Abstract
Early, accurate diagnosis of neurodegenerative dementia subtypes such as Alzheimer’s disease (AD) and frontotemporal dementia (FTD) is crucial for the effectiveness of their treatments. However, distinguishing these conditions becomes challenging when symptoms overlap or the conditions present atypically. Resting-state fMRI (rs-fMRI) studies have demonstrated condition-specific alterations in AD, FTD, and mild cognitive impairment (MCI) compared to healthy controls (HC). Here, we used machine learning to build a diagnostic classification model based on these alterations. We curated all rs-fMRIs and their corresponding clinical information from the ADNI and FTLDNI databases. Imaging data underwent preprocessing, time course extraction, and feature extraction in preparation for the analyses. The imaging features data and clinical variables were fed into gradient-boosted decision trees with fivefold nested cross-validation to build models that classified four groups: AD, FTD, HC, and MCI. The mean and 95% confidence intervals for model performance metrics were calculated using the unseen test sets in the cross-validation rounds. The model built using only imaging features achieved 74.4% mean balanced accuracy, 0.94 mean macro-averaged AUC, and 0.73 mean macro-averaged F1 score. It accurately classified FTD (F1 = 0.99), HC (F1 = 0.99), and MCI (F1 = 0.86) fMRIs but mostly misclassified AD scans as MCI (F1 = 0.08). Adding clinical variables to model inputs raised balanced accuracy to 91.1%, macro-averaged AUC to 0.99, macro-averaged F1 score to 0.92, and improved AD classification accuracy (F1 = 0.74). In conclusion, a multimodal model based on rs-fMRI and clinical data accurately differentiates AD-MCI vs. FTD vs. HC.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Differentiating between Alzheimer’s disease (AD) and frontotemporal dementia (FTD) remains a significant clinical challenge. Current standard procedures for diagnosing different subtypes of dementia depend mostly on patient clinical history, cognitive assessments, and neuropsychological tests while structural neuroimaging studies (MRI) are also routinely performed when available [1]. However, overlapping symptoms, early onset of AD, or atypical presentations and disease courses make accurate diagnosis using such tools more challenging [1,2,3]. For example, a considerable subset of behavioral-variant FTD patients may show memory deficits similar to AD [4, 5], while AD patients may atypically present with poor executive functioning that may even exceed that of behavioral-variant FTD (bvFTD) [6, 7] and show less marked memory impairment [8].
The accurate distinction of dementia subtypes has important implications on all facets of patient management. One aspect pertains to the administration of pharmacological and psychological care [9]. For instance, acetylcholine-esterase inhibitors demonstrate modest yet discernible cognitive improvements in Alzheimer’s disease (AD) patients, while exhibiting no such effects in frontotemporal dementia (FTD) patients [10, 11]. On the other hand, intranasal oxytocin has shown efficacy in ameliorating neuropsychiatric symptoms in FTD cases, though its efficacy in AD remains underexplored [11, 12]. More generally, the distribution of associated neuropsychiatric conditions varies between FTD and AD, necessitating tailored care strategies [3]. Furthermore, with the advent of disease-modifying treatments such as the recent FDA approval of lecanemab for early-stage AD [13], early and precise diagnosis of specific dementia subtypes has become more important than ever before as treatments increasingly target underlying disease etiologies rather than nonspecific symptoms.
Another aspect concerns the varying progressions and prognoses of dementia subtypes. For example, FTD patients, particularly those with the ALS variant, experience more rapid progression and shorter life expectancies compared to other subtypes [14, 15]. Differences between dementia subtypes should also be considered when evaluating the heritability risk of these conditions. Up to 50% of FTD cases may have a hereditary component (particularly associated with MAPT gene), and an autosomal dominant pattern of inheritance can be identified in up to 20% of the patients. However, the hereditary component is less significant in AD, with fewer than 5% of cases showing such a component, primarily due to mutations in the PSEN1 and PSEN2 genes [16]. Lastly, the projected threefold increase in the worldwide population of individuals living with dementia—from about 57 million in 2019 to an estimated 153 million by 2050—further highlights the escalating impact of this health concern and the necessity of achieving precise diagnoses as the foundation for effective disease management [17].
Given the significance and complexity of diagnosing dementia subtypes, investigators and clinical trial sponsors attempting to develop new treatments for these conditions often find it necessary to employ additional diagnostic techniques such as positron-emission tomography (PET) or cerebrospinal fluid analysis to achieve a homogeneous and accurately diagnosed patient cohort [13, 18]. Even though these novel methods can indeed diagnose various dementia subtypes, sometimes even before the presentation of clinical signs and symptoms, their cost and time-intensive nature have hindered their integration into routine clinical practice and pose significant financial and temporal burdens on research studies [18]. Therefore, there is a pressing need to develop automated diagnostic procedures with high accuracy that could simplify clinical research studies and potentially evolve into routine clinical diagnostic techniques in the future.
The power of machine learning models to recognize the complex patterns and relationships characteristic of biomedical data is well known [19]. Consequently, it is unsurprising that several studies have attempted to utilize machine learning methods on various clinical and paraclinical data to build tools that complement the diagnostic process for dementia. These data sources encompass demographic information, clinical presentation, past medical history, results of neuropsychological assessments, lab biomarkers, and findings from structural and functional (PET) imaging [20,21,22,23,24,25,26]. Surprisingly, however, none of these studies has leveraged data from resting-state fMRI (rs-fMRI) scans to train their models. This is despite previous reports indicating condition-specific alterations in rs-fMRI signals in dementia [27, 28]. For instance, impairment of connectivity in the default mode network in AD patients, impairment of the salience network in bvFTD patients, and an increase in default mode network connectivity in bvFTD patients have been consistently reported [29]. Furthermore, conducting an rs-fMRI study is more cost-effective and less time-consuming than a PET study, and unlike PET, rs-fMRIs can be safely repeated (e.g., in follow-up studies for assessing disease progression) since the technique does not utilize radioactive isotopes [30]. The significance of this lies in the important role that PET plays in current attempts to definitively diagnose dementia subtypes [18] and that PET images have already been used to create machine learning models to classify different dementia subtypes, albeit with limited accuracy [26].
