can be improved using a much larger flip angle (90°) at 1.5T with an extended scan time (~7 min) [30], or using administration of blood pool contrast agent [22], which showed excellent images with submillimeter isotropic resolution, good SNR, relatively short scan time of 4–6 min and so on. It should be noted that their study benefited greatly from the use of Ferumoxytol that is an intravascular contrast agent with a long half-life time. We were able to achieve higher resolution imaging of the atrium and the aorta at 3T (Fig. 6b, c) in ~2.5 min acquired 4–5 min after administration of Gadolinium contrast.
Only young healthy volunteers were involved in the current study. Thus 2D breath-hold imaging was well tolerated and provided excellent scores (all 4 s), while 3D free-breathing method was challenged by variations in breathing patterns. Although 3D imaging had a slightly higher in-plane encoded spatial resolution (along k x : 1.3 vs 1.5 mm), our results showed that the measured 2D sharpness was better than that with 3D imaging (Table 1), which was likely due to the residual respiratory motion in 3D data compared to the consistent breath-holds from our young, healthy volunteers.
This study has several limitations. While improvements to the proposed methods are feasible, they have not yet been implemented. Here we selected the same gating efficiency (25%) for all the subjects regardless of the specific respiratory pattern for different individuals. The potential exists to utilize self-gating motion signals to derive subject-specific gating efficiencies. Similarly, there is a possibility that motion correction methods could be implemented to provide benefits in either image quality or scan time.
A semi-automated region-growing algorithm was applied for segmenting the left ventricular chamber, which could introduce potential variability related to operator performance. Exploration of reliable and robust automatic segmentation algorithms will be investigated in the future. The 2D CINE imaging benefits from the in-flow effect which provides high blood-to-myocardium contrast. That effect is reduced in 3D imaging, especially toward the apex of the heart [46]. This provides a challenge to conventional segmentation methods that rely on a simple image intensity threshold. Improvements in blood-to-myocardium contrast are possible by employing contrast agents or using variable flip angle acquisition schemes [47]. Current image reconstruction for 3D cine imaging was implemented offline in MATLAB and required more than an hour to complete one data set. In future developments, optimization of the reconstruction algorithms and the possibility of implementing online reconstruction will be explored.
Conclusion
In conclusion, a highly-accelerated, self-gated, free-breathing, 3D cine imaging approach has been demonstrated in this volunteer study. It provides results that are comparable to those obtained with multi-slice 2D breath-hold CINE imaging in terms of qualitative and quantitative image quality measurements and cardiac function assessments. Advantages of this method are ease of prescription and acquisition, reduced demands on patient compliance, and reduced total scan time. Comparisons between bellows-gating and self-gating methods with 3D imaging demonstrated the feasibility of deriving reliable gating signal from the acquired k-space data itself instead of using external bellows. Results from repeated 3D imaging scans further demonstrated reproducibility of the proposed 3D imaging method.
References
Uribe S, Muthurangu V, Boubertakh R, Schaeffter T, Razavi R, Hill DL, Hansen MS (2007) Whole-heart cine MRI using real-time respiratory self-gating. Magn Reson Med 57:606–613
Manka R, Buehrer M, Boesiger P, Fleck E, Kozerke S (2010) Performance of simultaneous cardiac-respiratory self-gated three-dimensional MR imaging of the heart: initial experience. Radiology 255:909–916
Liu J, Spincemaille P, Codella NC, Nguyen TD, Prince MR, Wang Y (2010) Respiratory and cardiac self-gated free-breathing cardiac CINE imaging with multiecho 3D hybrid radial SSFP acquisition. Magn Reson Med 63:1230–1237
Spincemaille P, Liu J, Nguyen T, Prince MR, Wang Y (2011) Z intensity-weighted position self-respiratory gating method for free-breathing 3D cardiac CINE imaging. Magn Reson Imaging 29:861–868
Henningsson M, Chan RH, Goddu B, Goepfert LA, Razavi R, Botnar RM, Schaeffter T, Nezafat R (2013) Contrast-enhanced specific absorption rate-efficient 3D cardiac cine with respiratory-triggered radiofrequency gating. J Magn Reson Imaging 37:986–992
