Abstract
The dahlia isolate of Potato spindle tuber viroid (PSTVd-D) shares 97 % sequence homology with PSTVd-intermediate (PSTVd-I), but differs at eight positions in the nucleotide sequence from PSTVd-I: five substitutions at positions 42, 43, 127, 202, and 311, two insertions at 63/64 and 312/313, and one deletion at 119. PSTVd-D accumulates slowly and induces very mild symptoms in tomato (cv. Rutgers) plants. In contrast, PSTVd-I propagates faster and induces severe symptoms. Here we used deep-sequencing analysis of PSTVd-specific small RNAs (PSTVd-sRNA) that accumulate in PSTVd-I- and PSTVd-D-infected tomato plants to reveal that the number of PSTVd-sRNA reads extensively decreased in PSTVd-D-infected leaf and stem tissues, especially those derived from the regions containing the nucleotides 119, 127, and 202, in which the nucleotide sequence differed between the severe and mild symptom-inducing isolates. In comparison with healthy controls, relative expression levels (i.e., number of reads by deep sequencing) of various host microRNAs changed after infection with PSTVd-I and PSTVd-D. The relative abundance of miR159 and miR162 in PSTVd-I- and PSTVd-D-infected leaf and stem tissues decreased to nearly 50 % of that in healthy tissues. In PSTVd-I- and PSTVd-D-infected stem tissues, miR319, which is approximately five times more abundant in stem tissues than in leaves, also decreased to 33–63 % of that in healthy controls.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Viroids are single-stranded, circular RNA molecules and the smallest known plant pathogens, ranging from 246 to 401 nucleotides (nt) long (Diener 1987). The replication of these molecules is completely reliant on the host transcription machinery (Darós and Flores 2004), and their accumulation in host cells causes various degrees of symptoms from asymptomatic to mild to severe. To date, more than 32 species of viroids, classified into eight genera of two families, have been reported (Flores et al. 2005).
Potato spindle tuber viroid (PSTVd) is a species of the Pospiviroidae family that infects a wide host range mainly in the Solanaceae and Asteraceae (or Compositae) families, resulting in various degrees of systemic dwarfing and leaf curling in highly sensitive host species, but not in tolerant species (Singh, 1973). Recently, a new PSTVd variant was isolated from dahlia (Dahlia × hybrida) (Tsushima et al. 2011). As compared with isolate PSTVd-I (Intermediate) (accession no: M16826), PSTVd-D (Dahlia) (accession no: AB623143) asymptomatically infects dahlia but produces mild symptoms in tomato plants (Solanum lycopersicum cv. Rutgers). PSTVd-D and -I share the same central conserved region (CCR) and have over 97 % sequence homology. However, the nucleotide sequence of PSTVd-D differs from PSTVd-I at eight positions: five substitutions at nucleotide positions 42, 43, 127, 202, and 311, two insertions at 63/64 and 312/313, and one deletion at 119.
Viroids are strong inducers of RNA silencing, and a large amount of viroid small RNAs (sRNAs) accumulates in viroid-infected plants. Viroid-induced RNA silencing targets viroid replication; however, replication can continue against RNA silencing, leading to the development of disease symptoms. Viroid sRNAs, ranging in size from 21 to 24 nucleotides, are derived from the entire molecule of genomic (positive) and antigenomic (negative) strands; however, they are not produced evenly, but rather known to be generated from some hotspot regions (Itaya et al. 2007; Machida et al. 2007). Wang et al. (2004) suggested that viroid-induced RNA silencing is involved in viroid pathogenicity, and further support for this hypothesis was provided by studies of peach “calico” disease in which mRNA encoding the chloroplastic heat shock protein 90 can be targeted by specific homologous sRNA induced by infection of a specific variant of Peach latent mosaic viroid (Navarro et al. 2012). However, major components of the molecular mechanisms of symptom expression of morphological abnormalities such as dwarfism, leaf curling, and fruit malformation and discoloration, which are commonly caused by viroid infection, remain unknown. Here we performed multiplex deep sequencing analysis of viroid-specific sRNAs and microRNAs (miRNAs) that accumulate in tomato plants after infection with severe and mild symptom-inducing PSTVd isolates to discern the relevance between pathogenicity and viroid-induced RNA silencing.
Materials and Methods
Inoculum sources of PSTVd-I and PSTVd-D
Plasmid DNA containing infectious cDNA clones of either PSTVd-I (pTZ18R-Rz6-PSTV; accession no. M16826) or PSTVd-D (pBS-PSTVd-Dahlia-4S-14D; accession no. AB623143) were used to prepare in vitro transcripts. The plasmid DNA (~ 2 μg) was digested at 37 °C overnight with the NotI restriction enzyme (Takara Bio, Otsu, Shiga, Japan), cleaving downstream from the 3′-end of the cDNA insert. After incubation, the linearized plasmid DNA was extracted by phenol–chloroform, recovered by ethanol precipitation and dissolved in distilled water for in vitro transcription reaction. Transcription was performed at 37 °C for 2 h with T7 RNA polymerase (Invitrogen, Carlsbad, CA, USA) in a 20-μL reaction mixture according to the manufacturer’s instruction. After transcription, the reaction was stopped by adding 2 μL 0.1 M EDTA (pH 7.0), 2.5 μL 4 M LiCl, and 75 μL 99.5 % ethanol, and stored for 1 h at –30 °C. The RNA transcripts were recovered by ethanol precipitation, dissolved in 50 μL distilled water, and quantified by UV spectrophotometry. Inocula were prepared at concentrations of 100 ng RNA transcripts/μL in 50 mM sodium phosphate buffer (pH 7.5), 1 mg/mL bentonite. A leaf on 5 tomato seedlings at the first true-leaf stage was dusted with carborundum (600 mesh), then a 10-μL sample of inoculum was placed on the leaf and gently rubbed against the leaf 20 times using a sterile glass rod. Plants were then rinsed immediately with tap water and incubated in a growth chamber controlled at 25 °C, with a 16-h day and high light intensity (fluorescent, 40 W × 4).