Another shared characteristic among the previous studies employing machine learning for diagnosing dementia subtypes is that none of them built models that simultaneously classified AD, mild cognitive impairment (MCI), FTD, and healthy controls (HC). Furthermore, the studies that classified FTD focused mainly on bvFTD, resulting in the underrepresentation of other clinical subsets of FTD (primary progressive aphasias [semantic and nonfluent-variants]) even though they may constitute up to 28% of the FTD patient population [31].
Considering these areas for improvement, in this study, we have utilized resting-state fMRI (rs-fMRI) data to build a multi-class classification model that could simultaneously identify HC, MCI, AD, and FTD patients. In addition, while we do not separately classify the different subtypes of FTD, we have included data from all subtypes of FTD in our FTD class. Unlike previous studies on rs-fMRIs, however, we did not follow the commonly used pathway of functional connectivity analysis and used raw, time-course data instead. Utilization of different approaches and techniques for functional connectivity analysis (e.g., graph-theory network analysis, independent component analysis, seed-based analysis) makes the reproducibility of findings difficult and might lead to divergent conclusions, while analyzing raw (time-course) data may be more conducive to achieving reproducibility and widespread use [32].
As described in the “Methods” section, we compared three different relatively interpretable machine learning algorithms to choose the model structure for our study. We opted for relatively interpretable methods since many of the most powerful machine learning models, especially those utilizing deep learning, are viewed as black boxes due to the difficulty in interpreting their decision-making process [33]. Given that machine learning models are unlikely to be perfect, interpretability and easily understandable visualization of the decision-making process by the model is key in any application of machine learning to medicine [19]. In our experiment for model selection, gradient-boosted decision trees (XGBoost) showed superior classification performance compared to the two other algorithms. This was not unexpected as XGBoost is widely regarded as the state-of-the-art in numerous machine learning tasks involving tabular data, frequently outperforming deep learning models [34]. Moreover, XGBoost strikes a balance within the continuum of machine learning algorithms, offering the ability to robustly extract nonlinear relationships while maintaining relative interpretability in its decision-making process. Hence, we used XGBoost to create the models for our study.
Methods
Databases and Imaging (fMRI)
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (adni.loni.usc.edu) and the Frontotemporal Lobar Degeneration Neuroimaging Initiative (FTLDNI) databases. The ADNI was launched in 2003 as a public-private partnership led by Principal Investigator Michael W. Weiner, MD. FTLDNI launched in 2010 under the leadership of Dr. Howard Rosen, MD, at the University of California, San Francisco, with funding from the National Institute of Aging. For up-to-date information regarding these databases, please see www.adni-info.org and http://memory.ucsf.edu/research/studies/nifd. This study was exempt from IRB review due to the public availability of ADNI and FTLDNI and the strict deidentification of data within them.
According to the ADNI acquisition protocol, resting-state fMRI data were acquired with a gradient echo planar imaging (EPI) sequence (TR = 3000 ms; TE = 30 ms; matrix = 64 × 64; flip angle = 80°; voxel size = 3.313 mm × 3.313 mm × 3.313 mm; 48 slices) on a 3 T Philips scanner [35]. FTLDNI was launched based on the infrastructure established by ADNI and shares similar acquisition protocols.
This study used all the available rs-fMRIs in these databases (1351 fMRI scans [AD, 101; FTD, 396; HC, 470; MCI, 384] obtained from 434 patients [AD, 32; FTD, 151; HC from ADNI, 51; HC from FTLDNI, 96; MCI, 103]) and their linked clinical data.
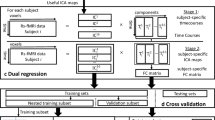
Preprocessing of Imaging Data
Resting-state fMRI data from the ADNI and FTLDNI databases were preprocessed using CONN [36] (RRID:SCR_009550) release 20.b and SPM [37] (RRID:SCR_007037) release 12.7771. Functional and anatomical data were preprocessed using CONN’s automated preprocessing pipeline. Then, the functional data was denoised using CONN’s standard denoising pipeline. The details of these pipelines are presented in Online Resource 1.
After this step, whole-brain gray matter was parcellated into 200 regions of interest (ROIs) based on a voxel-scale functional connectivity parcellation atlas by Schaefer et al. [38]. The time course of each ROI was expressed as the first eigenvariate of the processed time series and averaged across all voxels in the ROI [38, 39].
After completing CONN preprocessing and extracting time courses, our dataset still contained data from the 1351 fMRI scans described before. In preparation for our analysis, we proceeded to exclude the fMRI scans with more than 50% of their volumes identified as outliers (e.g., due to excessive motion artifacts) by CONN’s preprocessing pipelines, leaving 1084 scans for further analysis. Of the 267 excluded scans, 11 (11%), 133 (34%), 86 (18%), and 37 (10%) were from the AD, FTD, HC, and MCI classes, respectively. The number of patients remained the same after excluding scans with a disproportionately high percentage of outlier volumes. A summary of the clinical characteristics associated with the remaining scans is presented in Table 1. Finally, scans with more than 140-time points were truncated so that the final form of the time-course data became a 1084 * 200 * 140 array, corresponding to data recorded from 200 brain parcels (time series channels) over 140-time points in 1084 scans.
It is important to note that many patients in ADNI and FTLDNI underwent multiple rs-fMRI scans over a span of several years. In our final dataset, all patients with repeated scans retained their initial diagnosis (MCI, AD, FTD, HC) in subsequent studies and no instances of progression from MCI to AD were recorded. Consequently, scans from the same patient would be expected to have a higher correlation than those from different patients. High correlation between data elements in training and test data poses the risk of overfitting and artificial inflation of model performance metrics, respectively.
As explained later, we chose the XGBoost algorithm to construct our models. XGBoost incorporates regularization both in the objective function that it optimizes and by virtue of being an ensemble of weak learners [40], enabling it to counter the overfitting challenge. Concerning the issue of potentially inflated metrics, we present the unseen test-set metrics before and after excluding repeat scans (retaining only the initial scan for each patient) from the test sets in the models discussed in the text.
Feature Extraction
Following these steps, the time series data underwent feature extraction. To this end, we used a number of features with relevance to time series data from the tsfresh package in Python [41]. The features that were calculated from the data are presented in Table 2. This fMRI features dataset had 1084 rows (for the 1084 scans) and 75,400 columns (features that were calculated).
