Abstract
Microbial production of cis,cis-muconate (ccMA) from phenolic compounds obtained by chemical depolymerization of lignin is a promising approach to valorize lignin. Because microbial production requires a large amount of carbon and energy source, it is desirable to establish a ccMA-producing strain that utilizes lignin-derived phenols instead of general sources like glucose. We isolated Pseudomonas sp. strain NGC7 that grows well on various phenolic compounds derived from p-hydroxyphenyl, guaiacyl, and syringyl units of lignin. An NGC7 mutant of protocatechuate (PCA) 3,4-dioxygenase and ccMA cycloisomerase genes (NGC703) lost the ability to grow on vanillate and p-hydroxybenzoate but grew normally on syringate. Introduction of a plasmid carrying genes encoding PCA decarboxylase, flavin prenyltransferase, vanillate O-demethylase, and catechol 1,2-dioxygenase into NGC703 enabled production of 3.2 g/L ccMA from vanillate with a yield of 75% while growing on syringate. This strain also produced ccMA from birch lignin-derived phenols. All these results indicate the utility of NGC7 in glucose-free ccMA production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lignin is the second most abundant natural polymer resulting from oxidative coupling of three types of monolignols, p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol [28]. Lignin content in wood ranges between 15% and 40% [33], and gymnosperm (softwood) lignins are composed of guaiacyl (G) units with low levels of p-hydroxyphenyl (H) units, whereas angiosperm (hardwood) lignins are composed of G-units and syringyl (S) units [6, 39]. Grass lignins contain G- and S-units and more H-units than gymnosperm lignins [6, 39]. Despite the large amount of lignin on earth, its industrial use has been considerably limited mainly due to its heterogeneity [27]. One of the promising strategies to valorize lignin is microbial conversion of such heterogeneous lignin-derived phenols, obtained through chemical depolymerization of lignin, into particular value-added metabolites such as cis,cis-muconate (ccMA) [4, 35, 44]. ccMA is a platform chemical for synthesis of a variety of polymers including nylon 6,6 through adipic acid, and its annual market value has been estimated to be more than $22 billion [43].
To date, the microbial ccMA production from lignin-derived phenols has been vigorously investigated using engineered Pseudomonas putida, Escherichia coli, Amycolatopsis, and Corynebacterium glutamicum [2, 3, 36, 40, 43]. Among these strains, P. putida KT2440 has been particularly optimized for ccMA production from p-coumarate. In KT2440 cells, phenolic compounds derived from G-lignin including vanillate and ferulate, and from H-lignin including p-hydroxybenzoate (HBA) and p-coumarate are catabolized to protocatechuate (PCA) and further degraded through the PCA 3,4-cleavage pathway [12, 29]. To produce ccMA from lignin-derived phenols, the catabolic pathway of KT2440 was basically engineered as follows: (i) the PCA 3,4-dioxygenase gene (pcaHG) was inactivated and the exogeneous PCA decarboxylase gene (aroY) was expressed to convert PCA into catechol. (ii) the ccMA cycloisomerase gene (catB) was inactivated to accumulate ccMA [40]. The fed-batch conversion using an engineered KT2440 produced 13.5 g/L of ccMA from p-coumarate. Based on an observation that coexpression of aroY and kpdB enhanced the PCA decarboxylase activity [36], Johnson et al. achieved the production of 15.6 g/L of ccMA from p-coumarate using an engineered KT2440 expressing both aroY and ecdBD (ecdB is a homolog of kpdB) [14]. Later, kpdB was found to encode flavin prenyltransferase, which generates prenylated flavin necessary for the PCA decarboxylation [41]. Furthermore, the elimination of a global regulator of carbon catabolite repression and expressing aroY and ecdBD enabled the production of 50 g/L ccMA from p-coumarate using fed-batch and high pH feeding strategy [13, 32].
Although the ccMA titer has reached high levels close to the lethal toxicity limit in the bacterium [32], almost all the ccMA-producing strains constructed to date require glucose or other carbon and energy sources for cell growth. Thus, it is desirable to create a ccMA production system that utilizes lignin-derived phenols for both cell growth and feedstock for ccMA production. For this purpose, we previously created two types of ccMA-producing strains applicable to softwood lignin and hardwood lignin, respectively [37]. One is a KT2440 strain deficient in pcaHG and catB transformed with a plasmid carrying pcaHG and aroY. Since the metabolic flux of PCA is divided to the catabolism through tricarboxylic acid cycle and the accumulation of ccMA, this strain is able to produce ccMA from vanillate and/or HBA while growing on the same substrates. Since KT2440 is unable to grow on S-lignin-derived phenols such as syringate, we employed Sphingobium sp. strain SYK-6 that can grow on syringate in addition to HBA and vanillate to create another type of glucose-free ccMA-producing strain. An SYK-6 mutant of the PCA 4,5-dioxygenase gene (ligAB) harboring a plasmid that carries the vanillate O-demethylase gene (vanAB), the catechol 1,2-dioxygenase gene (catA), aroY, and kpdB produced ccMA from 5 mM vanillate with > 96% yield (mol ccMA/mol vanillate) while growing on 5 mM syringate. However, this strain was found to require a small amount of tryptone when growing on syringate in the presence of vanillate due to unknown reason.