Infection assay
Four to six weeks later, when plant showed symptoms, symptomatic leaves were harvested to extract low molecular weight RNA (LMW-RNA) (Sano et al. 2004).
Before the start of the infection assay, samples of LMW-RNA (~ 100 ng/μL) extracted from tomato plants infected with PSTVd-I or PSTVd-D were used for reverse transcription (RT)–polymerase chain reaction (PCR), followed by cloning and sequencing. RT was performed for 60 min at 37 °C with a random hexamer primer (final conc. 0.5 μM) and M-MuLV reverse transcriptase (Invitrogen) in a reaction mixture with a final volume of 25 μL, which included 2 μL of RT solution containing 20 pmol each of PSTVd-specific primer set: i.e., PS88 M (5′-CCCTGAAGCGCTCCTCCGAG-3′) and PS89P (5′-ATCCCCGGGGAAACCTGGAGCGAAC-3′) (Levy et al. 1994; Weidemann and Buchta 1998). Double-stranded cDNAs amplified by RT-PCR were ligated directly into pGEM-T vector (Promega, Madison, WI, USA) and used to transform Escherichia coli DH5α Competent Cells (Takara Bio). Ten cDNA clones were selected for sequencing and confirming that both PSTVd-I and -D replicated stably in tomato plants. Sequencing was performed by ABI 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) or by the Solgent Co. (Seoul, Korea).
The concentration of PSTVd was quantified by a dot-blot hybridization using a digoxigenin (DIG)-labeled PSTVd-D cRNA probe, the the final PSTVd (-I or -D) inoculum was prepared at ca. 5 µg/mL in 50 mM sodium phosphate buffer (pH 7.5) containing 1 mg/mL bentonite. Five tomato seedlings at the first true-leaf stage were inoculated with 20 μL of the inoculum per plant and incubated in a growth chamber controlled at 25 °C with a 16-h day.
Northern blot analysis
Samples of leaves or stems (~ 1 g) were homogenized in 5 mL of 2 × CTAB buffer (10 mL 1 M Tris–HCl [pH 9.5], 46.6 mL 3 M NaCl, 10 mL 0.2 M disodium EDTA [pH 7.0], 2 g CTAB powder, 0.5 mL 2-mercaptoethanol, and 28.4 mL distilled water; final volume 100 mL). Total nucleic acids were extracted once with an equal volume of phenol–chloroform (1:1), precipitated with ethanol, and dissolved in 100–400 μL distilled water. LMW-RNA soluble in 2 M LiCl was further fractionated with 2 M LiCl and RNase-free DNase I (RQ1 DNase; Promega) treatments, and dissolved in 50–100 μL distilled water. LMW-RNA (ca. 100 ng) was denatured (Sano et al. 2004) and fractionated by electrophoresis in 1.5 % (w/v) agarose gels (AGE, containing formaldehyde at a final concentration 0.66 M) in 1 × MOPS buffer. RNA was then transferred to nylon membrane (Biodyne; Pall Corp., Port Washington, NY, USA) and hybridized with DIG-labeled PSTVd-specific probe (Tsushima et al. 2011). The hybridized signals were visualized using the Chemidoc-XRS (Bio-Rad Laboratories, Hercules, CA, USA) imaging system and quantified using the Quantity One (version 4.6.2) software package.
Small RNA preparation and deep sequencing analysis
Samples of leaves and stems (~ 1 g) were separately collected from symptomatic tomato plants infected with PSTVd-I and PSTVd-D at 4 weeks post inoculation (wpi). Samples were frozen in liquid nitrogen and crushed into fine powders by sterilized motor and pestle, and RNA was extracted by TRI Reagent (Molecular Research Center, Cincinnati OH, USA) according to the manufacturer’s instruction. A sample (200 μg/100 μL) of LMW-RNA soluble in 2 M LiCl was prepared and mixed with 0.9 volume 50 % (w/v) urea solution and 0.1 volume marker dye solution [0.02 % (w/v) bromophenol blue, 0.02 % (w/v) xylene cyanol, 50 % (v/v) glycerol], and heat-denatured at 68 °C for 15 min. RNA was fractioned by 12 % (w/v) polyacrylamide gel (PAGE, acrylamide–bisacrylamide = 19:1) containing 1 × Tris–borate-EDTA (TBE) buffer, 8 M urea. After electrophoresis, RNA was briefly (~ 1 h) blotted to nylon membrane (Biodyne Plus) by contact with the gel surface. The membrane was hybridized with DIG-PSTVd cRNA probe as described already and the accumulation of PSTVd-specific small RNA (PSTVd-sRNA) confirmed. By referring to the RNA size maker, a gel slice containing small RNAs with the size 15–35 nucleotides was selected and excised, crushed into small pieces, added to 1 mL RNA gel elution buffer (0.5 M ammonium acetate, 1 mM EDTA [pH 8.0], 0.1 % (w/v) SDS), then shaken gently overnight at 37 °C. Gel debris was removed by passage through a TAKARA suprec-01 spin column (Takara Bio) according to the manufacturer’s instruction and small RNAs were recovered by ethanol precipitation, air-dried, and dissolved in 10 μL RNase-free distilled water.
