Abstract
Time-of-flight magnetic resonance angiography (TOF-MRA) is a non-contrast technique used to visualize neurovascular. However, manual reconstruction of the volume render (VR) by radiologists is time-consuming and labor-intensive. Deep learning-based (DL-based) vessel segmentation technology may provide intelligent automation workflow. To evaluate the image quality of DL vessel segmentation for automatically acquiring intracranial arteries in TOF-MRA. A total of 394 TOF-MRA scans were selected, which included cerebral vascular health, aneurysms, or stenoses. Both our proposed method and two state-of-the-art DL methods are evaluated on external datasets for generalization ability. For qualitative assessment, two experienced clinical radiologists evaluated the image quality of cerebrovascular diagnostic and visualization (scoring 0–5 as unacceptable to excellent) obtained by manual VR reconstruction or automatic convolutional neural network (CNN) segmentation. The proposed CNN outperforms the other two DL-based methods in clinical scoring on external datasets, and its visualization was evaluated by readers as having the appearance of the radiologists’ manual reconstructions. Scoring of proposed CNN and VR of intracranial arteries demonstrated good to excellent agreement with no significant differences (median, 5.0 and 5.0, P ≥ 12) at healthy-type scans. All proposed CNN image quality were considered to have adequate diagnostic quality (median scores > 2). Quantitative analysis demonstrated a superior dice similarity coefficient of cerebrovascular overlap (training sets and validation sets; 0.947 and 0.927). Automatic cerebrovascular segmentation using DL is feasible and the image quality in terms of vessel integrity, collateral circulation and lesion morphology is comparable to expert manual VR without significant differences.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Time-of-flight magnetic resonance angiography (TOF-MRA) is a non-contrast imaging technique of intracranial arteries, which is commonly used clinically to evaluate cerebral arterial disease [1]. Volume render reconstruction is the main post-processing technique, usually performed by experienced radiologists, allowing three-dimensional (3D) visualization of vascular structures to aid in early identification and diagnosis [2, 3]. However, as the number of TOF-MRA examinations increases, technicians are overwhelmed by time-consuming manual procedures. Consequently, there is growing interest in automated solutions for 3D visualization of intracranial arteries in clinical workflows, and deep learning (DL)-based automatic vessel segmentation may provide an effortless alternative. In addition, the 3D structure of vessels extracted by segmentation is a prerequisite for further digitization of cerebrovascular [4].
Recent studies of DL-based models have shown significant potential in various medical image-processing tasks [5,6,7]. Particularly, the convolutional neural network (CNN), one of the mainstream DL technologies, the design of CNN is inspired by the human visual system and is well suited for processing image tasks with local spatial correlations [8, 9]. U-Net [10], as a specifically CNN designed for segmentation tasks, has become the preferred method for object segmentation, including cerebrovascular segmentation, and has shown promising results in recent applications [11,12,13,14,15,16,17].
However, although these DL approaches can achieve high segmentation accuracy, it relies on a large amount of labeled data for model training [18]. Where labels refer to the intracranial artery contours hand-drawn by experts, which are also considered as the ground truth for segmentation. As an important component of CNN model construction and evaluation, labeling requires extreme time and labor-costly, so it is almost impossible to implement in large-scale studies.
Therefore, the lack of studies to verify the feasibility and accuracy of DL-based vessel segmentation models in external independent datasets with large sample sizes and comprehensive types [19,20,21]. This is one of the most important obstacles to the clinical application and development of automatic segmentation schemes in TOF-MRA [22, 23]. Assessment methods that combine expert image analysis with diagnosis for scoring may represent a new turning point in addressing the above barriers from a more clinical perspective [24,25,26,27].
This study proposed a method for extracting the 3D structure of intracranial arteries based on an attention mechanism and multi-level feature extraction, and compared and evaluated 3D U-Net [11] and 3D Brave-Net [12], which have performed well in the field of cerebrovascular segmentation in recent years [15]. A large external independent dataset of 408 subjects was used for qualitative evaluation. The dataset contains healthy subjects and the two most common cerebrovascular lesions: cerebral aneurysms and stenosis. By combining the visualization of the segmented vascular structure with manual VR, which is widely used in clinical practice, combined with clinical scoring results and diagnostic skills, the practical application of the proposed model was verified from a clinical perspective.