Based on the above background, we isolated a novel platform bacterium that grows well on HBA, vanillate, syringate, and ferulate to construct a ccMA production system that does not require any carbon source other than lignin-derived phenols. Using the isolated bacterium, we created an engineered ccMA-producing strain and evaluated its ccMA productivity from lignin-derived phenols and those obtained from birch lignin.
Materials and methods
Bacterial strains and culture conditions
Pseudomonas sp. strain NGC7, its mutants (NGC702 and NGC703), and isolated bacteria in this study were grown at 30 °C with shaking (160 rpm) in lysogeny broth (LB) or Wx minimal medium [18] containing lignin-derived phenols. E. coli NEB 10-beta was grown in LB at 37 °C. The media for E. coli transformants carrying antibiotic resistance markers were supplemented with 100 mg of ampicillin/L, 25 mg of kanamycin (Km)/L, or 12.5 mg of tetracycline (Tc)/L. The media for NGC703 harboring pTS084 carrying vanAB, aroY, kpdB, and catA in pJB866 [5, 37] were supplemented with 12.5 mg of Tc/L. Lignin-derived phenols; syringaldehyde, syringate, vanillin, vanillate, p-hydroxybenzaldehyde, HBA, PCA, and ferulate; were purchased from Tokyo Chemical Ind., Co., Ltd.; Sigma-Aldrich Co., LLC.; and FUJIFILM Wako Pure Chemical Corporation.
Screening of bacteria capable of growing on G-, H-, and S-lignin-derived phenols
Soil samples were collected from 40 different locations in Japan. Bacterial strains capable of growing on syringate were screened by the following three methods. (i) Soil samples (100 mg) were added to 10 mL of Wx medium containing 5 mM syringate and 0.1% yeast extract, and incubated with shaking for 24–72 h. The cultures (100 μL) growth observed were spread on Wx-medium agar plates containing 5 mM syringate, and then single colonies were isolated. (ii) Soil samples (100 mg) were added to 10 mL of Wx medium containing 5 mM syringate for 24 h, and portions of cultures were then subcultured four times in the same fresh media. Single colonies were picked up in the same way as described above. (iii) Syringate (0.1 g) was mixed with 500 g of soil every 24 h for 4–32 weeks. The resultant soil (100 mg) was added to 10 mL of Wx medium containing 5 mM syringate and 0.1% yeast extract, incubated for 24–72 h, and then single colonies were obtained. For the 186 strains obtained by the above methods (100, 6, and 80 strains, respectively), susceptibility to Tc and Km, which are antibiotic resistance markers of plasmids to be introduced, was evaluated on LB agar plates containing the antibiotics. The growth of the resulting 43 strains on Wx medium containing 10 mM syringate was evaluated. Seven strains that grew faster than others on syringate were selected, and their ability to grow on Wx medium containing 10 mM of syringaldehyde, vanillin, vanillate, ferulate, HBA, p-hydroxybenzaldehyde, PCA, and glucose was examined. For finally selected strain NGC7 isolated by the method described in (ii), the growth in Wx medium containing a 0.6% solution of birch lignin extracts described below was assessed. All growth was monitored by optical density measurements at 600 nm (OD600) or 660 nm (OD660) using a spectrophotometer (V-630 Bio; JASCO Co., Ltd.) and a TVS062CA biophotorecorder (Advantec Co., Ltd.), respectively.
Preparation of hardwood lignin-derived phenols
Hardwood lignin-derived phenols were prepared by the alkaline-nitrobenzene oxidation method [37]. Birch powder (1.5 g) treated with ethanol–benzene (1:2, v/v) was suspended in a solution consisting of 50 mL of 2 N NaOH and 3 mL of nitrobenzene (FUJIFILM Wako Pure Chemical Corporation) in a 200 mL stainless-steel vessel (Taiatsu Techno Co.) and heated at 170 °C for 2.5 h with stirring (500 rpm). The supernatant was extracted three times with diethyl ether to remove unreacted nitrobenzene. The alkaline aqueous phase obtained by performing the above-mentioned treatment twice was acidified with HCl and extracted three times with diethyl ether. The organic phase was collected and evaporated, and the resulting extract residue (0.771 g) was dissolved in 4 mL of 2 N NaOH and adjusted to pH 9 with HCl. The resulting solution (bNBL) was used as the hardwood lignin-derived phenols.
Sequencing of the 16SrRNA gene
The partial 16S rRNA gene of NGC7 was PCR amplified using total DNA of NGC7 as a template and conserved eubacterial primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1525R (5′-AAAGGAGGTGATCCAGCC-3′). The nucleotide sequence of the amplified fragment separated on 0.8% agarose gel electrophoresis was determined by Eurofins Genomics (Tokyo, Japan). Sequence analysis was performed with the MacVector program (MacVector, Inc.). Sequence similarity searches were carried out using the BLAST program [15]. Pairwise alignments were performed with EMBOSS alignment tool [30].
Identification of intermediate metabolites
NGC7 cells were grown in different Wx media, one containing 10 mM syringate and other 10 mM vanillate, and the cells were collected by centrifugation (5000×g for 5 min). The cells grown on syringate and vanillate were suspended in 50 mM Tris–HCl (pH 7.5) buffer (OD600 = 0.5) followed by incubation with 1 mM of each syringate and vanillate, respectively, at 30 °C with shaking. Portions of the reaction mixtures were periodically collected, and reactions were stopped by centrifugation. Samples were diluted with water, filtrated, and analyzed by HPLC.