Clinical and Demographic Data
We attempted to extract the clinical and demographic variables associated with the imaging studies to bolster the fMRI data. We identified the following variables to be present and similarly measured in both ADNI and FTLDNI databases: date of birth (enabling the calculation of age), sex, education, Mini-Mental State Examination (MMSE) total score, Clinical Dementia Rating (CDR) total score, forward and backward memory span tests, Boston Naming Test (BNT) score, letter verbal fluency test score, the 15-item Geriatric Depression Scale (GDS) total score, and Functional Activities Questionnaire scores. Throughout the paper, we will refer to these variables as clinical (instead of clinical and demographic) variables. We used an algorithm in R to assign values from neuropsychological tests to imaging studies if performed within 1 year of the fMRI scan. The forward and backward memory span tests were excluded from the data due to a very high percentage (> 60%) of missing data. All other variables had ≤ 25% missing data. However, the Functional Activities Questionnaire scores were also excluded due to the high discrepancy of data missing rate between the two databases, with 40% of FTD scans not having associated Functional Activities Questionnaire scores (FTLDNI). In contrast, less than 3.4% of AD scans did not have these results (ADNI).
Model Selection
To select the optimal machine learning model for our study, we compared three well-known and relatively interpretable methods: multinomial logistic regression (LR) and decision trees (DT) from the scikit-learn module and gradient-boosted decision trees from the XGBoost (Extreme Gradient Boosting) module in Python.
To compare the three models, we used a fivefold cross-validation procedure where the data was split into five folds with nearly identical percentages of all classes based on patient IDs (i.e., the set divisions were performed on the patients, not the fMRI scans). Even though the final classifications were performed on each individual fMRI scan (regardless of the patient it belonged to) and not the patients, this was done to prevent the inclusion of fMRI scans from a single patient in both sets, thus preventing information leakage and bias in model performance metrics. In each round of the CV, one of the folds (~ 20% of the scans) was used as the unseen hold-out test set, while the remaining four folds (~ 80% of the scans) were used as the train set. We did not perform hyperparameter optimization in this experiment as we intended to compare baseline model performances. As shown in Tables 1 and 2 in Online Resource 1, the XGBoost model achieved significantly better metrics compared to the LR and DT models. As a result, we used and optimized the XGBoost model for the remainder of this study.
A notable feature of XGBoost is its built-in regularization, making it suitable for the overfitting challenge posed by the repeated scans in our dataset. The ensemble nature of XGBoost contributes to its generalizability and robustness against overfitting [42]. Furthermore, XGBoost primarily optimizes the following objective function:
where \(n\) is the number of training examples, \({y}_{{\text{i}}}\) is the true target value for the \(i{\text{th}}\) example, \({\widehat{y}}_{i}^{t}\) is the predicted value in the \(t{\text{th}}\) iteration, \(L\) is the loss function, \(m\) is the number of trees (or boosting rounds) in the ensemble, \({g}_{t}\) is the \(m{\text{th}}\) tree in the ensemble, and \(\Omega \left({g}_{t}\right)\) is the regularization term applied to each tree. [42] \(\Omega \left({g}_{t}\right)\) itself is given by the following formula:
where \(\gamma\) is a parameter that controls the overall complexity of the tree, \({\rm T}\) is the number of leaves in the tree \({g}_{{\text{t}}}\), \({\omega }_{{\text{j}}}\) is the weight associated with the \(j{\text{th}}\) leaf of the tree, \({\lambda }_{1}\) is the regularization parameter controlling the strength of the L1 (Lasso) penalty, and \({\lambda }_{2}\) is the regularization parameter controlling the strength of the L2 (Ridge) penalty. In this study, we found no enhancements in model performance when incorporating L1 and L2 penalties. As a result, \({\lambda }_{1}\) and \({\lambda }_{2}\) were set to zero and we only optimized the \(\gamma\) hyperparameter.
Nested K-Fold Cross-Validation Procedure
To build the classification models, we used stratified, nested k-fold cross-validation (CV). The nested approach involves a two-layered technique with an external and an internal CV process. This approach enables model hyperparameter optimization while preventing model overfitting to the data and the presentation of overly optimistic metrics [43]. The external CV process was conducted identically to the model selection experiment described above and had five rounds. The internal CV was performed on the train set (and not on the whole dataset) created in each round of the external CV and was used to optimize model hyperparameters. Furthermore, variable selection in the imaging features data, imputation of missing values in clinical data, and standardization of the clinical data and selected imaging features were performed based on the train set defined by the external CV process. These steps ensured that no information leakage occurred between the train and unseen test sets during each external CV round.
For variable selection, the imaging features data were used to create an initial classification model on the train set using XGBoost [40]. Then, the model was used to determine variable importances based on the average improvement (gain) in the training set loss attributed to each feature. These importance values were normalized to achieve a unit sum, and the features with an importance of at least 0.01 or higher were selected. A summary of the selected features based on their corresponding cortical regions, including the number of times they were selected during the external CV process and their average ranks in variable importance across the folds, is presented in Table 3. A more complete analysis, including summaries specifically based on brain region or mathematical feature, is presented in Online Resource 2.
After this step, missing values in clinical data were imputed. The mean (for quantitative variables) and mode (for patient sex, the only qualitative variable in the clinical data) were calculated from the external CV round’s train set and were used to replace missing values in both the train and test sets. The imaging features data did not require any imputation as all values were available. Next, the clinical data and the selected features were standardized. In each round, means and standard deviations for each variable were calculated from the external CV’s train set and used to standardize that variable’s values in both the train and test sets.
Finally, the internal CV process within the train set of each external CV round was used to create that round’s final classification model with the preprocessed selected imaging features and clinical variables. For the internal CV, the RandomizedSearchCV function from the scikit-learn module was used to perform a threefold CV and find the optimal set of model hyperparameters. The final optimized model was then used to generate predictions and performance metrics on the external CV round’s unseen test set. Furthermore, the relative importances of the features fed into each model were calculated and are presented in Online Resource 4.
Reported Metrics and Statistical Analysis
In this paper, we report the mean and 95% confidence interval for each metric as calculated from the unseen hold-out test sets of the five external cross-validation rounds.