We expect that the developed model can be seamlessly integrated into the radiology workflow to realize the automatic segmentation process based on TOF-MRA images, which can not only achieve fine automatic visualization of cerebral arteries and save manual work steps, but also further promote the research progress of future automated quantitative analysis of vessels, and further improve the accuracy and efficiency of cerebral arterial disease diagnosis.
Methods
Patients
Data for this retrospective study were obtained from three institutions, in which a retrospective search of two independent radiology datasets from two tertiary hospitals identified TOF-MRA examinations developed between January 2007 and December 2023 for the assessment of intracranial artery-related status. Another academic dataset, collected by Centre of Advanced Studies and Innovation Lab (CASILab) and publicly available online (URL: https://marron.web.unc.edu/brain-artery-tree-data/) contains TOF-MRA images of 109 healthy volunteers from five age groups [3]. The Hilbert researchers randomly selected four scans from each age group and determined ground truth by manual recording of vessel contours and cross-validation by three radiologists [12]. Ultimately, a total of 20 scans with ground truth were included in this study.
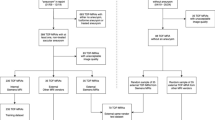
Demographic characteristics are detailed in Table 1. The overall experimental design and data flow of the workflow diagram is shown in Fig. 1.
MR Acquisition
The TOF-MRA scans involved in this study were performed on three different 3 T MR scanners with different imaging acquisition parameters, which were summarized in Table 2.
Segmentation Model
We implemented an integrated segmentation pipeline using a CNN architecture with an optimization strategy, as shown in Fig. 2. The addition of the CBAM module to the input and output parts of the network can increase the attention of the network to the target object while maintaining the U-shaped structure [28]. The MSFE module contains different convolutional kernel sizes that imply different perceptual field sizes, enabling the creation of feature maps containing convolutional features with different granularity, and feature concatenation implies the fusion of multi-scale features [29]. To ensure that the MSFE module performs well, we inserted it after each convolutional feature map with a pixel size of less than 20 [30]. The combination of 1D convolution and residual connection helps to achieve the fusion of each module and avoid possible degradation due to network upscaling.
Overview of the proposed CNN framework for automatic cerebrovascular segmentation methods. A Detailed flowchart of data preprocessing before model training and prediction. B The architecture of the proposed 3D CNN architecture. The deep learning convolutional neural network is trained on 64 × 64 × 8 voxel training patches from pre-processed images. Using 1 × 1 × 1 convolution as the last layer to calculate the DSC loss, and applied the sigmoid activation function to map the feature map to the segmentation probability map, then set the pixels with probability greater than 0.5 to 1 and the rest to 0, thus obtaining the final binary vessel segmentation results. Model’s learning rate of 0.0001 and maximum feasible batch size of 16. C Implementation foundation of advanced CBAM and MSFE technology. CBAM, convolutional machine attention mechanism; MSFE, multi-scale feature extraction; DSC, dice similarity coefficient; ReLU, rectified linear unit; Conv, convolutional layer; BN, batch normalization; AvgPool, average pooling; MaxPool, Max pooling
The network is trained by inputting 3D image patches and corresponding vessel labels, learning vascular features from TOF-MRA image data, and then predicting the probability that each pixel belongs to a vascular region. Finally, outputting a binary image of the spatial structure of cerebral arteries. To evaluate the performance advantage of our approach in recent DL-based vascular segmentation studies, we replicated two state-of-the-art CNN models, 3D U-Net [11] and 3D Brave-Net [12], as comparators. Figure 1 provides details of the data allocation during model training and testing, and all test data were obtained from independent external datasets.
Quantitative Metrics of Model Effectiveness
The performance of a vessel segmentation model can be evaluated from many different perspectives. To provide a broader understanding of the performance of our model, we report three different metrics: DSC, sensitivity, and accuracy. We use DSC to assess the spatial overlap between predictions and ground truth. Sensitivity is used to measure the ability to segment the vascular region. Accuracy represents the proportion of correct prediction results in the total predicted value.
Qualitative Clinical Assessment of Image Quality
To evaluate the diagnostic confidence of the CNN models for segmenting cerebral arteries, we conducted a multi-reader study. Chief physicians with 22 (S.Q.J.) and 20 (Y.H.L.) years of experience respectively reviewed the historical TOF-MRA scans of their tertiary hospitals, and randomly selected scans evaluated for different intracranial artery-related status (healthy, aneurysm, stenosis), which were then included in the reader study.
