Abstract
Berkeleyomyces rouxiae, a causal agent of black root rot of lettuce, is morphologically and physiologically indistinguishable from the closely related species B. basicola. PCR primers specific for these species were designed based on the nucleotide sequence of genes specific to the species. The primer pair specific to B. rouxiae did not amplify DNA of other plant pathogens that invade root or stem base of lettuce and was useful for diagnosis and quantitating the pathogen from lettuce root tissue.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
When black root rot of lettuce (Lactuca sativa) was reported for the first time in Japan by our laboratory group (Nakane et al. 2018, 2019b), the causal agent of this disease was initially described as Thielaviopsis basicola (Nakane et al. 2018). However, further investigation indicated that T. basicola belongs to a distinct genetic lineage from the genus Thielaviopsis (type species: T. ethacetica) and other related genera (de Beer et al. 2014; Nel et al. 2018). Moreover, isolates of T. basicola comprise two distinct lineages, so T. basicola was reclassified into two species of a new genus, as Berkeleyomyces basicola and B. rouxiae (Nel et al. 2018). Japanese isolates of lettuce black root rot pathogen were reidentified as B. rouxiae (Nakane et al. 2019b).
The two species of Berkeleyomyces are indistinguishable in terms of their morphological and physiological characteristics, but they differ in the nucleotide sequences for the genes for the ribosomal large subunit (LSU), the 60 s ribosomal protein RPL10 (60S), the internal transcribed spacer region (ITS), and the minichromosome maintenance complex component 7 (MCM7) differ between the species (Nel et al. 2018). Currently, these sequences have provided the most effective means for distinguishing the two species. Although B. basicola has not been found to date in Japan, its distribution in Japan and its pathogenicity on lettuce and other plant species globally remain unclear. Tools for reliable and rapid identification of Berkeleyomyces species should facilitate resolving such issues.
Roots of lettuce plants in fields are often invaded simultaneously by B. rouxiae and other pathogens such as Fusarium oxysporum f. sp. lactucae and root lesion nematode. Furthermore, black root rot symptoms are often similar to the disease caused by these pathogens (Nakane et al. 2019b, c). A molecular tool to identify and specifically detect Berkeleyomyces species is expected to be helpful for diagnosing black root rot disease. Therefore, after designing PCR primers from the nucleotide sequences of the respective Berkeleyomyces species, we investigated their potential for species identification, disease diagnostics, and quantitative detection of the pathogen.
Materials and methods
Species-specific PCR primers
Sequences of 60S and MCM7 downloaded from DDBJ/EMBL/GenBank databases were compared using ClustalW ver. 2.1 (Larkin et al. 2007) via the website of the DNA Data Bank of Japan (https://www.ddbj.nig.ac.jp/). Primers presented in Table 1 were designed from nucleotide sequences specific to each species. Fungal genomic DNA was extracted from a mycelial disk (5 mm diameter) from a 7-day-old culture on potato dextrose agar (PDA: BD, Franklin Lakes, NJ, USA) in 500 μl of DNA extraction buffer (0.5 M NaCl, 10 mM Tris–HCl pH 7.5, 10 mM EDTA, 1% [w/v] SDS), 250 μl of TE-saturated phenol, 250 μl of chloroform using glass beads in a 2 ml microtube. After bead beating (4800 rpm for 30 s) and centrifugation (6000×g for 5 min), DNA in the aqueous phase was precipitated using ethanol. The 20-μl PCR mixture contained 10 ng of genomic DNA of B. basicola CBS 430.74 (from the CBS-KNAW collections) or B. rouxiae LT1 (deposited at the National Agriculture and Food Research Organization Genetic Resources Center in Japan as a strain MAFF 246781), 1 μM of each primer, and 10 μl of Premix Ex Taq Hot Start Version (Takara Bio, Kusatsu, Japan). The PCR was run in a TaKaRa PCR Thermal Cycler Dice Gradient (Takara Bio) at 95 °C for 5 min; (95 °C for 30 s, 60 or 50 °C for 30 s, and 72 °C for 30 s) × 35 cycles; and 72 °C for 5 min. Annealing temperatures for the respective primer pairs are presented in Table 1.