The calculated metrics include balanced accuracy (with class-balanced sample weights according to the inverse prevalence of each target class), area under the receiver operating characteristic curve (AUC), sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), F1 score, positive likelihood ratio (LR+), negative likelihood ratio (LR−), and area under the precision-recall (PPV-sensitivity) curve (AUC-PR). We used balanced accuracy instead of accuracy due to the imbalanced class proportions in our dataset. For balanced accuracy, F1 score, AUC, and AUC-PR, two sets of metrics were calculated: one-vs-rest metrics for each class (turning the multi-class classification into a binary classification) and a macro-averaged metric (unweighted average of the metric between all classes to maintain the influence of infrequent classes) to represent the overall performance of that model.
Ultimately, the performance metrics of the models were compared using t-tests. In case of multiple testing (pairwise comparisons of the imaging features-only, best clinical-only, best combined clinical and imaging features, imaging features + CDR, and imaging features + MMSE models), the reported p-values are adjusted for multiple testing using the Bonferroni method. The reported effect sizes were calculated using Cohen’s D method.
All analyses in this study were performed using the pandas (version 1.5.3), numpy (version 1.25.2), scipy (version 1.11.4), researchpy (version 0.3.6), scikit-learn (version 1.2.2), and XGBoost (version 1.7.6) modules in Python (version 3.10.12). All figures were created using the matplotlib (version 3.7.1), seaborn (version 0.13.1), and dtreeviz (version 2.2.2) modules in Python and Inkscape (version 1.3.2).
Results
Model with fMRI Features Data
The fMRI features data were fed into the XGBoost algorithm to create a classification model. As explained in the “Methods” section, the features underwent a selection process before being used to build the final classification model. This model achieved a mean balanced accuracy of 74.4% (95% CI, 72.1–76.7%) and average classification accuracy of 99.2% (95% CI, 97.9–100.00%) in FTD, 99.0% (95% CI, 98.3–99.7%) in HC, and 93.9% (95% CI, 88.3–99.5%) in MCI scans in the unseen test sets of the five external CV rounds. However, the model only exhibited an average accuracy of 5.5% (95% CI, 0.0–15.8%) in classifying AD scans, misclassifying the rest as MCI (Fig. 1). These classification accuracies were reflected in the model’s F1 scores: 0.08 (95% CI, 0.00–0.21) in AD, 0.99 (95% CI, 0.99–1.00) in FTD, 0.99 (95% CI, 0.99–1.00) in HC, and 0.86 (95% CI, 0.82–0.89) in MCI scans; and 0.73 (95% CI, 0.69–0.77) overall (macro-averaged). Exclusion of the repeat scans from the unseen test sets did not affect the model’s metrics (overall balanced accuracy: 74.6% [95% CI, 71.2–78.0%; t(8) = 0.30, p = 0.770, d = 0.19]; overall F1 score: 0.73 [95% CI, 0.69–0.78; t(8) = 0.00, p = 1.000, d = 0.00]). The complete set of metrics and feature importances for this model may be viewed in Online Resource 4.
XGBoost classification model using selected features from fMRI scans. a Confusion matrix of the model’s predictions vs ground truth. b One-vs-Rest classification ROC curves and their corresponding AUC values of individual classes and their unweighted averages (macro-average). AD, Alzheimer’s disease; FTD, frontotemporal dementia; HC, healthy control; MCI, mild cognitive impairment; OvR, one-versus-rest
Model with Clinical Data
Next, we used all available clinical data to create a classification model. This model reached 70.6% (95% CI, 63.6–77.7%) balanced accuracy (Fig. 2a) in the unseen test sets. The F1 scores were 0.68 (95% CI, 0.60–0.76), 0.79 (95% CI, 0.73–0.85), 0.68 (95% CI, 0.56–0.80), and 0.72 (95% CI, 0.61–0.83) for the AD-, FTD-, HCI-, and MCI-scan-associated clinical data, respectively, and 0.72 (95% CI, 0.62–0.81) overall. In terms of variable importance, the CDR score was ranked first in all cross-validation rounds (average variable importance = 0.29, average rank = 1.2), followed by education (0.20, 2.2), MMSE score (0.17, 2.8), letter verbal fluency test score (0.11, 3.8), BNT score (0.06, 5.8), age (0.06, 6.2), sex (0.06, 6.8), and GDS (0.05, 7.2). Exclusion of the repeat scans from the unseen test sets yielded significantly better metrics (overall balanced accuracy: 74.5% [95% CI, 69.2–79.9%; t(8) = 2.73, p = 0.026, d = 1.73]; overall F1 score: 0.76 [95% CI, 0.72–0.81; t(8) = 2.36, p = 0.046, d = 1.49]).
XGBoost classification models using clinical and demographic information. a Confusion matrix and One-vs-Rest classification ROC curves of the model built using all clinical variables (included variables: age, sex, education, CDR score, MMSE score, BNT score, letter verbal fluency test score, and GDS score). b Confusion matrix and One-vs-Rest classification ROC curves of the best model built using clinical variables (included variables: CDR score, MMSE score, BNT score, letter verbal fluency test score, and GDS score). AD, Alzheimer’s disease; BNT, Boston Naming Test; CDR, Clinical Dementia Rating; FTD, frontotemporal dementia; GDS, 15-item Geriatric Depression Scale; HC, healthy control; MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination; OvR, one-versus-rest
Furthermore, we assessed the performance of models built with all possible combinations (subsets) of clinical variables (Online Resource 3). Of these models, the best performance was achieved using CDR score, MMSE score, BNT score, letter verbal fluency test score, and GDS score. As shown in Fig. 2b, a model using these variables achieved 71.6% (95% CI, 64.9–78.2%) balanced accuracy in the unseen test sets. Exclusion of the repeat scans from the unseen test sets yielded significantly better overall balanced accuracy: 75.9% (95% CI, 68.5–83.4%; t(8) = 2.67, p = 0.028, d = 1.69). The complete set of metrics for these models may be viewed in Online Resource 4.