The amount of small RNA was quantified by UV spectrophotometry and ~ 1 μg was sent to Hokkaido System Science Co. (Sapporo, Japan) for deep sequencing analysis by Genetic Analyzer IIx (Illumina, San Diego, CA, USA). Samples were quantified using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), and processed simultaneously in the Illumina system using an index-sequencing or multiplex strategy. Indexes were CGATGT for healthy leaf, TGACCA for healthy stem, ACAGTG for PSTVd-D-infected leaf, GCCAAT for PSTVd-D-infected stem, CAGATC for PSTVd-I-infected leaf, and CTTGTA for PSTVd-I-infected stem.
Adapter sequences were removed from the ends of the resulting raw short-read data based on the presence of an exact 10-nt match with the termini of the respective adapters, and identical short reads were grouped according to read size (15–37 nt). In this way, adapter-trimmed short-read data were converted to a nonredundant “short-read-sequence occurrence” format. These nonredundant data were then mapped to either the genomic or anti-genomic strand of PSTVd-D and PSTVd-I using hssmap, a specially written C language program that processes data from circular molecules by adding an appropriate extension to the 3′-terminus of a linear reference sequence.
MicroRNAs were analyzed using CLC Genomics Workbench (http://www.clcbio.com/products/clc-genomics-workbench/) version 6.5.1, which contains a databank of the latest miRBase (Release 20), according to the manufacturer’s instruction. Information on a list of sequences that includes precursor miRNAs together with annotations for mature regions in microRNAs was downloaded from miRBase (http://www.mirbase.org/).
Results
Symptoms and systemic accumulation of severe and mild symptom-inducing isolates of PSTVd
Five tomato seedlings were inoculated with approximately 5 µg/mL PSTVd (-I or -D) and then incubated for 6 weeks. Plants infected with PSTVd-I exhibited primary symptoms of leaf curling at approximately 3 wpi and were stunted by 4 wpi.
In contrast, those infected with PSTVd-D appeared normal at 4 wpi with only mild leaf curling. Symptom severity contrasted greatly between plants infected with PSTVd-D and PSTVd-I; those with PSTVd-I were notably stunted (Fig. 1a).
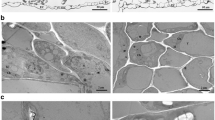
Symptoms caused by Potato spindle tuber viroid (PSTVd) and PSTVd-sRNA and their relative concentrations in PSTVd-infected Rutgers tomato. a Diseased plant at 4 wpi and healthy control. PSTVd-I caused epinasty and severe stunting (middle), but PSTVd-D caused only mild epinasty in the upper portion. b Samples (100 ng) of LMW-RNA were fractionated by denaturing AGE and hybridized with DIG-labeled PSTVd-cRNA probe. Five plants (1–5) infected with PSTVd-I started to show positive signals at 3 wpi, but PSTVd-D caused only a faint positive at 4 wpi. c Samples (2 μg) of LMW-RNA were fractioned by denaturing PAGE (upper panel) and hybridized with a DIG-labeled PSTVd-cRNA probe (lower panel). Lanes 1; healthy leaf, 2; PSTVd-D leaf, 3; PSTVd-I leaf, 4; healthy stem, 5; PSTVd-D stem, and 6; PSTVd-I stem. Arrow indicates the location of PSTVd-sRNA. Note the presence of strong positive signals in PSTVd-I-infected samples (lanes 3 and 6) and weak signals in PSTVd-D-infected samples (lanes 2 and 5)
Northern blot analyses of weekly viroid titers from 2 to 5 wpi showed that PSTVd-I reached detectable levels at approximately 3 wpi, whereas PSTVd-D was undetectable until approximately 4–5 wpi. Specifically, PSTVd-I began to accumulate approximately 1 week before PSTVd-D, consistent with the development and severity of symptoms (Fig. 1b). Collectively, these results indicate that the mild symptoms in tomato plants induced by PSTVd-D infection were attributable to the reduced accumulation, influenced by differences in their nucleotide sequences (Fig. 2).