Pathogen specificity of the primers
The PCR using primers roux60s_F and roux60s_R described above were then tested against B. rouxiae LT1 and several plant pathogens that invade the roots or stem base of lettuce were used: Pratylenchus penetrans MAFF 108086, F. oxysporum f. sp. lactucae (race 1) MAFF 244120, Verticillium dahliae 09100-1 (Usami et al. 2012), Plectosphaerella pauciseptata NBRC 109008, Pythium irregulare L1 (isolated from lettuce for this work), Py. aphanidermatum C1 (isolated from cucumber for this work), and Globisporangium ultimum MAFF 241945. Genomic DNA was extracted from the fungal pathogens as described above. For Pr. penetrans, DNA was extracted from approximately 5000 nematodes provided by the National Agriculture and Food Research Organization (NARO) Genetic Resources Center. The control PCR used primers for the internal transcribed spacer region of ribosomal DNA (rDNA-ITS). Primers ITS5 and ITS4 (White et al. 1990) were used for fungal pathogens and the primers reported by Ferris et al. (1993) were used for nematodes in the control PCR. The PCR conditions were identical to those described in the respective reports from earlier studies.
Assays using root samples from lettuce fields
Root samples of four lettuce plants (cv. Universe, susceptible to black root rot) were collected from a commercial lettuce field (field A) in Ibaraki Prefecture (Japan) in October 2018. Root samples of eight lettuce plants (cv. Blurush, resistant to black root rot) were collected from a commercial lettuce field (field B) in Ibaraki Prefecture (Japan) in September 2019. Root discoloration (including black root rot-like symptom) was observed on each sample collected from both lettuce fields. Stunting of lettuce plants was observed only in field A. Discolored tissues from lettuce roots were sampled for pathogen isolation as described by Nakane et al. (2019b). DNA was extracted using an ISOSPIN Plant DNA (Nippon Gene, Tokyo, Japan) from 50 mg of root tissue homogenized with a pestle (BioMasher II; Nippi, Tokyo, Japan) and used for the PCR with primers roux60s_F and roux60s_R as described above.
Quantitative detection of the pathogen
Lettuce plants (cv. Summer guy) were inoculated with B. rouxiae as described by Nakane et al. (2019b). Inoculated and noninoculated plants were maintained in a growth chamber (25 °C, 12-h photoperiod) for up to 4 weeks until roots had various symptom severities. Roots were then washed in running tap water for 1 h and symptom severity rated on a scale of 1–3, where 1 = slight, 2 = moderate, and 3 = severe. Symptom severity of noninoculated roots was rated as 0. Four root samples per severity rating were tested in a real-time PCR assay by extracting DNA from 50 mg of homogenized root tissue using ISOSPIN Plant DNA. The 25 μl PCR reaction mixture included 10 ng of DNA, 0.4 μM of each primer (roux60s_F and roux60s_R), and 12.5 μl of TB Green Premix Ex Taq II (Takara Bio). The PCR was done using a Thermal Cycler Dice Real Time System III (Takara Bio) at 95 °C for 30 s and (95 °C for 5 s, 60 °C for 30 s) × 35 cycles. The mass of B. rouxiae DNA was estimated using the standard curve method. Statistical analysis and box plot construction were performed using EZR ver. 1.32 (Kanda 2013; Saitama Medical Center, Jichi Medical University; https://www.jichi.ac.jp/saitama-sct/SaitamaHP.files/statmedEN.html).
Results and discussion
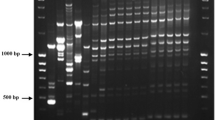
In the PCR assay using primers designed for this study (Table 1), amplicons of 88 bp and 331 bp were amplified, respectively, by primers basi60s (F and R) and basiMCM7 (F and R) specifically for B. basicola (Fig. 1). PCR products of the same size as those of B. basicola were amplified specifically for B. rouxiae by primers roux60s (F and R) and rouxMCM7 (F and R). Thus, the primers were useful for differentiating the two Berkeleyomyces species. Additionally, 19 isolates of B. rouxiae obtained from various plants in Japan were identified successfully using these primer pairs (data not shown).