HPLC analysis
HPLC analysis was performed with the Acquity UPLC system (Waters) using a TSKgel ODS-140HTP column (2.1 by 100 mm; Tosoh) with a flow rate of 0.5 mL/min. The mobile phase was a mixture of solution A (acetonitrile containing 0.1% formic acid) and solution B (water containing 0.1% formic acid) under the following conditions: 0–3 min, 1% A; 3–6 min, linear gradient 1–25% A; 6–7 min, decreasing gradient 25–1% A. Syringaldehyde and vanillin were detected at 310 nm, and other compounds were detected at 270 nm.
Estimation of the catabolic pathways of PCA and catechol
NGC7 cells were grown in Wx medium containing 10 mM glucose plus 10 mM PCA and 10 mM glucose plus 10 mM benzoate at 30 °C for 12 h. The resulting cells were washed with 50 mM Tris–HCl buffer (pH 7.5), suspended in the same buffer, and broken by an ultrasonic disintegrator. The supernatants of cell lysates were obtained as cell extracts after centrifugation (19,000×g for 15 min). Protein concentrations were determined by the Bradford method using the Bio-Rad protein assay kit (Bio-Rad Laboratories). Cell extracts (5 μg of protein/mL) were incubated with 100 μM of PCA and catechol in 50 mM Tris–HCl buffer (pH 7.5) for 30 min, and the UV–visible (VIS) spectral changes were periodically measured using a spectrophotometer.
Cloning of pcaHG and catBCA and their disruptions in NGC7
PCR primers for amplification of pcaHG and catBCA were prepared based on the regions highly conserved between these genes in P. putida KT2440 (AE015451.2), P. aeruginosa PAO1 (AE004091.2), P. oryzihabitans NBRC 102199 (NZ_BBIT01000011.1 and NZ_BBIT01000025.1), P. denitrificans ATCC 13867 (CP004143.1), and P. fulva 12-X (CP002727.1). The partial sequences of pcaHG and catBCA were PCR amplified using the total DNA of NGC 7 as a template and primer pairs of pcaH-F and pcaG-R (5′-TGATTACGCCAAGCTTCGTGATCGCACCTGGCATCC-3′ and 5′-GACGGCCAGTGAATTCAGATGTCGAAGAAGACCGTTTC-3′) and catB-F and catA-R (5′-TGATTACGCCAAGCTTACCATCCGCCCGCACAAGCTGGC-3′ and 5′-GACGGCCAGTGAATTCTCGCGGGTGGCGTAGGCGAAGTC-3′), respectively. The nucleotide sequences of the amplified fragments were determined by Eurofins Genomics.
A 1.3-kb pcaHG fragment and a 2.2-kb catBCA fragment were digested with EcoRI and HindIII, which cut the sites added to the PCR primers, and the resulting fragments were respectively cloned into the same sites of pK19mobsacB [34] to generate pPcaHG and pCatBCA. Internal regions of pcaHG and catB were deleted by digestion of these plasmids by ApaI-DraIII and EcoRV-ScaI, respectively, to generate pPcaHGd and pCatBd. pPcaHGd was introduced into NGC7 by triparental mating, and candidate mutants were isolated as described previously [23]. The disruption of gene was examined by colony PCR using the primer pairs of pcaH-F and pcaG-R. Subsequently, pCatBd was introduced in the resulting pcaHG mutant (NGC702), and a catB mutant of NGC702 (NGC703) was obtained by the same method described above. The growth of NGC703 on lignin-derived phenols and benzoate was tested using Wx medium containing 10 mM vanillate, HBA, PCA, syringate, or benzoate. Conversion of PCA and catechol by NGC703 was examined by incubating NGC703 cells with 5 mM glucose in the presence of 5 mM PCA or catechol for 24–48 h. Portions of the cultures were periodically collected, and reactions were stopped by centrifugation. Samples were diluted with water, filtrated, and analyzed by HPLC.
ccMA production using NGC703 harboring pTS084
pTS084 was introduced into NGC703 by electroporation. ccMA production by NGC703(pTS084) cells pre-grown in LB was evaluated by the following methods. (i) NGC703(pTS084) cells were incubated in a tube containing 10 mL Wx medium with 10 mM syringate plus 5 mM vanillate, 10 mM syringaldehyde plus 5 mM vanillin, or 8 mM syringaldehyde plus 5 mM vanillin plus 1 mM syringate plus 1 mM vanillate. (ii) NGC703(pTS084) cells were incubated in a 500 mL shake-flask containing 50, 100, or 200 mL of Wx medium with 10 mM syringate plus 5 mM vanillate. (iii) NGC703(pTS084) cells were incubated in a 500 mL shake-flask using 200 mL of medium by feeding 10 mM syringate plus 5 mM vanillate every 12 h for six times. (iv) NGC703(pTS084) cells were incubated in a tube containing 5 mL Wx medium by feeding 0.24% or 0.6% bNBL every 12 h for five times. Portions of the reaction mixtures were periodically collected for the measurement of cell growth (OD600) and substrate conversions. The reactions were stopped by centrifugation, and the resultant supernatants were filtrated, and analyzed by HPLC.