Comparison of computer-predicted secondary structures of Potato spindle tuber viroid (PSTVd)-I (a) and -D (b). Broken lines divide the five structural regions (from left to right, TL terminal left, P pathogenicity, C central, V variable, TR terminal right). Central conserved region (CCR) is boxed. The positions of nucleotide changes found in PSTVd-D, i.e., five substitutions, three insertions and one deletion, are shown by arrows. Abbreviation, for example, C42U means the C nucleotide at the position 42 in PSTVd-I changed to U in PSTVd-D
Accumulation of PSTVd sRNA in PSTVd-I- and -D-infected tomato plants
Since viroid-specific sRNA are believed to play key roles in viroid pathogenicity through RNA silencing (Navarro et al. 2012; Wang et al. 2004), multiplex deep sequencing analysis was performed to assess the accumulation of PSTVd sRNA in tomato leaf and stem tissues infected with PSTVd-I and -D at 4 wpi.
Approximately 5.5 to 6 million reads of sRNA, ranging from 15 to 43 nt long, were obtained from leaf and stem samples of healthy plants and those infected with PSTVd-I or PSTVd-D. Thus, almost the same number of sRNA reads was obtained from each sample by the analysis. Further analysis of the raw data revealed 489,133 reads from PSTVd sRNA in PSTVd-I-infected leaves, 673,834 in PSTVd-I-infected stems, 105,757 in PSTVd-D-infected leaves, and 522,648 in PSTVd-D-infected stems (Table 1). These results were consistent with the density of hybridization signals of PSTVd sRNA detected by northern blot analysis (Fig. 1c). The analysis was repeated under redundant conditions allowing 1 mismatch; however, only the data obtained under stringent conditions are presented here because there were no significant differences between them, i.e., PSTVd-small RNAs contained those with a mismatch, but present only in a minor fraction.
The quantity of PSTVd sRNA was obviously lower in leaves than in stems and lower in those infected with PSTVd-D than with PSTVd-I. The viroid titer was higher in stem tissues and PSTVd-I-infected plants, indicating that the total number of PSTVd sRNA reads was correlated with viroid titer. In accordance with the RNA gel blot analysis of two PSTVd strains of mild and severe (RG1) (Itaya et al. 2001) or lethal (AS1) and mild (QFA) (Matoušek et al. 2007), our deep sequencing analysis also revealed that severe strains induced a higher level of PSTVd sRNA than the mild strains did.
The size distribution of PSTVd-sRNA was concentrated in the 21–24 nt range (Fig. 3a, b), consistent with the results reported previously for various viroid–host combinations (Di Serio et al. 2009; Martinez et al. 2010; Navarro et al. 2009; Owens et al. 2012; St-Pierre et al. 2009; Wang et al. 2011). Therefore, the analysis was focused on species 21–24 nt long. However, it should be noted that a small peak found in all samples indicated the presence of 17-nt species, in which a specific PSTVd sRNA molecule with the sequence 5′-AGGCGGCTCGGAGGAGC-3′, derived from nucleotides 70–86 of the positive strand, was predominant in the population. However, the function and role of this PSTVd sRNA awaits further analysis.
Size distribution of Potato spindle tuber viroid (PSTVd)-sRNA in leaf (a) and stem (b) obtained by deep sequencing. Horizontal axis indicates length of the small RNA and vertical axis indicates the number of reads. Red bars indicate PSTVd-I plus strand (leaf and stem), similarly, yellow indicate PSTVd-I minus, blue indicate PSTVd-D plus, and green indicate PSTVd-D minus
Among PSTVd sRNA derived from positive strands, the 22-nt species was the most abundant, followed by the 21-, 24-, and 23-nt species. While in the negative strand, the 21-nt species had the largest population, followed by the 22-, 24-, and 23-nt species. The positive/negative ratios in the leaf and stem tissues of PSTVd-I-infected tomato were both 3:1, whereas the ratios were 2.8 and 5.9 in the leaf and stem tissues of PSTVd-D-infected tomato plants, respectively (Table 1). The 5′-end nucleotide was predominantly a C.
In a further analysis focused on the 2 major 21- and 22-nt species, we identified changes in the number of reads of individual PSTVd sRNA species in PSTVd-I- and PSTVd-D-infected tissues. The results are summarized in Fig. 4a and b and presented as scatter diagrams with the number of reads of PSTVd-D on the vertical axis and those of PSTVd-I on the horizontal. In general, the number of reads was lower in PSTVd-D- than in PSTVd-I-infected tissues (50 %–90 % and 15 %–28 % of PSTVd-I in stem and leaf tissues, respectively). Notably, the quantity of PSTVd sRNA was significantly reduced in PSTVd-D-infected tissues. For example, among the 21-nt species derived from positive strands in stem tissues, the regression line between the number of reads in PSTVd-D (y-axis) and PSTVd-I (x-axis) fit the formula y = 0.5086x + 28.492 (Fig. 4b), the one from nucleotides 185–205 (designated hereafter as PST-sRNA21P-185) resulted in 14,925 reads in PSTVd-I infections, but only 1,870 (12.5 %) in PSTVd-D infections. More drastically, among the relatively abundant species (i.e., those exceeding 500 reads) of the positive strand-coded 21-nt species in stem tissues, PST-sRNA21P-49, -50, -51, -59, -69, -107, -109, -110, -111, -113, and -202 accounted for less than 10 % of PSTVd-I- in PSTVd-D-infected plants (Fig. 4b, “red” dots in the upper right panel). As a brief but detailed description, PST-sRNA21P-49 resulted in 501 reads in PSTVd-I and only 1 (0.2 %) in PSTVd-D, similarly PST-sRNA21P-50 resulted in 1,981 and 1 (0.1 %), PST-sRNA21P-51 resulted in 642 and 23 (3.6 %), and PST-sRNA21P-69 resulted in 1,563 and 92 (5.9 %) (Table 2). Notably, 27 of the 44 listed contained at least one of the nucleotide changes found in PSTVd-I and -D, suggesting that these nucleotide changes significantly influenced PSTVd-sRNA accumulation.