The image quality of 3D visualization in manually VR and automatic CNN was scored, with vessels displayed in the original TOF-MRA slice and MIP as the gold standard of reference. The clinically score was independently evaluated by two radiologists with experience in cerebrovascular imaging (T.Z., with 10 years of experience, and J.L., with 13 years of experience) according to diagnostic of cerebral arterial integrity, visualization of collateral circulations, and description of lesion morphology (types of aneurysms and stenosis). The readers independently graded the images on a five-point Likert-type score (5, excellent; 4, good; 3, acceptable; 2, poor; and 1, unacceptable) from the above three categories, where all categories score greater than 2 were defined as clinically acceptable (see Text, Supplemental Digital Content 1, which demonstrates scoring details). We also calculated the summed visual score (SVS) as the sum of scores [22].
The same TOF-MRA scans were independently reviewed by two readers (Y.L. and S.J.), and the scores of all samples were finally cross-validated to obtain a unified result, for mitigating the influence of subjective bias. If the two diverged, a third party (S.Q.J. and Y.H.L) to discuss until a consensus is reached.
As shown in Fig. 1, a total of three independent datasets were randomized and de-identified for visual scoring, with the reader having no knowledge of clinical history. Multiple datasets were used for the following reasons, firstly, as the data for model training was sourced from local dataset institution-1, 58 of 89 were randomly selected from the remaining scans for comparing the performance of the CNN models and manual VR. Then, the generalization performance of three CNN models and manual VR was tested on the external hospital-1 dataset. Finally, a further comparison between the proposed CNN and manual VR was conducted using the external hospital-2 dataset.
Statistical Analysis
The Shapiro-Wilk test was used to test the normal distribution of all indicators; median values and IQR were calculated for nonnormally distributed data. We use reader clinically scores to compare two types methods of cerebrovascular 3D visualization: CNN automatic segmentation and expert manual VR. For scores from the local dataset of institution-1 and the external dataset of hospitals-1, the Friedman test was applied to compare characteristics derived from 3D U-Net, 3D Brave-Net, proposed CNN, and VR (at hospitals-1 dataset). For the external hospitals-1 and hospitals-2 datasets, pairwise comparisons between proposed CNN and manual VR scores were performed by the Wilcoxon paired signed rank test. All statistical analyzes were performed by one researcher (Y.Q.M.) using Prism version 9. P < 0.05 was considered to indicate a statistically significant difference for all previously mentioned tests.
Results
Patient Characteristics
A total of 394 patients (mean age ± SD, 59 years ± 13; 141 males) were included in the final analysis (Table 1), which included 230, 79, and 85 TOF-MRA scans with three common clinical diagnoses of cerebral arterial: healthy, aneurysm, and stenosis, respectively.
Ablation Studies for Proposed Network
In this subsection, the contributions of the different components of the proposed network are evaluated. First, 20 TOF-MRA scans with ground truth labels were randomly divided into training, validation, and test sets in a 12:4:4 ratio to quantitatively evaluate the effects of CBAM and MSFE. The image quality of the segmentation results was then qualitatively assessed using independent data sets from hospitals. The segmentation performance of each model on the test data is summarized in Fig. 3.
Contribution of different components on segmentation performance of proposed network. A, B The comparison of TP, FP, FN, DSC, and sensitivity values for each model on 4 labeled test data. Where the TP values are based on subtracting 45,000 to show the difference between the models more significantly. A show the effect of each optimization method on the recognition ability of the blood vessels and background. The addition of the CBAM attention mechanism to the 3D U-Net backbone network can effectively improve the background recognition rate and reduce FP; however, the ability to distinguish vessels is inhibited. In contrast, the addition of the MSFE module has the opposite effect, even though TP is improved, and the probability of the background being mistaken for a vessel increases significantly, leading to an increase in FP. The proposed network model combines both optimization strategies to improve TP while suppressing the rise in FP and FN. C One representative visual example shown, yellow arrows indicate regions that are easily misidentified or segmented as vessels. TP, true positives; FP, false positive; FN, false negatives; DSC, dice similarity coefficient; CBAM, convolutional machine attention mechanism; MSFE, multi-scale feature extraction
The DSC value of the 3D U-Net-based extracted vessel mask is 88.1%. When the CBAM module is integrated into the U-Net backbone network, the DSC value increases to 88.7%, while the integration of the MSFE module increases the sensitivity value from 85.1 to 86.0% (Fig. 3B). Subsequently, the best sensitivity and the second highest DSC value were obtained for the proposed network. Although the number of FPs was slightly higher than other models, the proposed network was more in line with the clinical need to obtain the most complete vessels while controlling the number of FPs. Qualitative assessment of segmented vessel morphology in the hospital test data yielded results consistent with those described above (Fig. 3C). Due to the small size of the labeled dataset, the difference in the evaluation indicators between the networks is very small, but in terms of visualization, our model clearly reflects the excellent effect of vessel segmentation.