Nucleotide sequence accession numbers
The nucleotide sequences of the 16S rRNA gene, pcaHG, and catBCA of Pseudomonas sp. strain NGC7 were deposited in the DDBJ/EMBL/GenBank databases under accession numbers LC466004, LC466005, and LC466006, respectively.
Results and discussion
Isolation of a bacterial strain capable of growing on G-, H-, and S-lignin-derived phenols
To isolate a bacterial strain that can grow on phenolic compounds derived from G-, H-, and S-lignin, we first screened candidates using Wx minimal medium containing 5 mM syringate (an S-lignin-derived phenol) from soil samples of 40 different locations in Japan. Among 186 strains obtained, we selected a strain named NGC7 with high growth rate and high final cell yield. NGC7 grew well on syringate, syringaldehyde, vanillate, vanillin, ferulate, HBA, p-hydroxybenzaldehyde, PCA, and glucose (Fig. S1a-i). In addition, we examined the ability of NGC7 to grow on hardwood lignin-derived phenols prepared from birch by alkaline-nitrobenzene oxidation (bNBL, as described in the Materials and methods section). bNBL contained 112 mM syringaldehyde, 9.8 mM syringate, 48.6 mM vanillin, and 7.4 mM vanillate. NGC7 grew well in Wx medium containing a 0.6% bNBL solution (Fig. S1j).
To identify the genus of NGC7, the nucleotide sequence of the 16S rRNA gene of NGC7 was determined. The nucleotide sequence of the 16S rRNA gene of NGC7 showed 99.2–99.5% identity with the strains of Pseudomonas putida NBRC 14164 (NC_021505.1, PP4_RS00780), P. plecoglossicida FPC951 (AB009457.1), P. taiwanensis BCRC 17751 (NR_116172.1), and P. monteilii CIP 104883 (NR_024910.1), indicating that NGC7 belongs to genus Pseudomonas.
Estimation of the catabolic pathways of lignin-derived phenols in Pseudomonas sp. strain NGC7
To estimate the catabolic pathways of syringate and vanillate in NGC7, resting cells (OD600 = 0.5) of NGC7 grown on syringate and vanillate were incubated with 1 mM of each syringate and vanillate, respectively, and the culture supernatants were analyzed by HPLC. Syringate was completely degraded within 60 min, and a faint peak with a retention time of 4.3 min was observed after 15 min (Fig. S2a–d). This compound was identified as 3-O-methylgallate based on the comparison of the retention time and UV–Vis spectrum of the authentic compound (Fig. S2e, f). Therefore, syringate appears to be subjected to O demethylation (Fig. 1). Vanillate was also completely degraded after 60 min, however, no intermediate metabolite was observed (Fig. S2g–i).
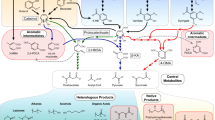
Proposed catabolic pathways of lignin-derived phenols in Pseudomonas sp. strain NGC7 and engineered route for ccMA production. Pathways presumed to comprise multiple enzymatic reactions are indicated by dotted arrows. HBN, p-hydroxybenzaldehyde; HBA, p-hydroxybenzoate; PCA, protocatechuate; ccMA, cis,cis-muconate. Enzymes: VanAB, vanillate O-demethylase; PcaHG, PCA 3,4-dioxygenase; CatA, catechol 1,2-dioxygenase; CatB, ccMA cycloisomerase; AroY, PCA decarboxylase; KpdB, flavin prenyltransferase. To produce ccMA from H- and G-lignin-derived phenols, pcaHG and catB were disrupted by homologous recombination, and pTS084 carrying vanA (PP_3736) and vanB (PP_3737) of Pseudomonas putida KT2440 (accession number, NC_002947.4), aroY of Klebsiella pneumoniae subsp. pneumoniae A170-40 (AB479384.2), kpdB of K. pneumoniae subsp. pneumoniae NBRC14940 (AB920346.1), and catA (PP_3713) of P. putida KT2440 (NC_002947.4) was introduced
In general, vanillate and HBA are converted to PCA through O demethylation and hydroxylation, respectively, and the resulting PCA is catabolized via one of the PCA 2,3-, 3,4-, or 4,5-cleavage pathways in bacteria [9, 11, 16, 17]. In Gram-negative bacteria, PCA 3,4- and 4,5-cleavage pathways are major catabolic routes of PCA. To estimate the PCA catabolic pathway of NGC7, the cell extract (5 μg of protein/mL) of NGC7 grown on PCA was incubated with 100 μM PCA, and the UV–Vis spectra of the reaction mixture were periodically monitored. As a result, the absorbance at 252 nm and 290 nm decreased, however, any other absorbance was not observed (Fig. S3a). As the reaction product of PCA 4,5-dioxygenase, 4-carboxy-2-hydroxymuconate-6-semialdehyde, has an absorbance at 410 nm [20], NGC7 appears to catabolize PCA via the PCA 3,4-cleavage pathway. However, catechol is generally degraded through catechol 1,2- or 2,3-cleavage pathway in bacteria [7, 12, 42]. Similarly, the cell extract (5 μg of protein/mL) of NGC7 grown on benzoate was incubated with 100 μM catechol, and the UV–Vis spectra of the reaction mixture were monitored. Since the absorbance at 375 nm derived from the reaction product of catechol 2,3-dioxygenase, 2-hydroxymuconate-6-semialdehyde [25], was not observed (Fig. S3b), NGC7 seems to degrade catechol via the catechol 1,2-cleavage pathway, which generates ccMA as an intermediate metabolite.