Scatter diagrams of Potato spindle tuber viroid (PSTVd)-sRNA of 21- and 22-nt in tomato leaf (a) and stem (b) infected with PSTVd-D (x-axis) versus PSTVd-I (y-axis). P plus strand, M minus strand. Among the relatively abundant species (>500 reads), those accounting for <10 % of PSTVd-I- in PSTVd-D-infected plants are shown as red dots
The differences in the accumulation patterns of the PSTVd-sRNA species described above can be overlooked by plotting PSTVd sRNA on the PSTVd genome. The number of reads of the 21- and 22-nt species of PSTVd-sRNAs with positive polarity derived from the nucleotide positions around 100–140 (i.e., the boundary of the upper CCR to V region), consisting of the mutations at positions 119 and 127 (Figs. 2b and 5a) and positions 180–220 (i.e., the lower portion of TR to the boundary of the V region) including the mutation at position 202 (Figs. 2b and 5a) was reduced significantly in both leaf and stem tissues of PSTVd-D-infected tomato plants. Therefore, the three changes in nucleotides (at positions 119, 127, and 202) changed the PSTVd sRNA hotspot pattern.
a Sequence profiles of Potato spindle tuber viroid (PSTVd)-sRNA populations recovered from leaf and stem tissues infected with PSTVd-I (upper panels, 1–4) and PSTVd-D (lower panels, 5–8). From left to right, (1, 5): 21-nt species from leaf, (2, 6): 22-nt species form leaf, (3, 7): 21-nt species from stem, and (4, 8): 22-nt species from stem. The upper half of each panels shows the profile of PSTVd-sRNA derived from a positive strand, and the lower half shows those from a negative strand. Specific peaks were significantly lower in PSTVd-D infected samples are shown by red arrows. b Histogram of 5′-end of PSTVd-sRNA of 21- and 22-nt with positive polarity on PSTVd genomic RNA. Blue bars indicate those from PSTVd-I; red bars indicate those from PSTVd-D. Five PSTVd structural regions are indicated at the top of the panel. The arrows indicate nucleotide positions 49–68 (upper strand of the P region) and 294–312 (lower strand of the P region), in which PSTVd-sRNA were significantly reduced to essentially 0 in PSTVd-D-infected tomato leaf and stem tissues
In addition, the 21- and 22-nt species of PSTVd-sRNA derived from positive strands with the 5′-end ranging from nucleotides 49 to 68 (the upper strand of the P region) and from nucleotides 294 to 312 (the lower strand of the P region) were significantly reduced to virtually 0 in PSTVd-D-infected tomato leaf and stem tissues (Fig. 5b). For example, the total number of 21- and 22-nt PSTVd sRNAs with positive polarity starting from nucleotide 49 to 68 was 8,018 in PSTVd-I, but only 30 in PSTVd-D-infected leaf tissues. Similarly, the number was 9,918 and 128 in PSTVd-I- and PSTVd-D-infected stem tissues. The significant changes in the PSTVd sRNA population can be partly attributed to the nucleotide changes found either in the upper strand of the P-region (i.e., A insertion between 63/64) and/or in the opposite strand (i.e., A to C substitution in 311 and 2 U insertions between 312/313).
Analysis of miRNA expression in PSTVd-I- and PSTVd-D-infected tomato plants
MiRNAs play a vital role in the life cycle of plants. Recent studies have shown that relative expression levels of certain host miRNAs can change in viroid-infected plants (Diermann et al. 2010; Ivanova et al. 2014; Owens et al. 2012). Therefore, it would be notable to analyze sRNA data sets for potential differences in miRNA expression levels in plants infected with viroids of different pathogenicities. Based on the sRNA deep sequencing data, we further analyzed how miRNA expression was influenced by infection with the severe and mild symptom-inducing PSTVd isolates. The number of miRNA reads in the PSTVd-I- and PSTVd-D-infected tomato tissues was extracted using the miRBase database of published miRNA sequences and annotations (http://www.mirbase.org/) as a reference and is summarized after normalization in Table 3 with annotations. The number of reads between leaf and stem tissues was almost the same, but with some noticeable differences (i.e., the abundance of miR319 was approximately 50-fold greater in stem tissues, whereas the abundance of miR6023 and miR6024 was approximately fourfold greater in leaf tissues). The influence of PSTVd-I and PSTVd-D infection was visualized by scatter diagrams using the number of reads in healthy controls as the horizontal axis and those in PSTVd-I- and PSTVd-D-infected tissues as the vertical axis (Fig. 6). Because the number of reads was considerably low for some of the miRNAs, those exceeding 100 reads/million in at least 1 treatment were selected for evaluation.