Model with Combined Imaging and Clinical Information
In pursuit of better model performance, we built an XGBoost classifier using all the available clinical variables and fMRI features. The balanced accuracy for this model was 90.4% (95% CI, 87.6–93.2%) in the unseen test sets (Fig. 3a). Exclusion of the repeat scans from the unseen test sets did not affect overall balanced accuracy: 90.6% (95% CI, 87.0–94.2%; t(8) = 0.27, p = 0.792, d = 0.17).
XGBoost classification models using a combination of clinical and imaging data. a Confusion matrix and One-vs-Rest classification ROC curves of the model built using fMRI features and all clinical variables. b Confusion matrix and One-vs-Rest classification ROC curves of the model built using fMRI features and all clinical features except for age. AD, Alzheimer’s disease; CDR, Clinical Dementia Rating; FTD, frontotemporal dementia; HC, healthy control; MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination; OvR, one-versus-rest
Interestingly, a slightly higher balanced accuracy in the test sets was achieved when combining all clinical variables except for age with imaging features (balanced accuracy = 91.1% [95% CI, 87.1–95.1%]). This model was our best-performing model and had an average accuracy of 98.8% (95% CI, 96.5–100.0%) for FTD, 98.8% (95% CI, 97.2–100.0%) for HC, and 95.3% (95% CI, 90.2–100.0%) for MCI scans. Once again, the lowest class-specific accuracy was observed in AD scans, where the accuracy was only 71.6% (95% CI, 56.8–86.4%), with the remaining scans being misclassified as MCI. The F1 scores were 0.74 (95% CI, 0.62–0.87), 0.99 (95% CI, 0.98–1.00), 0.99 (95% CI, 0.98–1.00), and 0.93 (95% CI, 0.90–0.97) for the AD-, FTD-, HCI-, and MCI-scan-associated clinical data, respectively, and 0.92 (95% CI, 0.87–0.96) overall (Fig. 3b). Exclusion of the repeat scans from the unseen test sets did not affect the metrics (overall balanced accuracy: 91.1% [95% CI, 87.1–95.1%; t(8) = 0.00, p = 1.000 d = 0.00]; overall F1 score: 0.92 [95% CI, 0.88–0.95; t(8) = 0.00, p = 1.000 d = 0.00]). The complete set of metrics and feature importances for these models may be viewed in Online Resource 4.
The balanced accuracy, F1 score, and AUC of the best combined model were significantly better than their corresponding values from both the best clinical (t(8) = 15.45, p < 0.001, d = 9.77; t(8) = 12.34, p < 0.001, d = 7.80; t(8) = 21.73, p < 0.001, d = 13.74) and the imaging features-only (t(8) = 14.15, p < 0.001, d = 22.37; t(8) = 19.59, p < 0.001, d = 12.39; t(8) = 20.69, p < 0.001, d = 13.09) models.
Minimizing the Number of Variables Used in the Combined Model
To find the most minimal model with acceptable accuracy, we built XGBoost classifiers using all combinations (subsets) of clinical variables and fMRI features. The balanced accuracy and AUC metrics (as calculated in the unseen tests of the external CV rounds) for a selection of these models are presented in Table 4. The metrics for all the models may be viewed in Online Resource 3.
For example, a model using a smaller subset of clinical variables (education, MMSE score, CDR score, and BNT score) achieved very similar metrics (balanced accuracy = 91.1% [95% CI, 87.6–94.6%]) to the best-performing model mentioned in the previous section. On the other hand, the smallest model with a mean balanced accuracy above 90% only used the CDR score in combination with imaging features (balanced accuracy = 90.7% [95% CI, 84.2–97.2%]) (Fig. 4a). Exclusion of the repeat scans from the unseen test sets did not affect overall balanced accuracy: 89.3% (95% CI, 83.1–95.4%; t(8) = −0.97, p = 0.360, d = 0.61). In addition, the balanced accuracy and F1 score metrics of the imaging features + CDR model were not significantly different from the best combined model (t(8) = 0.00, p = 1.000, d = 0.00; t(8) = 0.00, p = 1.000, d = 0.00) but its AUC was significantly lower (t(8) = −4.14, p = 0.032, d = −2.62).
Minimizing the number of clinical variables used in the XGBoost classification models. a Confusion matrix and One-vs-Rest classification ROC curves of the model built using fMRI features and CDR score. b Confusion matrix and One-vs-Rest classification ROC curves of the model built using fMRI features and MMSE score. AD, Alzheimer’s disease; CDR, Clinical Dementia Rating; FTD, frontotemporal dementia; HC, healthy control; MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination; OvR, one-versus-rest
Furthermore, while achieving slightly lower metrics, simpler models which might be more practical in the clinical setting (regarding the time and expertise required to gather the clinical information) still demonstrated acceptable performance. For example, the model using sex, education, and MMSE score alongside imaging features achieved a balanced accuracy of 89.1% (95% CI, 84.7–93.5%). Similarly, the model using only the MMSE score alongside imaging features achieved a balanced accuracy of 88.7% (95% CI, 86.3–91.1%) (Fig. 4b). Exclusion of the repeat scans from the unseen test sets did not affect the overall balanced accuracy (88.7% [95% CI, 85.1–92.3%]; t(8) = 0.00, p = 1.000, d = 0.00) of this model. In addition, the balanced accuracy, F1 score, and AUC metrics of the imaging features + MMSE model were not significantly different from the best combined model ((t(8) = − 2.63, p = 0.301, d = 1.66; t(8) = 2.06, p = 0.731, d = 1.30; t(8) = 0.00, p = 1.000, d = 0.00). Figure 5 shows one of the estimator trees in this model and the complete set of metrics for the models shown in Fig. 4 and their feature importances may be viewed in Online Resource 4.