Isolation of pcaHG and catB and disruption of these genes in NGC7
According to the estimation of the catabolic pathways of PCA and catechol, NGC7 was suggested to possess pca and cat genes. The amplification primers for partial sequences of pcaHG encoding PCA 3,4-dioxygenase and catBCA encoding ccMA cycloisomerase, muconolactone Δ-isomerase, and catechol 1,2-dioxygenase, were designed based on the highly conserved regions among Pseudomonas strains including P. putida KT2440 and P. aeruginosa PAO1. PCR using these primers successfully amplified the fragments (Fig. S4). The nucleotide sequencing of these fragments revealed the presence of pcaHG and catBCA whose deduced amino acid sequences showed 94.7–98.1% identities with those of KT2440 pcaHG and 94.7–97.3% identities with those of KT2440 catBCA, respectively.
For the construction of an NGC7 mutant of both pcaHG and catB, pcaHG was first disrupted by homologous recombination using pK19mobsacB carrying a pcaHG fragment whose internal region was deleted and then catB was inactivated in a similar manner (Fig. S5). The resulting strain NGC703 could no longer grow on vanillate, HBA, and PCA in addition to benzoate but was able to grow on syringate as well as the wild type (Fig. S6). These results indicate that pcaHG is essential for NGC7 to grow on vanillate, HBA, and PCA, and the former two compounds are suggested to be catabolized via PCA (Fig. 1). In addition, these results indicate that an aromatic ring cleavage enzyme gene different from pcaHG is involved in syringate catabolism. To date, the syringate catabolic genes have been clarified only in Sphingobium sp. SYK-6, and recently also reported in Novosphingobium aromaticivorans DSM 12444 [8, 21]. In SYK-6, syringate is mainly degraded through the O demethylation of syringate to generate gallate via 3-O-methylgallate [1, 22] and the subsequent ring cleavage of gallate by gallate dioxygenase (DesB) whose substrate specificity is restricted [19, 38]. Since KT2440 also carries the gallate dioxygenase gene (galA) [24], which shows 58% amino acid sequence identity with DesB, syringate may be degraded in NGC7 via a similar pathway involving gallate dioxygenase shown in SYK-6. However, it was suggested that benzoate is catabolized through the catechol 1,2-cleavage pathway, and catB is essential for the catabolism.
To confirm whether NGC703 lost the ability to convert PCA and ccMA, this strain was grown in 5 mM glucose plus 5 mM PCA and 5 mM glucose plus 5 mM catechol, and the supernatant of the cultures were analyzed by HPLC. After 24 h, PCA was not transformed, whereas catechol was completely converted to ccMA (Fig. 2). Furthermore, the accumulated ccMA was not converted even after 48 h. These results indicated that pcaHG and catB are essential for the conversion of PCA and ccMA in NGC7.
Conversion of PCA and catechol by NGC703 cells. NGC703 cells were grown in Wx medium containing 5 mM glucose with 5 mM PCA (a) or 5 mM catechol (b). Portions of the cultures were collected at the start and after 24 h and 48 h (for conversion of catechol) of incubation and then analyzed by HPLC. Retention times of PCA, catechol, and ccMA were 3.2, 3.3, and 3.6 min, respectively
Construction of an NGC703-based ccMA-producing strain and its ccMA productivity
To confer PCA decarboxylation activity and reinforce the vanillate/syringate O demethylation and catechol conversion, pTS084, which carries aroY, kpdB, vanAB, and catA, was introduced to NGC703 (Fig. 1). Based on the ratio of S-lignin-derived phenols versus G-lignin-derived phenols in bNBL, cell growth and ccMA productivity of NGC703(pTS084) were examined using 10 mL Wx medium containing 10 mM syringate plus 5 mM vanillate. This strain grew well in this medium and produced ccMA with a yield of 87% (mol ccMA/mol vanillate) (Fig. 3a, b). Since the bNBL contains high amount of syringaldehyde and vanillin, cell growth and ccMA yield were evaluated by incubating NGC703(pTS084) cells in 10 mL Wx-medium containing 10 mM syringaldehyde plus 5 mM vanillin. The growth was somewhat retarded compared to the conditions of 10 mM syringate plus 5 mM vanillate but the maximum cell yields after 36 h were almost the same (Fig. 3a). This strain produced ccMA with a yield of 83% (mol ccMA/mol vanillin), which is equivalent to the value when using vanillate and syringate (Fig. 3c). Next, based on the ratio of lignin-derived phenols contained in bNBL, NGC703(pTS084) cells were incubated in Wx medium containing 8 mM syringaldehyde plus 5 mM vanillin plus 1 mM syringate plus 1 mM vanillate. The cell growth was almost the same as that incubated with 10 mM syringaldehyde plus 5 mM vanillin, and the strain produced ccMA (65% mol ccMA/mol vanillin plus vanillate) (Fig. S7). However, significant PCA accumulation was observed. To examine the effect of aeration on ccMA production, NGC703(pTS084) cells were grown in a 500 mL shake-flasks containing 50, 100, or 200 mL of Wx medium with 10 mM syringate plus 5 mM vanillate (Fig. 4a–d). When using 200 mL medium, the ccMA yield was the highest (80% mol ccMA/mol vanillate), and the accumulation of PCA was considerably lower than that in other conditions (Fig. 4c). PCA decarboxylase is known to be oxygen sensitive, and its half-life was 5–8 min under aerobic conditions [26]. In contrast, oxygen is necessary for the generation of active form of prenylated flavin, a cofactor of PCA decarboxylase [26]. Therefore, PCA decarboxylase activity appears to be determined by the relationship between the level of inactivation of AroY and the availability of active prenylated flavin. Dissolved oxygen in the 200 mL medium may have been optimum among the conditions examined. Subsequently, ccMA production by NGC703(pTS084) was examined in a 500 mL shake-flask using 200 mL of medium by feeding 10 mM syringate plus 5 mM vanillate every 12 h for six times. The ccMA titer was 3.2 g/L (22.5 mM) with a yield of 75% (mol ccMA/mol vanillate) (Fig. 4e). In a previous study, we achieved glucose-free ccMA production using an engineered strain of Sphingobium sp. SYK-6 harboring pTS084 [37]. However, this strain has the disadvantage of requiring small amount of tryptone during ccMA production. NGC703(pTS084) enables glucose-free ccMA production from lignin-derived phenols without the addition of any other organic compounds. Moreover, the ccMA titer of 3.2 g/L is equivalent to that achieved by production from p-coumarate by an engineered KT2440 carrying aroY and ecdBD (KT2440-CJ184) in a shake-flask culture using glucose as the carbon and energy source [14].