Quantitative Evaluation of Models’ Effectiveness
Twenty pairs of TOF-MRA scans from Institution-1 and corresponding ground-truth labels were included in the dice similarity coefficient (DSC) evaluation. The performance of three CNN models for automatic vessel segmentation achieved DSC of 0.937 vs 0.942 vs 0.947 and 0.922 vs 0.928 vs 0.927 (3D U-Net vs 3D Brave-Net vs proposed CNN) on the training sets and validation sets, respectively.
A total of 60,000 3D image patches extracted from the 20 TOF-MRA data were used for model training and validation. In addition, a 3D U-Net model and 3D Brave-Net were built separately in this study for comparison as existing state-of-the-art techniques. Figure 4 presents the evolution of the DSC for the three models on the training and validation sets, with all curves reaching convergence before the 15th epoch.
The validation DSCs of the 3D U-Net model, 3D Brave-Net, and proposed models were 0.922, 0.928, and 0.927, respectively, which initially shows the improvement achieved by both optimization models based on 3D U-Net. Proposed network achieved the highest accuracies of 0.9982 and 0.9974 for the training and validation datasets, respectively. It outperformed the original network (with accuracies of 0.9977 and 0.9972, respectively) and Context U-Net (with accuracies of 0.9980 and 0.9973, respectively).
Because the scarcity of vascular-labeled data resulted in a small test set that was insufficient to support a reliable qualitative assessment of the segmentation model performance, an independent, comprehensive, and hospital-based dataset was used to visually validate the segmented vessel of each model. Figure 5B shows two representative examples from the validation and test datasets. Proposed network outperforms the other two methods in terms of segmentation completeness for major vessels and side branches, whereas the 3D U-Net model exhibited a robust segmentation performance with good generalization capabilities. In contrast, from the segmentation results of Brave-Net, the model may overfit the training dataset in practical applications but with weak generalization ability.
Evaluation results showing the visualization performance of three CNN models on healthy intracranial arteries in local and external datasets. A Bar chart shows the score of artery score on three different CNN segmentation models. B Three-dimensional visualization of intracranial arteries acquired with three CNN models. There is no gap in the actual effect of all models on the local dataset, and the cerebrovascular are relatively complete. However, on the external dataset, except that the proposed CNN still maintains robust segmentation performance, the visualization results of the other two models lack most of the main vessels, which cannot meet the diagnostic needs. SVS: summed visual score
Clinical Assessment of Image Quality
As summarized in Table 3, the qualitative image quality scores of U-Net, Brave-Net, and proposed CNN to be of diagnostic quality (acceptable, good, or excellent) (P ≥ 0.20) on the entire local dataset institution-1. However, on the external comprehensive dataset hospitals-1, less than half of the U-Net and Brave-Net scoring results were considered diagnostically acceptable (25 of 50 [36%] vs 28 of 58 [40%]). The two readers considered 316 of 316 (100%) of scans processed by the proposed CNN and expert manual VR were rated as good to excellent (P ≥ 0.12) on the two external datasets. The clinical scoring results of three CNN models and manual VR on the three datasets are detailed below.
First, the local dataset institution-1 contains 78 TOF-MRA scans from healthy subjects, 58 of which are used for reader clinical scoring to evaluate the vascular image quality of the four methods. Median reader scores for two individual score of cerebrovascular integrity and collateral circulation and one visual total score of SVS were no significantly different between U-Net, Brave-Net, and proposed CNN (5.0 [IQR, 4.0–5.0]; P ≥ 0.02). This qualitatively assessment is consistent with the results of the DSC quantitative evaluation (Fig. 5). These 58 healthy-type scans processed by the three CNNs were diagnostically acceptable (two individual score > 2).