A representative decision tree from the fMRI features + MMSE model. The final model for this round of cross-validation is an aggregate of 400 trees similar to the one shown in the figure. The number 400 was derived during hyperparameter optimization. AD, Alzheimer’s disease; FTD, frontotemporal dementia; HC, healthy control; MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination
Discussion
In this study, we have developed a multimodal machine learning model aimed at classifying three subtypes of dementia (MCI, AD, FTD) along with healthy controls. This model exhibits a balanced accuracy of ~90% when tested on previously unseen data. It utilizes an average of 15 features extracted from rs-fMRI time course data, combined with minimal clinical information (only the MMSE or CDR scores). One exciting aspect of our findings was that the model built using only fMRI features accurately classified FTD, HC, and MCI scans. However, it encountered challenges in identifying AD scans, with nearly all being misclassified as MCI. This is in line with previous studies, which had shown significant differences between the rs-fMRIs of FTD and AD [28], while rs-fMRIs from MCI patients were shown to have similar alterations to AD patients but only of a lower magnitude [44]. Nevertheless, the low classification accuracy of the AD group may also stem from the comparatively fewer scans in this class compared to the other classes. This explanation is supported by the high variability (manifested as wide confidence intervals) of AD classification accuracies across the cross-validation rounds.
The features selected using XGBoost’s variable importance measure may also provide valuable insights (Table 3). For example, the most consistently selected features belonged to the left visual cortex. Functional connectivity changes in the visual networks and occipital cortex may indeed be early differentiators of FTD (particularly, bvFTD) and AD [28]. Alongside the left visual cortex, features from the prefrontal cortices in both hemispheres were frequently selected. Functional connectivity studies have consistently demonstrated dysfunctions in the anterior (ventromedial, anteromedial, and dorsal prefrontal cortex) default mode network (DMN) in the brains of AD and MCI patients [45]. These alterations can differentiate AD from healthy patients [46] and the decline in DMN connectivity is associated with MCI to AD progression [47]. Decreased coherent activity in the dorsal prefrontal component of the anterior DMN has also been observed in behavioral variant FTD [48]. The temporal and somatomotor cortices were also common among the selected features. Alterations of temporal lobe activity and functional connectivity are widespread in both FTD [49] and AD [50]. Somatomotor cortex alterations possibly underlie the motor dysfunctions observed in AD, even during simple tasks in its early stages [51, 52], and contribute to motor signs in FTD [53].
Adding clinical information to the rs-fMRI features helped the model better classify AD and MCI scans. In our study, we were limited by the clinical tests that were similarly available in ADNI and FTLDNI, and future studies may show that tests other than MMSE or CDR better complement the fMRI features data. For example, the short form of the Montreal Cognitive Assessment (MoCA) would be an interesting candidate because it has a similar sensitivity to the full version and takes less time to perform than either MMSE or CDR in the clinic [54].
In conclusion, our study demonstrates that a multimodal model based on features derived from rs-fMRI time course data along with minimal clinical information offers an automated and highly accurate approach for classifying AD-MCI vs. HC vs. FTD. However, it is important to acknowledge the limitations that impact our study and conclusions. The class imbalance within our dataset, particularly the relatively fewer scans in the AD class, and the greater proportion of repeat scans in the AD and MCI classes compared to the FTD class may have constrained the model’s ability to differentiate AD and MCI scans. Another limitation of our model is that XGBoost’s enhanced performance relative to traditional decision trees comes at the cost of decreased transparency. While XGBoost effectively quantifies the overall significance of each feature in its classifications, the randomness inherent in the boosting process complicates the comprehension of how each specific feature influences the model’s prediction in each instance of classification. Finally, although we utilized data from two separate databases (ADNI and FTLDNI) to train our model, both were predominantly curated by the same teams, employing identical equipment and protocols at the same institutions. Therefore, to assess the broader generalizability of our model, it is essential to test it on entirely external datasets containing rs-fMRIs obtained from different machines by diverse teams in future studies. Future research could also delve deeper into the neural correlates of the rs-fMRI features that were selected using XGBoost’s variable importance measure and elucidate the physiological relevance of the brain parcels from which these features were derived.
Data Availability
All data used in this study were obtained from the ADNI and FTLDNI databases. Access to these databases is contingent on adherence to their respective data use agreements and publications policies outlined on their corresponding web pages on the LONI Image and Data Archive (IDA) website (https://ida.loni.usc.edu/). The final time course data extracted from the fMRI scans obtained from these databases is available from the corresponding author on reasonable request for researchers whose data access has been approved by ADNI and FTLDNI.
Abbreviations
- AD:
-
Alzheimer’s disease
- ADNI:
-
Alzheimer’s Disease Neuroimaging Initiative
- BNT:
-
Boston Naming Test
- CDR:
-
Clinical Dementia Rating
- CV:
-
Cross-validation
- FTD:
-
Frontotemporal dementia
- FTLDNI:
-
Frontotemporal Lobar Degeneration Neuroimaging Initiative
- GDS:
-
15-Item Geriatric Depression Scale
- HC:
-
Healthy control
- MCI:
-
Mild cognitive impairment
- MMSE:
-
Mini-Mental State Examination
- PET:
-
Positron-emission tomography
- rs-fMRI:
-
Resting-state functional magnetic resonance imaging
References
Arvanitakis Z, Shah RC, Bennett DA: Diagnosis and Management of Dementia: Review. JAMA 322:1589-1599, 2019
Sutovsky S, et al.: Clinical accuracy of the distinction between Alzheimer's disease and frontotemporal lobar degeneration. Bratisl Lek Listy 115:161-167, 2014