Growth of NGC703(pTS084) on S-lignin-derived phenols and ccMA production from G-lignin-derived phenols. NGC703(pTS084) cells were incubated in Wx medium containing 10 mM syringate plus 5 mM vanillate (a, b) or 10 mM syringaldehyde plus 5 mM vanillin (a, c). OD600 and concentrations of substrates and metabolites were periodically monitored. a Cell growth on syringate plus vanillate and syringaldehyde plus vanillin. b, c Concentrations of substrates and metabolites. 3-O-methylgallate was under the detection limit (0.5 μM). Each value is the average ± the standard deviation from three independent experiments
Optimization of ccMA production at flask level. a–d NGC703(pTS084) cells were incubated in 500 mL shake-flasks containing 50 mL (a), 100 mL (b), and 200 mL (c) of Wx medium with 10 mM syringate plus 5 mM vanillate. OD600 and concentrations of substrates and metabolites were monitored during incubation. a–c Concentrations of substrates and metabolites. d Cell growth in 50 mL, 100 mL, and 200 mL cultures. e ccMA production with repeated feeding of syringate and vanillate. NGC703(pTS084) cells were incubated in a 500 mL shake-flask using 200 mL of Wx medium by feeding 10 mM syringate plus 5 mM vanillate every 12 h for six times. 3-O-methylgallate was under the detection limit (0.5 μM). Each value is the average ± the standard deviation from three independent experiments
Finally, ccMA production from bNBL by NGC703(pTS084) was examined in a tube containing 5 mL Wx medium by feeding 0.24% or 0.6% bNBL (0.27/0.67 mM syringaldehyde, 0.023/0.059 mM syringate, 0.12/0.29 mM vanillin, and 0.018/0.044 mM vanillate) every 12 h for five times. ccMA was produced with titers of 58.4 mg/L (411 μM) and 144 mg/L (1013 μM), respectively, when feeding 0.24% and 0.6% bNBL (Fig. 5). Although the accumulation of PCA during ccMA production needs to be improved, this study also demonstrates the applicability of NGC703(pTS084) in glucose-free ccMA production from real woody biomass. Here, we employed bNBL for ccMA production as a model system. In future, combining the ccMA production system by this strain with more substantial lignin depolymerization techniques such as base-catalyzed depolymerization and formic-acid-induced hydrolytic depolymerization should be considered [10, 31].
ccMA production from bNBL. NGC703(pTS084) cells were incubated in a tube containing 5 mL Wx medium by feeding 0.24% bNBL (a, c) or 0.6% bNBL (b, c) every 12 h for five times. OD600 and concentrations of substrates and metabolites were monitored during incubation. a, b Concentrations of substrates and metabolites. 3-O-methylgallate was under the detection limit (0.5 μM). c Cell growth when feeding 0.24% bNBL and 0.6% bNBL. Each value is the average ± the standard deviation from three independent experiments
Conclusion
We isolated a novel platform bacterium, Pseudomonas sp. strain NGC7, and succeeded in constructing an engineered NGC7 which utilizes lignin-derived phenols for cell growth and ccMA production. Engineered NGC7 will prove to be a promising strain for glucose-free production of various value-added substances from lignin-derived phenols. For ccMA production, it is necessary to improve the decarboxylation activity and investigate the optimum culture conditions using a fed-batch bioreactor.