Then, we verified the practical application performance of each model and expert manual VR in the external comprehensive dataset hospitals-1, which contains 69 scans of cerebrovascular health, aneurysm, or stenosis type. Manual VR reconstruction commonly used in clinical practice. As shown in Fig. 5A, there are significant differences among the three CNN methods in the SVS and three individual scores of cerebrovascular: integrity, collateral circulation, and lesion morphology (P < 0.0001).
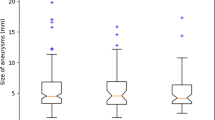
The U-Net and Brave-Net showed low performance of scores (median, 2.0 [IQR, 1.0–3.0]). A group comparison of the proposed CNN and manual VR showed no significant difference (P ≥ 0.12). Among them, the IQR range of proposed CNN is better than VR for the score of lesion morphology in patients with aneurysm (Fig. 6), although there was no significant difference (median, 5 [IQR, 5–5] vs 5 [IQR, 4–5], P = 0.12). Notably, the proposed CNN for automated segmentation and visualization of vessels was rated as having an expert manual VR reconstruction appearance (intraclass correlation coefficient, 0.992–0.993; P < 0.001).
MIP of raw images and segmented vessel of three model in the two healthy samples. The first column is the original TOF-MRA MIP image, the second column is U-Net, the third column is Brave-Net, and the fourth column is the proposed method. These two samples are from the training dataset (rows 1 to 5) and the independent dataset (rows 6 to 8), where poorly segmented regions are indicated by red arrows. In the training data, the left posterior cerebral artery (PCA), anterior cerebral artery (ACA) and middle cerebral artery (MCA) long-segment branch segments are missing in 3D U-Ne, and ACA and PCA branch segments are missing in Brave-Net. In independent external data, 3D U-Net missing bilateral vertebral arteries (VA), right MCA, and bilateral posterior inferior cerebellar arteries (PICA). Brave-Net is missing most blood vessels, the arrows are not used for graphic indication here. The proposed CNN is missing the left VA and MCA branches
Finally, to further compare our method with manual VR for clinical applicability, reader scores were incorporated into an external comprehensive database Hospitals-2. Scores from both techniques had the same median score and IQA in most scans (179 of 247 [72%]), and there were no significant differences between groups (P ≥ 0.07), except for SVS of aneurysm (median of CNN and VR, 15[IQA, 15–15] vs 15[IQA, 15–15]; P = 0.01) and SVS of stenosis (median of CNN and VR 15 [IQA, 14–15] vs 15 [IQA, 15–15], P = 0.02).
Discussion
There is growing interest in alternatives to expert manual VR reconstruction for cerebrovascular visualization. Our results demonstrate the feasibility of using DL to automatically segment and visualize cerebral vessels 3D structure from TOF-MRA scans, with a high cerebrovascular overlap (mean, 0.927), a high degree of diagnosis quality (median scores, 5 [IQA, 4–5]; P > 0.05) and a few seconds of scans processing time (mean seconds, 10 ± 3.7), the hardware platform used is detailed in the code environment on the last page. The proposed CNN has promising generalization applicability (No. of acceptable diagnoses (percentages), 25 [36%] vs 28 [40%] vs 69 [100%]), with reproduces good vessel segmentation performance (median scores range; [2, 3] vs [2, 3] vs [5]) on external datasets hospital-1. Furthermore, in our multi-reader study, image quality scores from the proposed CNN to be diagnostically acceptable on scans at two external datasets (316 of 316).
In the hospital-1 datasets, proposed CNN for automatic vessel segmentation and visualization were rated as having expert manual reconstruction of VR appearance (intraclass correlation coefficient, 0.992–0.993; P < 0.001). Unlike the pseudo-3D visualization of VR, the segmented cerebral vessels can be extracted their 3D structure from TOF-MRA data, which can provide a prerequisite for the subsequent automated measurement of morphological features, such as vessel radius, and can also assist in the intelligently assisted diagnosis of CVD [31], providing researchers with richer tools for cerebrovascular analysis [32].