PCR assay results obtained for Berkeleyomyces basicola (lanes 1, 3, 5, and 7) and B. rouxiae (lanes 2, 4, 6, and 8) using primers specific to the respective species. Primer pairs basi60s_F and basi60s_R (lanes 1 and 2) and basiMCM7_F and basiMCM7_R (lanes 5 and 6) are specific to B. basicola. Primer pairs roux60s_F and roux60s_R (lanes 3 and 4) and rouxMCM7_F and rouxMCM7_R (lanes 7 and 8) are specific to B. rouxiae. M: DNA size marker 100 bp ladder
All previously reported Japanese isolates of black root rot pathogen of lettuce are B. rouxiae (Nakane et al. 2019a, b). Therefore, the potential of primer pair roux60s_F and roux60s_R (specific to B. rouxiae) for disease diagnosis was investigated. For the PCR assay, B. rouxiae LT1 and several plant pathogens known to invade the root or stem base of lettuce were used. Although amplification of rDNA-ITS was observed in all pathogens (Fig. 2, lanes 1–8), primers roux60s_F and roux60s_R produced amplicons only for the black root rot pathogen B. rouxiae (Fig. 2, lanes 9–16). The PCR assay was applied to lettuce samples from naturally infested commercial fields. For samples from field A, PCR products of 88 bp were amplified in 20 of 24 samples (lanes 1–24 except for lanes 7, 9, 15, and 20 in Fig. 3), whereas B. rouxiae was isolated from 4 of 48 root tissue samples on water agar plates. Because we could not isolate the pathogen from samples immediately after collection, sample degradation and contamination lowered the frequency of B. rouxiae isolation on agar plates. However, the frequency of pathogen detection by PCR was very high. B. rouxiae was not isolated from any of the 54 samples of lettuce root tissues from field B. Additionally, no PCR product was amplified from any of the 18 samples tested (lanes 25–42 in Fig. 3). Apparently, the root discoloration on lettuce plants in field B was not a symptom caused by B. rouxiae although lettuce plants in field A were infected with B. rouxiae. Lettuce plants cultivated in field B were of a cultivar that is resistant to black root rot. These results are consistent with the results of pathogen isolation and PCR assay.
PCR assay results of plant pathogens that invade the roots or stem base of lettuce. Universal primers for each pathogen amplifying ITS1-5.8S-ITS2 region were used in lanes 1–8. Primers roux60s_F and roux60s_R specific to Berkeleyomyces rouxiae were used in lanes 9–16. Lanes: 1 and 9, B. rouxiae; 2 and 10, Pratylenchus penetrans; 3 and 11, Fusarium oxysporum f. sp. lactucae; 4 and 12, Verticillium dahliae; 5 and 13, Plectosphaerella pauciseptata; 6 and 14, Pythium irregulare; 7 and 15, Pythium aphanidermatum; 8 and 16, Globisporangium uncinulatum; M, DNA size marker 100 bp ladder
PCR assay results of lettuce roots from naturally infested fields. Primers roux60s_F and roux60s_R specific to Berkeleyomyces rouxiae were used. Lanes: 1–24, root samples from field A (B. rouxiae was isolated); 25–42, root samples from field B (B. rouxiae was not isolated); 43, genomic DNA of B. rouxiae; M, DNA size marker 100 bp ladder
Subsequently, quantitative detection of B. rouxiae from infected lettuce roots was performed with real-time PCR using primers roux60s_F and roux60s_R. The relative mass of B. rouxiae DNA (a maximum amount = 1) detected from each root sample is presented in Fig. 4. Typical symptoms on lettuce roots are presented in Fig. 5. Results show that more DNA of B. rouxiae was detected from root tissues of plants with more severe symptoms. Thus, the species-specific PCR primers can be used to quantify the black root rot pathogen from diseased lettuce roots.