References
Abe T, Masai E, Miyauchi K, Katayama Y, Fukuda M (2005) A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J Bacteriol 187:2030–2037. https://doi.org/10.1128/JB.187.6.2030-2037.2005
Barton N, Horbal L, Starck S, Kohlstedt M, Luzhetskyy A, Wittmann C (2018) Enabling the valorization of guaiacol-based lignin: integrated chemical and biochemical production of cis,cis-muconic acid using metabolically engineered Amycolatopsis sp. ATCC 39116. Metab Eng 45:200–210. https://doi.org/10.1016/j.ymben.2017.12.001
Becker J, Kuhl M, Kohlstedt M, Starck S, Wittmann C (2018) Metabolic engineering of Corynebacterium glutamicum for the production of cis,cis-muconic acid from lignin. Microb Cell Fact 17:115. https://doi.org/10.1186/s12934-018-0963-2
Beckham GT, Johnson CW, Karp EM, Salvachua D, Vardon DR (2016) Opportunities and challenges in biological lignin valorization. Curr Opin Biotechnol 42:40–53. https://doi.org/10.1016/j.copbio.2016.02.030
Blatny JM, Brautaset T, Winther-Larsen HC, Karunakaran P, Valla S (1997) Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid 38:35–51. https://doi.org/10.1006/plas.1997.1294
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546. https://doi.org/10.1146/annurev.arplant.54.031902.134938
Burlage RS, Hooper SW, Sayler GS (1989) The TOL (pWW0) catabolic plasmid. Appl Environ Microbiol 55:1323
Cecil JH, Garcia DC, Giannone RJ, Michener JK (2018) Rapid, parallel identification of catabolism pathways of lignin-derived aromatic compounds in Novosphingobium aromaticivorans. Appl Environ Microbiol 84:e01185-18. https://doi.org/10.1128/AEM.01185-18
Crawford RL (1975) Novel pathway for degradation of protocatechuic acid in Bacillus species. J Bacteriol 121:531–536
Das A, Rahimi A, Ulbrich A, Alherech M, Motagamwala AH, Bhalla A, da Costa Sousa L, Balan V, Dumesic JA, Hegg EL, Dale BE, Ralph J, Coon JJ, Stahl SS (2018) Lignin conversion to low-molecular-weight aromatics via an aerobic oxidation-hydrolysis sequence: comparison of different lignin sources. ACS Sustain Chem Eng 6:3367–3374. https://doi.org/10.1021/acssuschemeng.7b03541
Harwood CS, Parales RE (1996) The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol 50:553–590
Jiménez JI, Miñambres B, García JL, Díaz E (2002) Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ Microbiol 4:824–841. https://doi.org/10.1046/j.1462-2920.2002.00370.x
Johnson CW, Abraham PE, Linger JG, Khanna P, Hettich RL, Beckham GT (2017) Eliminating a global regulator of carbon catabolite repression enhances the conversion of aromatic lignin monomers to muconate in Pseudomonas putida KT2440. Metab Eng Commun 5:19–25. https://doi.org/10.1016/j.meteno.2017.05.002
Johnson CW, Salvachua D, Khanna P, Smith H, Peterson DJ, Beckham GT (2016) Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab Eng Commun 3:111–119. https://doi.org/10.1016/j.meteno.2016.04.002
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9. https://doi.org/10.1093/nar/gkn201
Kamimura N, Masai E (2014) The protocatechuate 4,5-cleavage pathway: overview and new findings. In: Nojiri H, Tsuda M, Fukuda M, Kamagata Y (eds) Biodegradative bacteria: how bacteria degrade, survive, adapt, and evolve. Springer, Tokyo, pp 207–226. https://doi.org/10.1007/978-4-431-54520-0_10
Kasai D, Fujinami T, Abe T, Mase K, Katayama Y, Fukuda M, Masai E (2009) Uncovering the protocatechuate 2,3-cleavage pathway genes. J Bacteriol 191:6758–6768. https://doi.org/10.1128/JB.00840-09
Kasai D, Kamimura N, Tani K, Umeda S, Abe T, Fukuda M, Masai E (2012) Characterization of FerC, a MarR-type transcriptional regulator, involved in transcriptional regulation of the ferulate catabolic operon in Sphingobium sp. strain SYK-6. FEMS Microbiol Lett 332:68–75. https://doi.org/10.1111/j.1574-6968.2012.02576.x
Kasai D, Masai E, Miyauchi K, Katayama Y, Fukuda M (2005) Characterization of the gallate dioxygenase gene: three distinct ring cleavage dioxygenases are involved in syringate degradation by Sphingomonas paucimobilis SYK-6. J Bacteriol 187:5067–5074. https://doi.org/10.1128/JB.187.15.5067-5074.2005
Maruyama K, Ariga N, Tsuda M, Deguchi K (1978) Purification and properties of α-hydroxy-γ-carboxymuconic ε-semialdehyde dehydrogenase. J Biochem 83:1125–1134
Masai E, Katayama Y, Fukuda M (2007) Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotechnol Biochem 71:1–15
Masai E, Sasaki M, Minakawa Y, Abe T, Sonoki T, Miyauchi K, Katayama Y, Fukuda M (2004) A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J Bacteriol 186:2757–2765
Masai E, Shinohara S, Hara H, Nishikawa S, Katayama Y, Fukuda M (1999) Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J Bacteriol 181:55–62
Nogales J, Canales A, Jimenez-Barbero J, Garcia JL, Diaz E (2005) Molecular characterization of the gallate dioxygenase from Pseudomonas putida KT2440. The prototype of a new subgroup of extradiol dioxygenases. J Biol Chem 280:35382–35390