However, there are some significant differences between proposed CNN and VR in the types of aneurysms and stenosis scans at hospital-2 datasets (P = 0.001 vs P = 0.02), which may be related to the differences in scanner imaging parameters and equipment in different centers, and the model also lacks adequate large training sets with labels of relevant patient types for training to enhance its own performance. Overall, the proposed CNN method did not demonstrate substantial over-or underestimation of cerebral vessels 3D structure, because the individual scores were also not significantly different in the case of patients TOF-MRA (P > 0.07). With the inclusion of larger quantity, more comprehensive, and higher quality labeled data in the future, there is still a lot of room for improvement in the performance of deep learning models.
Our study has several limitations. First, this is a model construction on a single type of small training set with 20 TOF-MRA scans, although each scan is cropped into 3000 small cubes with or without vessels, which limits the development of DL model performance. Second, for a proof-of-concept study, we focused on only three common types of cerebrovascular status diagnoses. Third, we did not consider the reality degree of the tertiary circulation in the collateral circulation score, because, in clinical diagnosis, this type of microvessel is of less concern than the other larger vessel. Fourth, we did not assess the relationship between vessel diameter and lesion level, whereas these quantitative parameters may serve as risk factors for vascular events [33]. As shown in Fig. 7, although we found that the diameter of the vessel presented by automatic vessel segmentation was closer to the vessel diameter of the original TOF-MRA slice than that of VR, but we did not obtain relevant parameters for further statistical analysis, because automated vessel parameter measurement has not been implemented, and this part is placed in the next step.
Cerebrovascular image quality comparison between the proposed CNN and manual VR. The first column is the original TOF-MRA MIP image, the second column is manual VR, and the third column is proposed methods. In VR, the collateral circulation was not fully reconstructed, while the CNN model successfully segmented and reconstructed relatively complete collateral circulation. It is used as one of the reference standards for raw image MIP evaluation. Red arrows indicate the location of the patient’s cerebrovascular abnormalities
In conclusion, this proof-of-concept study demonstrated the feasibility of using DL to automatically segment and visualize cerebral vessels in 3D from raw TOF-MRA scans. The proposed CNN automated method and expert manual VR showed comparable image quality for vessel integrity, collateral circulation, and lesion morphology, with few significant qualitative differences. Although the CNN automated approach is neither intended nor likely to replace all expert manual processing, it has the potential to expand the accessibility of TOF-MRA-rich automated processing by relieving the labor involved, whereas the potential role of CNN automatic segmentation on TOF-MRA medical images needs further research and verification such as cerebrovascular digitization and intelligent disease-assisted diagnosis. These have gradually emerged in the related applications of CT angiography medical images in recent years [3, 34].
Data Availability
The TOF-MRA scans data and manual labeling of vessel contours for model training in this study are available in http://doi.org/https://doi.org/10.5281/zenodo.3968844. The two hospital datasets used for external testing of the model is not openly accessible due to current data protection laws.
Abbreviations
- TOF-MRA:
-
Time-of-flight magnetic resonance angiography
- VR:
-
Volume render
- 3D:
-
Three-dimensional
- CNN:
-
Convolutional neural network
- DSC:
-
Dice similarity coefficient
- SVS:
-
Summed visual score
References
MacDonald M E, Frayne R. Cerebrovascular MRI: a review of state-of-the-art approaches, methods and techniques. NMR Biomed 2015; 28(7): 767-791. https://doi.org/10.1002/nbm.3322
Mallouhi A, Felber S, Chemelli A, et al. Detection and characterization of intracranial aneurysms with MR angiography: comparison of volume-rendering and maximum-intensity-projection algorithms [J]. American Journal of Roentgenology, 2003, 180(1): 55-64. https://doi.org/10.2214/ajr.180.1.1800055
Bullitt E, Zeng D, Gerig G, et al. Vessel tortuosity and brain tumor malignancy: a blinded study1 [J]. Academic radiology, 2005, 12(10): 1232-1240.
Fu F, Wei J, Zhang M, et al. Rapid vessel segmentation and reconstruction of head and neck angiograms using 3D convolutional neural network [J]. Nature communications, 2020, 11(1): 4829. https://doi.org/10.1038/s41467-020-18606-2
Suganyadevi S, Seethalakshmi V, Balasamy K. A review on deep learning in medical image analysis [J]. International Journal of Multimedia Information Retrieval, 2022, 11(1): 19-38.