Scatter plots of miRNAs reads in tomato leaf (a) and stem (b) infected with Potato spindle tuber viroid (PSTVd)-D and PSTVd-I versus healthy controls. y-axis number of reads for PSTVd-I (red) and -D (blue), x-axis healthy. Major miRNA species described in the text are shown by arrows and dotted ovals
The changes in miRNA levels after PSTVd infection were classified into 3 categories: stable, downregulated, and upregulated. Among those examined (Table 3), no significant changes were observed in miR6022 levels in both leaf and stem tissues regardless of infection. Of the downregulated group, miR159 was abundant in both leaf and stem tissues, and the number of reads in PSTVd-infected leaf and stem tissues was reduced to 60 %–78 % and 43 %–47 %, respectively, of the number in healthy controls. Such was also the case for miR162; the number of reads in PSTVd-infected leaf and stem tissues was reduced to 61 %–76 % and 51 %–57 %, respectively, of the healthy controls. As described earlier, the abundance of miR319 was approximately 50-fold greater in stem tissues than in leaf tissues, and the number of reads in PSTVd-infected stem tissues was more significantly reduced, to 33 %–63 % of healthy controls (Fig. 6b), but no change was apparent in leaf tissues (Fig. 6a). In PSTVd-infected stem tissues, miR172a and miR172b were also abundant, but the level was only 67 %–94 % of that in healthy controls. It should be noted that the level was reduced even more after infection with PSTVd-I, the severe symptom-inducing isolate. The abundance of miR156a, miR156b, and miR156c was approximately fivefold greater in PSTVd-infected leaf tissues and reduced to 42 %–52 % of that the level in healthy controls. In addition, miR171a and miR171c were abundant in leaf tissues and slightly decreased after PSTVd infection. Of the miRNA species in both stem and leaf tissues, miR166a and miR166b were the most abundant, but their numbers were decreased in PSTVd-infected tissues with a reduction rate of only approximately 20 %.
On the contrary, in the upregulated group, miR6027 was approximately 1.5-fold higher only in PSTVd-I-infected leaf and stem tissues. The levels of miR482b, miR5301, miR6023, and miR6024 were also high, but only in PSTVd-I-infected stem tissues and low in PSTVd-D-infected stem tissues (Fig. 6b; dotted circle). Because miR6023, miR6024, miR6027, and miR482b were annotated as microRNA related to disease resistance (Table 3), the content of each may have increased by infection with a severe symptom-inducing isolate.
Discussion
Of the 2 PSTVd isolates analyzed in the present study, PSTVd-I accumulated faster and reached higher levels than PSTVd-D did. The plants infected with the severe symptom-inducing isolate accumulated more PSTVd sRNA in both leaf and stem tissues. These results indicated that accumulation of PSTVd sRNA was correlated with viroid titers in the tissue. Data on deep-sequencing analyses of viroid-specific sRNA have already been reported in host species infected with PSTVd, Citrus exocortis viroid, Hop stunt viroid, Grapevine yellow speckle viroid 1, and Peach latent mosaic viroid. However, individual viroid-specific sRNA species have not yet been fully evaluated. Therefore, we analyzed differences in the accumulated levels of each PSTVd sRNA species induced by infection with either the severe or mild symptom-inducing PSTVd isolate. As expected from northern hybridization analysis, the total number of PSTVd sRNA reads was confirmed to be lower in the tissues infected with the mild symptom-inducing PSTVd isolate. Notably, we found that the accumulated levels of PSTVd sRNA species were drastically downregulated in the mild symptom-inducing isolates including the nucleotide positions 119, 127, and 202, in which the nucleotide was changed between the severe and mild symptom-inducing isolates, i.e., mutations in viroid molecules can significantly affect the accumulation of viroid-specific sRNA.
In addition, another notable finding was that the accumulation of PSTVd-sRNA species with the 5′-end ranging from nucleotides 49–69 of the PSTVd genomic (or positive) strand were drastically downregulated and essentially nonexistent in those infected with the mild symptom-inducing isolate. This region is located in the upper portion of the PSTVd pathogenicity region and includes an A-rich sequence, which is a characteristic of the members of the Pospiviroidae family. Although it has long been predicted and discussed in some pioneering work that a possible link might exist between viroid pathogenicity and viroid-inducing RNA silencing (Itaya et al. 2001, 2007; Papaefthimiou et al. 2001; Wang et al. 2004), no evidence has been reported that suggests a relevant correlation between viroid pathogenicity and viroid-specific sRNA derived from the pathogenicity region. Therefore, the results obtained in the present study require further analysis.