Musa G, et al.: Alzheimer's Disease or Behavioral Variant Frontotemporal Dementia? Review of Key Points Toward an Accurate Clinical and Neuropsychological Diagnosis. J Alzheimers Dis 73:833-848, 2020
Pennington C, Hodges JR, Hornberger M: Neural correlates of episodic memory in behavioral variant frontotemporal dementia. J Alzheimers Dis 24:261-268, 2011
Bertoux M, et al.: So Close Yet So Far: Executive Contribution to Memory Processing in Behavioral Variant Frontotemporal Dementia. J Alzheimers Dis 54:1005-1014, 2016
Reul S, Lohmann H, Wiendl H, Duning T, Johnen A: Can cognitive assessment really discriminate early stages of Alzheimer's and behavioural variant frontotemporal dementia at initial clinical presentation? Alzheimers Res Ther 9:61, 2017
Ossenkoppele R, et al.: The behavioural/dysexecutive variant of Alzheimer's disease: clinical, neuroimaging and pathological features. Brain 138:2732-2749, 2015
Tellechea P, et al.: Early- and late-onset Alzheimer disease: Are they the same entity? Neurologia (Engl Ed) 33:244-253, 2018
Raamana PR, Rosen H, Miller B, Weiner MW, Wang L, Beg MF: Three-Class Differential Diagnosis among Alzheimer Disease, Frontotemporal Dementia, and Controls. Front Neurol 5:71, 2014
Seibert M, et al.: Efficacy and safety of pharmacotherapy for Alzheimer's disease and for behavioural and psychological symptoms of dementia in older patients with moderate and severe functional impairments: a systematic review of controlled trials. Alzheimers Res Ther 13:131, 2021
Huang M-H, et al.: Treatment Efficacy of Pharmacotherapies for Frontotemporal Dementia: A Network Meta-Analysis of Randomized Controlled Trials. The American Journal of Geriatric Psychiatry 31:1062-1073, 2023
Michaelian JC, et al.: Pilot Randomized, Double-Blind, Placebo-Controlled Crossover Trial Evaluating the Feasibility of an Intranasal Oxytocin in Improving Social Cognition in Individuals Living with Alzheimer's Disease. J Alzheimers Dis Rep 7:715-729, 2023
van Dyck CH, et al.: Lecanemab in Early Alzheimer's Disease. N Engl J Med 388:9-21, 2023
Kansal K, et al.: Survival in Frontotemporal Dementia Phenotypes: A Meta-Analysis. Dement Geriatr Cogn Disord 41:109-122, 2016
El-Wahsh S, et al.: Predictors of survival in frontotemporal lobar degeneration syndromes. Journal of Neurology, Neurosurgery & Psychiatry 92:425-433, 2021
Hinz FI, Geschwind DH: Molecular genetics of neurodegenerative dementias. Cold Spring Harbor perspectives in biology 9:a023705, 2017
Nichols E, et al.: Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: an analysis for the Global Burden of Disease Study 2019. The Lancet Public Health 7:e105-e125, 2022
Dokholyan NV, Mohs RC, Bateman RJ: Challenges and progress in research, diagnostics, and therapeutics in Alzheimer's disease and related dementias. Alzheimers Dement (N Y) 8:e12330, 2022
Goecks J, Jalili V, Heiser LM, Gray JW: How Machine Learning Will Transform Biomedicine. Cell 181:92-101, 2020
Moguilner S, et al.: Multi-feature computational framework for combined signatures of dementia in underrepresented settings. Journal of Neural Engineering 19:046048, 2022
Garcia-Gutierrez F, et al.: Diagnosis of Alzheimer's disease and behavioural variant frontotemporal dementia with machine learning-aided neuropsychological assessment using feature engineering and genetic algorithms. Int J Geriatr Psychiatry 37, 2021
García-Gutierrez F, et al.: GA-MADRID: design and validation of a machine learning tool for the diagnosis of Alzheimer's disease and frontotemporal dementia using genetic algorithms. Med Biol Eng Comput 60:2737-2756, 2022
Maito MA, et al.: Classification of Alzheimer's disease and frontotemporal dementia using routine clinical and cognitive measures across multicentric underrepresented samples: A cross sectional observational study. Lancet Reg Health Am 17, 2023
Gurevich P, Stuke H, Kastrup A, Stuke H, Hildebrandt H: Neuropsychological Testing and Machine Learning Distinguish Alzheimer's Disease from Other Causes for Cognitive Impairment. Front Aging Neurosci 9:114, 2017
Grueso S, Viejo-Sobera R: Machine learning methods for predicting progression from mild cognitive impairment to Alzheimer’s disease dementia: a systematic review. Alzheimer's Research & Therapy 13:162, 2021
Perovnik M, et al.: Automated differential diagnosis of dementia syndromes using FDG PET and machine learning. Front Aging Neurosci 14:1005731, 2022
Ibrahim B, et al.: Diagnostic power of resting-state fMRI for detection of network connectivity in Alzheimer's disease and mild cognitive impairment: A systematic review. Hum Brain Mapp 42:2941-2968, 2021
Hafkemeijer A, et al.: Resting state functional connectivity differences between behavioral variant frontotemporal dementia and Alzheimer's disease. Front Hum Neurosci 9:474, 2015
Hohenfeld C, Werner CJ, Reetz K: Resting-state connectivity in neurodegenerative disorders: Is there potential for an imaging biomarker? NeuroImage: Clinical 18:849–870, 2018
Raimondo L, et al.: Advances in resting state fMRI acquisitions for functional connectomics. NeuroImage 243:118503, 2021
Logroscino G, et al.: Incidence of Syndromes Associated With Frontotemporal Lobar Degeneration in 9 European Countries. JAMA Neurology 80:279-286, 2023
Lv H, et al.: Resting-State Functional MRI: Everything That Nonexperts Have Always Wanted to Know. AJNR Am J Neuroradiol 39:1390-1399, 2018
Zhang Y, et al.: Opening the black box: interpretable machine learning for predictor finding of metabolic syndrome. BMC Endocrine Disorders 22:214, 2022
Shwartz-Ziv R, Armon A: Tabular data: Deep learning is not all you need. Information Fusion 81:84-90, 2022
Jack CR, Jr., et al.: The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging 27:685-691, 2008
Whitfield-Gabrieli S, Nieto-Castanon A: Conn: A Functional Connectivity Toolbox for Correlated and Anticorrelated Brain Networks. Brain Connectivity 2:125-141, 2012
Penny W, Friston K, Ashburner J, Kiebel S, Nichols T: Statistical Parametric Mapping: The Analysis of Functional Brain Images, 2007
Schaefer A, et al.: Local-Global Parcellation of the Human Cerebral Cortex from Intrinsic Functional Connectivity MRI. Cerebral Cortex 28:3095-3114, 2017
Luna LP, et al.: Resting-state fMRI functional connectivity and clinical correlates in Afro-descendants with schizophrenia and bipolar disorder. Psychiatry Research: Neuroimaging 331:111628, 2023
Chen T, Guestrin C: XGBoost: A Scalable Tree Boosting System, San Francisco, California, USA: Association for Computing Machinery, 2016
Christ M, Braun N, Neuffer J, Kempa-Liehr AW: Time Series FeatuRe Extraction on basis of Scalable Hypothesis tests (tsfresh – A Python package). Neurocomputing 307:72-77, 2018