Goni M R, Ruhaiyem N I R, Mustapha M, et al. Brain vessel segmentation using deep learning-a review [J]. IEEE Access, 2022. https://doi.org/10.1109/ACCESS.2022.3214987
Li H, Tang Z, Nan Y, et al. Human treelike tubular structure segmentation: A comprehensive review and future perspectives [J]. Computers in Biology and Medicine, 2022: 106241. https://doi.org/10.1016/j.compbiomed.2022.106241
Salehi A W, Khan S, Gupta G, et al. A study of CNN and transfer learning in medical imaging: Advantages, challenges, future scope [J]. Sustainability, 2023, 15(7): 5930.
Sarvamangala D R, Kulkarni R V. Convolutional neural networks in medical image understanding: a survey [J]. Evolutionary intelligence, 2022, 15(1): 1-22.
O RonnebergerischerBroxU-net: Convolutional networks for biomedical image segmentation[C]//Medical image computing and computer-assisted intervention–MICCAI, 201518th international conference, Munich, Germany, October 5–9, 2015, proceedings, part III 18 Springer International Publishing 2015 234 241
Livne M, Rieger J, Aydin O U, et al. A U-Net deep learning framework for high performance vessel segmentation in patients with cerebrovascular disease. Front Neurosci 2019, 13: 97. https://doi.org/10.3389/fnins.2019.00097
Hilbert A, Madai V I, Akay E M, et al. BRAVE-NET: fully automated arterial brain vessel segmentation in patients with cerebrovascular disease. Frontiers in artificial intelligence 2020, 78. https://doi.org/10.3389/frai.2020.552258
Du G, Cao X, Liang J, et al. Medical image segmentation based on u-net: A review. J Imaging Sci and Techn 2020, 64: 1–12. 1062–3701/2020/64(2)/020508/12
Yang C, Li Y, Bai Y, et al. SS-Net: 3D Spatial-Spectral Network for Cerebrovascular Segmentation in TOF-MRA[C]//International Conference on Artificial Neural Networks. Cham: Springer Nature Switzerland, 2023: 149–159.
Chen C, Zhou K, Wang Z, et al. All answers are in the images: A review of deep learning for cerebrovascular segmentation [J]. Computerized Medical Imaging and Graphics, 2023, 107: 102229.
Yang C, Zhang H, Chi D, et al. Contour attention network for cerebrovascular segmentation from TOF-MRA volumetric images [J]. Medical Physics, 2024, 51(3): 2020-2031.
Simon A B, Hurt B, Karunamuni R, et al. Automated segmentation of multiparametric magnetic resonance images for cerebral AVM radiosurgery planning: a deep learning approach [J]. Scientific Reports, 2022, 12(1): 786.
Sun C, Shrivastava A, Singh S, et al. Revisiting unreasonable effectiveness of data in deep learning era[C]//Proceedings of the IEEE international conference on computer vision. 2017: 843–852.
Sanchesa P, Meyer C, Vigon V, et al. Cerebrovascular network segmentation of MRA images with deep learning[C]//2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019). IEEE, 2019: 768-771. https://doi.org/10.1109/ISBI.2019.8759569
Y Wang G Yan H Zhu JointVesselNet: Joint volume-projection convolutional embedding networks for 3D cerebrovascular segmentation[C]//Medical Image Computing and Computer Assisted Intervention–MICCAI, et al 2020 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part VI 23 Springer International Publishing 2020 106 116https://doi.org/10.1007/978-3-030-59725-2_11
Chen Y, Jin D, Guo B, et al. Attention-assisted adversarial model for cerebrovascular segmentation in 3D TOF-MRA volumes [J]. IEEE Transactions on Medical Imaging, 2022, 41(12): 3520-3532. https://doi.org/10.1109/TMI.2022.3186731
Yu A C, Mohajer B, Eng J. External validation of deep learning algorithms for radiologic diagnosis: a systematic review [J]. Radiology: Artificial Intelligence, 2022, 4(3): e210064. https://doi.org/10.1148/ryai.210064.
Moccia S, De Momi E, El Hadji S, et al. Blood vessel segmentation algorithms—review of methods, datasets and evaluation metrics [J]. Computer methods and programs in biomedicine, 2018, 158: 71-91. https://doi.org/10.1016/j.cmpb.2018.02.001.