Accumulation of miRNA, which are also small RNA species, was downregulated in general in PSTVd-infected leaf tissues, regardless of pathogenicity, although the accumulation levels slightly differed between leaf and stem tissues, and some were apparently upregulated. For instance, those related to disease resistance, such as miR482b, miR482c, miR6023, miR6024, and miR6027, and those related to transcription, such as miR172a, miR172b, miR172c, miR172d, and miR5031, were upregulated in stem tissues infected with PSTVd-I, which induces severe symptoms. This result might be due to strong resistance to infection because PSTVd-I induced severe stunting of the infected plants (i.e., shortened internodes). Among the miRNA downregulated by PSTVd infection, changes in miR319 and miR159 were notable. Although both were downregulated after infection with the severe and mild symptom-inducing PSTVd isolates, miR159 accumulated at high levels in both leaf and stem tissues, and miR319 accumulated at high levels in stem tissues. Particularly notable was the large downregulation of both miR159 and miR319 in stem tissues, although the reduction was greater in those infected with the severe symptom-inducing PSTVd isolate than in those infected with the mild symptom-inducing isolate (i.e., the reduced accumulation was consistent with the severity of symptoms). Diermann et al. (2010) performed deep-sequencing analysis of sRNA generated in tomato (cv. Rutgers) plants infected with PSTVd-AS1 (a lethal strain) and reported that the accumulation of miR319 and miR159 in PSTVd-infected tomato plants was reduced to approximately 1/3 that in healthy controls. Similarly, Owens et al. (2012) reported that miR159 levels in PSTVd-I infected tomato (cv. Rutgers) tissues were reduced to approximately 1/3 that in healthy controls by deep-sequencing analysis of sRNA. In addition, Ivanova et al. (2014) analyzed changes in miRNA expression levels in PSTVd-infected Phelipanche ramosa (parasite plant) using a microarray chip and found that all but two (miR395 and miR156) of the 43 miRNA species that showed significant signals were downregulated during PSTVd infection. Here, mirR159 and miR319 were reduced to less than approximately 1/4 and 1/8, respectively, of the levels in healthy controls. As presented here, miR319 levels in PSTVd-I- and PSTVd-D-infected tomato (cv. Rutgers) stem tissues were reduced to approximately 1/3 and 2/3, respectively, of that in healthy controls. Levels of miR159 in both PSTVd-I- and PSTVd-D-infected stem and leaf tissues were also reduced to approximately 1/2 of the healthy controls. Collectively, these findings provide strong evidence that PSTVd infection significantly affected levels of miRNAs, especially miR159 and mir319.
The MIR159 and MIR319 genes were first identified in Arabidopsis thaliana and are known as one of eight highly conserved miRNA families that are believed to have evolved from a common ancestor (Cuperus et al. 2011). The miR159 and miR319 can act to regulate siRNA pathways through posttranscriptional regulation (Li et al. 2011). The targets of the MIR159 gene family are members of the highly conserved GAMYB (MYB33) gene family, which play important roles as transcription factors in the development of aleurone cells, flowers, and seeds as well as in the growth of vegetative tissues (Allen et al. 2007). Overexpression of miR159 in Arabidopsis could reduce AtMyb33 levels, resulting in delayed flowering under short day conditions (Achard et al. 2004). In addition, Li et al. (2013) reported that miR159 is a negative regulator of floral transition under short photoperiods and can be used to control flowering time in gloxinia (Sinningia speciosa) by suppressing miR159 expression. When we consider the report that chrysanthemum (Chrysanthemum × morifolium Ramat) infected with Chrysanthemum stunt viroid blooms a few days to 3 weeks earlier than healthy plants of the same variety (Bouwen and van Zaayen 2003), the observation that miR159 is downregulated during PSTVd infection is extremely notable. Owens et al. (2012) used a microarray to analyze alterations in gene expression in PSTVd-infected tomato plants and found that the gibberellin β-hydroxylase gene, which is involved in gibberellin biosynthesis, was significantly downregulated in PSTVd infection. Because miR159 can target GAMYB, expression levels of GAI (DELLA), GAMYB, and LEAFY (genes in the central portion of the GA signaling pathway) were further analyzed; However, PSTVd infection had limited effect on these genes. In conclusion, microRNA analysis in the field of viroid research is still in its infancy; it is necessary to investigate the effects of viroid infection on levels of microRNA including miR159 and/or miR319 in relation to possible molecular mechanisms of viroid pathogenicity through GA signaling pathways.