Khan AA, Chaudhari O, Chandra R: A review of ensemble learning and data augmentation models for class imbalanced problems: Combination, implementation and evaluation. Expert Systems with Applications 244:122778, 2024
Wainer J, Cawley G: Nested cross-validation when selecting classifiers is overzealous for most practical applications. Expert Systems with Applications 182:115222, 2021
Binnewijzend MA, et al.: Resting-state fMRI changes in Alzheimer's disease and mild cognitive impairment. Neurobiol Aging 33:2018-2028, 2012
Tao W, et al.: Inflection Point in Course of Mild Cognitive Impairment: Increased Functional Connectivity of Default Mode Network. Journal of Alzheimer's Disease 60:679-690, 2017
Chen G, et al.: Classification of Alzheimer disease, mild cognitive impairment, and normal cognitive status with large-scale network analysis based on resting-state functional MR imaging. Radiology 259:213-221, 2011
Li Y, et al.: Abnormal Resting-State Functional Connectivity Strength in Mild Cognitive Impairment and Its Conversion to Alzheimer's Disease. Neural Plast 2016:4680972, 2016
Caminiti SP, et al.: Affective mentalizing and brain activity at rest in the behavioral variant of frontotemporal dementia. Neuroimage Clin 9:484-497, 2015
Reyes P, et al.: Functional Connectivity Changes in Behavioral, Semantic, and Nonfluent Variants of Frontotemporal Dementia. Behavioural Neurology 2018:9684129, 2018
Schwab S, et al.: Functional Connectivity Alterations of the Temporal Lobe and Hippocampus in Semantic Dementia and Alzheimer's Disease. J Alzheimers Dis 76:1461-1475, 2020
Vidoni ED, Thomas GP, Honea RA, Loskutova N, Burns JM: Evidence of altered corticomotor system connectivity in early-stage Alzheimer's disease. J Neurol Phys Ther 36:8-16, 2012
Whitwell JL: Atypical clinical variants of Alzheimer's disease: are they really atypical? Front Neurosci 18:1352822, 2024
Ferreri F, Pauri F, Pasqualetti P, Fini R, Dal Forno G, Rossini PM: Motor cortex excitability in Alzheimer's disease: a transcranial magnetic stimulation study. Ann Neurol 53:102-108, 2003
McDicken JA, et al.: Accuracy of the short-form Montreal Cognitive Assessment: Systematic review and validation. Int J Geriatr Psychiatry 34:1515-1525, 2019
Schreiber T, Schmitz A: Discrimination power of measures for nonlinearity in a time series. Physical Review E 55:5443-5447, 1997
Batista GEAPA, Keogh EJ, Tataw OM, de Souza VMA: CID: an efficient complexity-invariant distance for time series. Data Mining and Knowledge Discovery 28:634–669, 2014
Acknowledgements
Data collection and sharing for this project was funded by the Frontotemporal Lobar Degeneration Neuroimaging Initiative (National Institutes of Health Grant R01 AG032306). The study is coordinated through the University of California, San Francisco, Memory and Aging Center. FTLDNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Furthermore, data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Author information
Authors and Affiliations
Consortia
Contributions
Licia Luna conceptualized the study. Rohan Sanghera, Daniel Stevens, Richard Dagher, Licia Luna, and Mohammad Amin Sadeghi curated and prepared the data. Mohammad Amin Sadeghi, Shinjini Kundu, and Licia Luna designed the methodology of the study. Mohammad Amin Sadeghi implemented the computer code and visualized the results. Mohammad Amin Sadeghi, Daniel Stevens, and Licia Luna wrote the original draft of the manuscript. Shinjini Kundu, Vivek Yedavalli, Craig Jones, Haris Sair, and Licia Luna reviewed and edited the text. Licia Luna supervised the project.
Corresponding author
Ethics declarations
Ethics Approval
This study was exempt from IRB review due to the public availability of ADNI and FTLDNI and the strict deidentification of data within them.
Consent to Participate
The data used in this study was from the ADNI and FTLDNI databases which we obtained from the Laboratory of Neuroimaging at the University of Southern California. Data access was subject to data use agreements with ADNI and FTLDNI, both of which had obtained written informed consent forms from their participants regarding their participation in the studies and the use of their deidentified data in future studies by qualified investigators for research purposes. The contents of this manuscript were approved by these organizations before submission to this journal.
Consent for Publication
The data used in this study was from the ADNI and FTLDNI databases which we obtained from the Laboratory of Neuroimaging at the University of Southern California. Data access was subject to data use agreements with ADNI and FTLDNI, both of which had obtained written informed consent forms from their participants regarding the use of their deidentified data in future studies for research and scientific publication by qualified investigators. The contents of this manuscript were approved by these organizations before submission to this journal.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
For Alzheimer’s Disease Neuroimaging Initiative: Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wpcontent/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
For Frontotemporal Lobar Degeneration Neuroimaging Initiative: Data used in preparation of this article were obtained from the Frontotemporal Lobar Degeneration Neuroimaging Initiative (FTLDNI) database. The investigators at FTLDNI contributed to the design and implementation of FTLDNI and/or provided data, but did not participate in analysis or writing of this report (unless otherwise listed). The FTLDNI investigators included the following individuals: Howard Rosen; University of California, San Francisco (PI) Bradford C. Dickerson; Harvard Medical School and Massachusetts General Hospital Kimoko Domoto-Reilly; University of Washington School of Medicine David Knopman; Mayo Clinic, Rochester Bradley F. Boeve; Mayo Clinic Rochester Adam L. Boxer; University of California, San Francisco John Kornak; University of California, San Francisco Bruce L. Miller; University of California, San Francisco William W. Seeley; University of California, San Francisco Maria-Luisa Gorno-Tempini; University of California, San Francisco Scott McGinnis; University of California, San Francisco Maria Luisa Mandelli; University of California, San Francisco
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sadeghi, M.A., Stevens, D., Kundu, S. et al. Detecting Alzheimer’s Disease Stages and Frontotemporal Dementia in Time Courses of Resting-State fMRI Data Using a Machine Learning Approach. J Digit Imaging. Inform. med. (2024). https://doi.org/10.1007/s10278-024-01101-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10278-024-01101-1