Nozaki M, Kagamiyama H, Hayaishi O (1963) Metapyrocatechase I. Purification, crystallization and some properties. Biochemische Zeitschrift 338:582–590
Payer SE, Marshall SA, Barland N, Sheng X, Reiter T, Dordic A, Steinkellner G, Wuensch C, Kaltwasser S, Fisher K, Rigby SEJ, Macheroux P, Vonck J, Gruber K, Faber K, Himo F, Leys D, Pavkov-Keller T, Glueck SM (2017) Regioselective para-carboxylation of catechols with a prenylated flavin dependent decarboxylase. Angew Chem Int Ed Engl 56:13893–13897. https://doi.org/10.1002/anie.201708091
Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, Langan P, Naskar AK, Saddler JN, Tschaplinski TJ, Tuskan GA, Wyman CE (2014) Lignin valorization: improving lignin processing in the biorefinery. Science 344:1246843. https://doi.org/10.1126/science.1246843
Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, Boerjan W (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem Rev 3:29–60. https://doi.org/10.1023/B:PHYT.0000047809.65444.a4
Ravi K, Garcia-Hidalgo J, Gorwa-Grauslund MF, Liden G (2017) Conversion of lignin model compounds by Pseudomonas putida KT2440 and isolates from compost. Appl Microbiol Biotechnol 101:5059–5070. https://doi.org/10.1007/s00253-017-8211-y
Rice P, Longden I, Bleasby A (2000) EMBOSS: the european molecular biology open software suite. Trends Genet 16:276–277
Rodriguez A, Salvachúa D, Katahira R, Black BA, Cleveland NS, Reed M, Smith H, Baidoo EEK, Keasling JD, Simmons BA, Beckham GT, Gladden JM (2017) Base-catalyzed depolymerization of solid lignin-rich streams enables microbial conversion. ACS Sustain Chem Eng 5:8171–8180. https://doi.org/10.1021/acssuschemeng.7b01818
Salvachúa D, Johnson CW, Singer CA, Rohrer H, Peterson DJ, Black BA, Knapp A, Beckham GT (2018) Bioprocess development for muconic acid production from aromatic compounds and lignin. Green Chem 20:5007–5019. https://doi.org/10.1039/c8gc02519c
Sarkanen KV, Ludwig CH (1971) Lignins: occurrence, formation, structure and reactions. Wiley, New York
Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73
Shikinaka K, Otsuka Y, Nakamura M, Masai E, Katayama Y (2018) Utilization of lignocellulosic biomass via novel sustainable process. J Oleo Sci 67:1059–1070. https://doi.org/10.5650/jos.ess18075
Sonoki T, Morooka M, Sakamoto K, Otsuka Y, Nakamura M, Jellison J, Goodell B (2014) Enhancement of protocatechuate decarboxylase activity for the effective production of muconate from lignin-related aromatic compounds. J Biotechnol 192 Pt A:71–77. https://doi.org/10.1016/j.jbiotec.2014.10.027
Sonoki T, Takahashi K, Sugita H, Hatamura M, Azuma Y, Sato T, Suzuki S, Kamimura N, Masai E (2017) Glucose-free cis, cis-muconic acid production via new metabolic designs corresponding to the heterogeneity of lignin. ACS Sustain Chem Eng 6:1256–1264. https://doi.org/10.1021/acssuschemeng.7b03597
Sugimoto K, Senda M, Kasai D, Fukuda M, Masai E, Senda T (2014) Molecular mechanism of strict substrate specificity of an extradiol dioxygenase, DesB, derived from Sphingobium sp. SYK-6. PLoS One 9:e92249. https://doi.org/10.1371/journal.pone.0092249
Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153:895–905. https://doi.org/10.1104/pp.110.155119
Vardon DR, Franden MA, Johnson CW, Karp EM, Guarnieri MT, Linger JG, Salm MJ, Strathmann TJ, Beckham GT (2015) Adipic acid production from lignin. Energy Environ Sci 8:617–628. https://doi.org/10.1039/c4ee03230f
White MD, Payne KA, Fisher K, Marshall SA, Parker D, Rattray NJ, Trivedi DK, Goodacre R, Rigby SE, Scrutton NS, Hay S, Leys D (2015) UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis. Nature 522:502–506. https://doi.org/10.1038/nature14559
Williams PA, Sayers JR (1994) The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation 5:195–217. https://doi.org/10.1007/BF00696460
Wu W, Dutta T, Varman AM, Eudes A, Manalansan B, Loque D, Singh S (2017) Lignin valorization: two hybrid biochemical routes for the conversion of polymeric lignin into value-added chemicals. Sci Rep 7:8420. https://doi.org/10.1038/s41598-017-07895-1
Xu Z, Lei P, Zhai R, Wen Z, Jin M (2019) Recent advances in lignin valorization with bacterial cultures: microorganisms, metabolic pathways, and bio-products. Biotechnol Biofuels. https://doi.org/10.1186/s13068-019-1376-0
Acknowledgements
We thank the Advanced Low Carbon Technology Development (ALCA) program, Japan Science and Technology Agency (JPMJAL1506) for funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
N.K, T.S, and E.M. are inventors on a patent related to this work. The authors declare that they have no other conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shinoda, E., Takahashi, K., Abe, N. et al. Isolation of a novel platform bacterium for lignin valorization and its application in glucose-free cis,cis-muconate production. J Ind Microbiol Biotechnol 46, 1071–1080 (2019). https://doi.org/10.1007/s10295-019-02190-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-019-02190-6