Barkauskas KJ, Rajiah P, Ashwath R, Hamilton JI, Chen Y, Ma D, Wright KL, Gulani V, Griswold MA, Seiberlich N (2014) Quantification of left ventricular functional parameter values using 3D spiral bSSFP and through-time non-Cartesian GRAPPA. J Cardiovasc Magn Reson 16:65
Zhu Y, Liu J, Weinsaft JW, Spincemaille P, Nguyen T, Prince MR, Bao S, Xie Y, Wang Y (2015) Free-breathing 3D imaging of right ventricular structure and function using respiratory and cardiac self-gated cine MRI. BioMed Res Int 2015:9. doi:10.1155/2015/819102
Bhat H, Ge L, Nielles-Vallespin S, Zuehlsdorff S, Li D (2011) 3D radial sampling and 3D affine transform-based respiratory motion correction technique for free-breathing whole-heart coronary MRA with 100% imaging efficiency. Magn Reson Med 65:1269–1277
Moghari MH, Roujol S, Chan RH, Hong SN, Bello N, Henningsson M, Ngo LH, Goddu B, Goepfert L, Kissinger KV, Manning WJ, Nezafat R (2013) Free-breathing 3D cardiac MRI using iterative image-based respiratory motion correction. Magn Reson Med 70:1005–1015
Pang J, Sharif B, Fan Z, Bi X, Arsanjani R, Berman DS, Li D (2014) ECG and navigator-free four-dimensional whole-heart coronary MRA for simultaneous visualization of cardiac anatomy and function. Magn Reson Med 72:1208–1217
Santelli C, Nezafat R, Goddu B, Manning WJ, Smink J, Kozerke S, Peters DC (2011) Respiratory bellows revisited for motion compensation: preliminary experience for cardiovascular MR. Magn Reson Med 65:1097–1102
Leung AO, Paterson I, Thompson RB (2008) Free-breathing cine MRI. Magn Reson Med 60:709–717
Hu P, Hong S, Moghari MH, Goddu B, Goepfert L, Kissinger KV, Hauser TH, Manning WJ, Nezafat R (2011) Motion correction using coil arrays (MOCCA) for free-breathing cardiac cine MRI. Magn Reson Med 66:467–475
Feng L, Axel L, Chandarana H, Block KT, Sodickson DK, Otazo R (2016) XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing. Magn Reson Med 75:775–788
Mascarenhas NB, Muthupillai R, Cheong B, Pereyra M, Flamm SD (2006) Fast 3D cine steady-state free precession imaging with sensitivity encoding for assessment of left ventricular function in a single breath-hold. AJR Am J Roentgenol 187:1235–1239
Liu J, Wieben O, Jung Y, Samsonov AA, Reeder SB, Block WF (2010) Single breathhold cardiac CINE imaging with multi-echo three-dimensional hybrid radial SSFP acquisition. J Magn Reson Imaging 32:434–440
Coppo S, Piccini D, Bonanno G, Chaptinel J, Vincenti G, Feliciano H, van Heeswijk RB, Schwitter J, Stuber M (2015) Free-running 4D whole-heart self-navigated golden angle MRI: initial results. Magn Reson Med 74:1306–1316
Tsao J, Boesiger P, Pruessmann KP (2003) k–t BLAST and k–t SENSE: dynamic MRI with high frame rate exploiting spatiotemporal correlations. Magn Reson Med 50:1031–1042
Kozerke S, Tsao J, Razavi R, Boesiger P (2004) Accelerating cardiac cine 3D imaging using k–t BLAST. Magn Reson Med 52:19–26
Greil GF, Germann S, Kozerke S, Baltes C, Tsao J, Urschitz MS, Seeger A, Tangcharoen T, Bialkowsky A, Miller S, Sieverding L (2008) Assessment of left ventricular volumes and mass with fast 3D cine steady-state free precession k–t space broad-use linear acquisition speed-up technique (k–t BLAST). J Magn Reson Imaging 27:510–515
Wech T, Pickl W, Tran-Gia J, Ritter C, Beer M, Hahn D, Kostler H (2014) Whole-heart cine MRI in a single breath-hold—a compressed sensing accelerated 3D acquisition technique for assessment of cardiac function. Rofo 186:37–41
Han F, Zhou Z, Han E, Gao Y, Nguyen KL, Finn JP, Hu P (2016) Self-gated 4D multiphase, steady-state imaging with contrast enhancement (MUSIC) using rotating cartesian K-space (ROCK): validation in children with congenital heart disease. Magn Reson Med. doi:10.1002/mrm.26376
Donoho DL (2006) Compressed sensing. IEEE Trans Inf Theory 52:1289–1306
Candes EJ, Romberg J, Tao T (2006) Robust uncertainty principles: exact signal reconstruction from highly incomplete frequency information. IEEE Trans Inf Theory 52:489–509
Candes EJ, Tao T (2006) Near-optimal signal recovery from random projections: universal encoding strategies? IEEE Trans Inf Theory 52:5406–5425
Lustig M, Donoho D, Pauly JM (2007) Sparse MRI: the application of compressed sensing for rapid MR imaging. Magn Reson Med 58:1182–1195
Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P (1999) SENSE: sensitivity encoding for fast MRI. Magn Reson Med 42:952–962
Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A (2002) Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 47:1202–1210