Relative amounts of DNA of Berkeleyomyces rouxiae in lettuce roots with different symptom severities (0–3, portrayed in Fig. 5) were estimated using real-time PCR with primers roux60s_F and roux60s_R. Maximum amount = 1. DNA from four root samples per symptom severity was used
In Japan, black root rot of various plant species other than lettuce has also been reported (Horita and Ohgami 2015; Kobayashi and Kotani 1987; Nishikawa 2007). Causal agents of these diseases were reported initially as T. basicola or Chalara elegans (both are synonyms of B. basicola and B. rouxiae). On the basis of results using the microorganism search system in NARO Genebank, some of those isolates were reidentified as B. rouxiae (https://www.gene.affrc.go.jp/databases-micro_search.php). Therefore, primer pair roux60s_F and roux60s_R might be useful for the diagnosis of black root rot of other plant species in addition to lettuce. It is particularly interesting that all previously identified isolates of black root rot pathogen of plants (including lettuce and other species) in Japan were B. rouxiae. Because B. basicola has not been found to date in Japan, we need to test continually for its presence, so the four pairs of PCR primers designed here (Table 1) are expected to be useful for this monitoring. Although Berkeleyomyces species are distinguishable only by DNA sequences, these primers enable us to identify the causal agent of black root rot of plants confidently. In the present study, we showed that the primer pair specific to B. rouxiae is usable for real-time PCR. Quantitative detection of the pathogen using real-time PCR is expected to be useful for evaluating the resistance and tolerance of lettuce cultivars, which vary greatly in tolerance and susceptibility to black rot (Nakane et al. 2019a, c). Because B. basicola has not yet caused disease in Japan, we did not test plant tissues using the PCR and primers designed here, but trials to detect Berkeleyomyces species from soil samples will be done in the future.
References
de Beer ZW, Duong TA, Barnes I, Wingfield BD, Wingfield MJ (2014) Redefining Ceratocystis and allied genera. Stud Mycol 79:187–219
Ferris VR, Ferris JM, Faghihi J (1993) Variation in spacer ribosomal DNA in some cyst-forming species of plant parasitic nematodes. Fundam Appl Nematol 16:177–184
Horita H, Ohgami D (2015) Vegetable and ornamental crop diseases caused by Thielaviopsis species in Hokkaido (in Japanese). Hokunou 82:385–391
Kanda Y (2013) Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant 48:452–458
Kobayashi T, Kotani S (1987) Occurrence of black root rot of okura and its control (in Japanese). Proc Assoc Pl Protec Shikoku 22:47–55
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Nakane R, Miki S, Ikeda K, Sakai H, Usami T (2018) Black root rot of lettuce caused by Thielaviopsis basicola (in Japanese with English abstract). Jpn J Phytopathol 84:207
Nakane R, Ishiyama Y, Sasaki D, Usami T (2019a) Management of black root rot of lettuce in Japan using resistant cultivars (in Japanese with English abstract). Jpn J Phytopathol 85:253–254
Nakane R, Miki S, Ikeda K, Sakai H, Hayashi K, Usami T (2019b) First report of black root rot of lettuce in Japan caused by Berkeleyomyces rouxiae. J Gen Plant Pathol 85:436–439
Nakane R, Usami T, Miki S (2019c) Occurrence of black root rot of lettuce caused by Berkeleyomyces rouxiae in Japan (in Japanese). Plant Prot 73:425–428
Nel WJ, Duong TA, Wingfield BD, Wingfield MJ, de Beer ZW (2018) A new genus and species for the globally important, multihost root pathogen Thielaviopsis basicola. Plant Pathol 67:871–882
Nishikawa J (2007) Black root rot on pansy geranium caused by Thielaviopsis basicola in Japan (in Japanese with English abstract). Jpn J Phytopathol 73:309–310
Usami T, Itoh M, Morii S, Miyamoto T, Kaneda M, Ogawara T, Amemiya Y (2012) Involvement of two different types of Verticillium dahliae in lettuce wilt in Ibaraki Prefecture, Japan. J Gen Plant Pathol 78:348–352
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Acknowledgements
We thank Mr. Syuhei Fuda, Ms. Kanako Hayashi, Mr. Motonori Takagi, Dr. Takuya Miyamoto, and Mr. Takashi Ogawara (Ibaraki Agricultural Center) for providing experimental materials. This work was supported by JSPS KAKENHI Grant Number JP17H03955.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights statement
This article describes no study with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nakane, R., Usami, T. PCR primers to identify and detect Berkeleyomyces rouxiae, a causal agent of black root rot of lettuce. J Gen Plant Pathol 86, 335–339 (2020). https://doi.org/10.1007/s10327-020-00933-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10327-020-00933-3