Haji-Valizadeh H, Shen D, Avery R J, et al. Rapid reconstruction of four-dimensional MR angiography of the thoracic aorta using a convolutional neural network [J]. Radiology: Cardiothoracic Imaging, 2020, 2(3): e190205. https://doi.org/10.1148/ryct.2020190205.
Foreman S C, Neumann J, Han J, et al. Deep learning–based acceleration of Compressed Sense MR imaging of the ankle [J]. European Radiology, 2022, 32(12): 8376-8385. https://doi.org/10.1007/s00330-022-08919-9
Chung M, Calabrese E, Mongan J, et al. Deep learning to simulate contrast-enhanced breast MRI of invasive breast cancer [J]. Radiology, 2022, 306(3): e213199. https://doi.org/10.1148/radiol.213199.
Shi Z, Zhao X, Zhu S, et al. Time-of-Flight Intracranial MRA at 3 T versus 5 T versus 7 T: Visualization of Distal Small Cerebral Arteries. Radiology 2022: 220114. https://doi.org/10.1148/radiol.220114
Woo S, Park J, Lee J Y, et al. CBAM: Convolutional block attention module. Proceedings of the European conference on computer vision (ECCV) 2018, 3–19. dio: 0.1007/978–3–030–01234–2_1
Szegedy C, Ioffe S, Vanhoucke V, et al. Inception-v4, inception-resnet and the impact of residual connections on learning. Thirty-first AAAI conference on artificial intelligence 2017. https://doi.org/10.48550/arXiv.1602.07261
He F, Liu T, Tao D. Why resnet works? residuals generalize [J]. IEEE transactions on neural networks and learning systems, 2020, 31(12): 5349-5362.
Friedman J I, Tang C Y, de Haas H J, et al. Brain imaging changes associated with risk factors for cardiovascular and cerebrovascular disease in asymptomatic patients. JACC: cardiovascular imaging 2014; 7(10): 1039–1053. https://doi.org/10.1016/j.jcmg.2014.06.014
Yin X, Ye G, Wang D. Middle cerebral artery tortuosity associated with aneurysm development [J]. Journal of neurosurgery, 2018, 130(5): 1763-1764. https://doi.org/10.3171/2018.7.JNS181688
Gutierrez J, Cheung K, Bagci A, et al. Brain arterial diameters as a risk factor for vascular events [J]. Journal of the American Heart Association, 2015, 4(8): e002289. https://doi.org/10.1161/JAHA.115.002289
Yang Y, Huan X, Guo D, et al. Performance of deep learning-based autodetection of arterial stenosis on head and neck CT angiography: an independent external validation study [J]. La radiologia medica, 2023: 1–13. https://doi.org/10.1007/s11547-023-01683-w.
Funding
This work was supported by the Natural Science Foundation of Shanghai (Grant No. 20ZR1438300), the Key Program of the National Natural Science Foundation of China (Grant No.81830052), the Science and Technology Innovation Action Plan of Shanghai (Grant No.18441900500), Shanghai Key Laboratory of Molecular Imaging (Grant No. 18DZ2260400).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Consent to Participant
All study participants provided informed consent, and the study design was approved by the appropriate ethics review board. We have read and understood your journal’s policies, and we believe that neither the manuscript nor the study violates any of these.
Code Environment
The environment configurations used in this experiment is a 64-bit Windows 10 Professional operating system, Intel Xeon E5-2673 CPU 2.4 GHz processor, NVIDIA Ge Force GT 11 GB X GPU, using PyCharm 2020, programming environment and language is Python 3.6, and deep learning framework is TensorFlow-GPU 2.2.0. The time required to segment a TOF-MRA scans is about a few seconds.
Institutional Review Board
Institutional Review Board of two tertiary hospitals approved the study in compliance with the Health Insurance Circulation and Accountability Act and waived the written informed consent requirement.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Min, Y., Li, J., Jia, S. et al. Automated Cerebrovascular Segmentation and Visualization of Intracranial Time-of-Flight Magnetic Resonance Angiography Based on Deep Learning. J Digit Imaging. Inform. med. (2024). https://doi.org/10.1007/s10278-024-01215-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10278-024-01215-6