References
Achard P, Herr A, Baulcombe DC, Harberd NP (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131:3357–3365
Allen RS, Li J, Stahle MI, Dubroué A, Gubler F, Millar AA (2007) Genetic analysis reveals functional redundancy and the major target gene of the Arabidopsis miR159 family. Proc Natl Acad Sci USA 104:16371–16376
Bouwen I, van Zaayen A (2003) Chrysanthemum stunt viroid. In: Hadidi A et al (eds) Viroids. CSIRO Publishing, Collingwood, pp 218–223
Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23:431–442
Darós JA, Flores R (2004) Arabidopsis thaliana has the enzymatic machinery for replicating representative viroid species of the family Pospiviroidae. Proc Natl Acad Sci USA 101:6792–6797
Di Serio F, Gisel A, Navarro B, Delgado S, de Alba AEM, Donvito G, Flores R (2009) Deep sequencing of the small RNAs derived from two symptomatic variants of a chloroplastic viroid: implications for their genesis and for pathogenesis. PLoS One 4:e7539
Diener TO (ed) (1987) The viroids. Plenum Press, New York
Diermann N, Matoušek J, Junge M, Riesner D, Steger G (2010) Characterization of plant miRNAs and small RNAs derived from Potato spindle tuber viroid (PSTVd) in infected tomato. Biol Chem 391:1379–1390
Flores R, Hernández C, de Alba AEM, Daròs JA, Di Serio F (2005) Viroids and viroid–host interactions. Annu Rev Phytopathol 43:117–139
Itaya A, Folimonov A, Matsuda Y, Nelson RS, Ding B (2001) Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol Plant Microbe Interact 14:1332–1334
Itaya A, Zhong X, Bundschuh R, Qi Y, Wang Y, Takeda R, Harris AR, Molina C, Nelson RS, Ding B (2007) A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J Virol 81:2980–2994
Ivanova D, Milev I, Vachev T, Baev V, Yahubyan G, Minkov G, Gozmanova M (2014) Small RNA analysis of Potato spindle tuber viroid infected Phelipanche ramosa. Plant Physiol Biochem 74:276–282
Levy L, Lee IM, Hadidi A (1994) Simple and rapid preparation of infected plant tissue extracts for PCR amplification of virus, viroid and MLO nucleic acids. J Virol Methods 49:295–304
Li Y, Li C, Ding G, Jin Y (2011) Evolution of MIR159/319 microRNA genes and their post-transcriptional regulatory link to siRNA pathways. BMC Evol Biol 11:122
Li X, Bian H, Song D, Ma S, Han N, Wang J, Zhu M (2013) Flowering time control in ornamental gloxinia (Sinningia speciosa) by manipulation of miR159 expression. Ann Bot 111:791–799
Machida S, Yamahata N, Watanuki H, Owens RA, Sano T (2007) Successive accumulation of two size classes of viroid-specific small RNA in Potato spindle tuber viroid-infected tomato plants. J Gen Virol 88:3452–3457
Martinez G, Donaire L, Llave C, Pallas V, Gomez G (2010) High-throughput sequencing of Hop stunt viroid-derived small RNAs from cucumber leaves and phloem. Mol Plant Pathol 11:347–359
Matoušek J, Kozlová P, Orctova L, Schmitz A, Pešina K, Bannach O, Diermann N, Steger G, Riesner D (2007) Accumulation of viroid-specific small RNAs and increase in nucleolytic activities likned to viroid-caused pathogenesis. Biol Chem 388:1–13
Navarro B, Pantaleo V, Gisel A, Moxon S, Dalmay T, Bisztray G, Di Serio F, Burgyán J (2009) Deep sequencing of viroid-derived small RNAs from grapevine provides new insights on the role of RNA silencing in plant–viroid interaction. PLoS One 4:e7686
Navarro B, Gisel A, Rodio ME, Delgado S, Flores R, Di Serio F (2012) Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide degradation of a host mRNA as predicted by RNA silencing. Plant J 70:991–1003
Owens RA, Tech KB, Shao JY, Sano T, Baker CJ (2012) Global analysis of tomato gene expression during Potato spindle tuber viroid infection reveals a complex array of changes affecting hormone signaling. Mol Plant Microbe Interact 25:582–598
Papaefthimiou I, Hamilton AJ, Denti MA, Baulcombe DC, Tsagris M, Tabler M (2001) Replicating Potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res 29:2395–2400
Sano T, Yoshida H, Goshono M, Monma T, Kawasaki H, Ishizaki K (2004) Characterization of a new viroid strain from hops: evidence for viroid speciation by isolation in different host species. J Gen Plant Pathol 70:181–187
Singh RP (1973) Experimental host range of the Potato spindle tuber ‘virus’. Am Potato J 50:111–123
St-Pierre P, Hassen IF, Thompson D, Perreault JP (2009) Characterization of the siRNAs associated with Peach latent mosaic viroid infection. Virology 383:178–182
Tsushima T, Murakami S, Ito H, He YH, Raj APC, Sano T (2011) Molecular characterization of Potato spindle tuber viroid in dahlia. J Gen Plant Pathol 77:253–256
Wang MB, Bian XY, Wu LM, Liu LX, Smith NA, Isenegger D, Wu RM, Masuta C, Vance VB, Watson JM, Rezaian A, Dennis ES, Waterhouse PM (2004) On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc Natl Acad Sci USA 101:3275–3280
Wang Y, Shibuya M, Taneda A, Kurauchi T, Senda M, Owens RA, Sano T (2011) Accumulation of Potato spindle tuber viroid-specific small RNAs is accompanied by specific changes in gene expression in two tomato cultivars. Virology 413:72–83
Weidemann HL, Buchta U (1998) A simple and rapid method for the detection of Potato spindle tuber viroid (PSTVd) by RT-PCR. Potato Res 41:1–8
Acknowledgments
This work was supported in part by Japan Society for the Promotion of Science KAKENHI Grant No. 24380026.
Author information
Authors and Affiliations
Corresponding author
Additional information
Daiki Tsushima and Charith Raj Adkar-Purushothama have contributed equally to the research.
Rights and permissions
About this article
Cite this article
Tsushima, D., Adkar-Purushothama, C.R., Taneda, A. et al. Changes in relative expression levels of viroid-specific small RNAs and microRNAs in tomato plants infected with severe and mild symptom-inducing isolates of Potato spindle tuber viroid . J Gen Plant Pathol 81, 49–62 (2015). https://doi.org/10.1007/s10327-014-0566-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10327-014-0566-7