Lustig M, Pauly JM (2010) SPIRiT: iterative self-consistent parallel imaging reconstruction from arbitrary k-space. Magn Reson Med 64:457–471
Piccini D, Feng L, Bonanno G, Coppo S, Yerly J, Lim RP, Schwitter J, Sodickson DK, Otazo R, Stuber M (2016) Four-dimensional respiratory motion-resolved whole heart coronary MR angiography. Magn Reson Med. doi:10.1002/mrm.26221
Cheng JY, Zhang T, Ruangwattanapaisarn N, Alley MT, Uecker M, Pauly JM, Lustig M, Vasanawala SS (2015) Free-breathing pediatric MRI with nonrigid motion correction and acceleration. J Magn Reson Imaging 42:407–420
Prieto C, Doneva M, Usman M, Henningsson M, Greil G, Schaeffter T, Botnar RM (2015) Highly efficient respiratory motion compensated free-breathing coronary MRA using golden-step Cartesian acquisition. J Magn Reson Imaging 41:738–746
Zhu Y, Guo Y, Lingala SG, Lebel RM, Law M, Nayak KS (2016) GOCART: GOlden-angle CArtesian randomized time-resolved 3D MRI. Magn Reson Imaging 34:940–950
Piccini D, Littmann A, Nielles-Vallespin S, Zenge MO (2011) Spiral phyllotaxis: the natural way to construct a 3D radial trajectory in MRI. Magn Reson Med 66:1049–1056
Küstner T, Würslin C, Gatidis S, Martirosian P, Nikolaou K, Schwenzer NF, Schick F, Yang B, Schmidt H (2016) MR image reconstruction using a combination of compressed sensing and partial Fourier acquisition: ESPReSSo. IEEE Trans Med Imaging 35:2447–2457
Liu J, Saloner D (2014) Accelerated MRI with CIRcular Cartesian UnderSampling (CIRCUS): a variable density Cartesian sampling strategy for compressed sensing and parallel imaging. Quant Imaging Med Surg 4:57–67
Liu J, Pedoia V, Heilmeier U, Ku E, Su F, Khanna S, Imboden J, Graf J, Link T, Li X (2016) High-temporospatial-resolution dynamic contrast-enhanced (DCE) wrist MRI with variable-density pseudo-random circular Cartesian undersampling (CIRCUS) acquisition: evaluation of perfusion in rheumatoid arthritis patients. NMR Biomed 29:15–23
Liu J, Faraji F, Kefayati S, Haraldsson H, Saloner D (2015) Highly accelerated intracranial 4D flow MRI with CIRcular Cartesian Undersampling (CIRCUS). In: Proceedings of the 23rd Annual Meeting of ISMRM, Toronto, Canada, p 458
Otazo R, Kim D, Axel L, Sodickson DK (2010) Combination of compressed sensing and parallel imaging for highly accelerated first-pass cardiac perfusion MRI. Magn Reson Med 64:767–776
Feng L, Srichai MB, Lim RP, Harrison A, King W, Adluru G, Dibella EV, Sodickson DK, Otazo R, Kim D (2013) Highly accelerated real-time cardiac cine MRI using k–t SPARSE-SENSE. Magn Reson Med 70:64–74
Winkelmann S, Schaeffter T, Koehler T, Eggers H, Doessel O (2007) An optimal radial profile order based on the Golden Ratio for time-resolved MRI. IEEE Trans Med Imaging 26:68–76
Larson AC, Kellman P, Arai A, Hirsch GA, McVeigh E, Li D, Simonetti OP (2005) Preliminary investigation of respiratory self-gating for free-breathing segmented cine MRI. Magn Reson Med 53:159–168
Rosset A, Spadola L, Ratib O (2004) OsiriX: an open-source software for navigating in multidimensional DICOM images. J Digit Imaging 17:205–216
Wetzl J, Schmidt M, Zenge MO, Lugauer F, Lazar L, Nadar M, Maier A, Hornegger J, Forman C (2015) Isotropic 3-D CINE imaging with Sub-2 mm resolution in a single breath-hold. In: Proceedings of the 24th Annual Meeting of ISMRM, Singapore, May 2016, p 1011
Usman M, Cruz G, Prieto C (2016) Highly-efficient free breathing whole heart CINE MRI with self gated 3D CASPR-TIGER trajectory. In: Proceedings of the 24th Annual Meeting of ISMRM, Singapore, May 2016, p 1814
Nezafat R, Herzka D, Stehning C, Peters DC, Nehrke K, Manning WJ (2008) Inflow quantification in three-dimensional cardiovascular MR imaging. J Magn Reson Imaging 28:1273–1279
Srinivasan S, Ennis DB (2014) Variable flip angle balanced steady-state free precession for lower SAR or higher contrast cardiac cine imaging. Magn Reson Med 71:1035–1043
Acknowledgements
This work was supported in part by Grant from the NIH K25EB014914 (JL), NIH R56HL133663 (JL), GE Healthcare (JL), NIH R01NS059944 (DS), R01HL114118 (DS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This study was conducted under IRB approvals (#12-09781, #14-14769) at University of California San Francisco.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Liu, J., Feng, L., Shen, HW. et al. Highly-accelerated self-gated free-breathing 3D cardiac cine MRI: validation in assessment of left ventricular function. Magn Reson Mater Phy 30, 337–346 (2017). https://doi.org/10.1007/s10334-017-0607-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10334-017-0607-2