Abstract
Major hepatectomy causes a risk of postoperative liver dysfunction, failure, and infections like surgical site infection. Preoperative assessment of the liver volume and function of the remnant liver is a mandatory prerequisite before performing such surgery. The aim of this work is to develop and test a software application for evaluation of the residual function of the liver prior to the intervention of the surgeons. For this purpose, a technique for evaluation of liver volume from computed tomography (CT) images has been developed. Furthermore, the methodology algorithms were implemented and incorporated within a software tool with three basic functionalities: volume determination based on segmentation of liver from CT images, virtual tumour resection and estimation of the residual liver function and 3D visualisation. Forty-one sets of abdominal CT images consisting of different number of tomographic slice images were used to test and evaluate the proposed approach. Volumes that were obtained after manual tracing by two surgeon experts showed a relative difference of 3.5 %. The suggested methodology was encapsulated within an application with user-friendly interface that allows surgeons interactively to perform virtual tumour resection, to evaluate the relative residual liver and render the final result. Thereby, it is a tool in the surgeons’ hands that significantly facilitates their duties, saves time, and allows them to objectively evaluate the situation and take the right decisions. At the same time, the tool appears to be appropriate educational instrument for virtual training of young surgeon specialists.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Background
Liver resection is related to surgical removal of part of the liver due to liver or metastatic colorectal cancers. Major hepatectomy causes a risk of postoperative liver dysfunction, failure and infections like surgical site infection [1]. It has been shown that the incidence of severe hepatic dysfunction and infection following liver resection increased significantly with smaller percentage residual liver volume [2]. Among patients with normal liver, the limit for safe resection ranges from 20 to 30 % future remnant liver of total liver volume [3].
In spite of improvements in surgical and postoperative management, the parameters determining how much liver can be resected are still largely undefined. Literature review shows that preoperative assessment of the liver volume and function of the remnant liver is a mandatory prerequisite before major hepatectomy. This process is related to applying various methods for the extraction and evaluation of the liver volume, as well as the volume of the remnant liver. One approach is to perform manually tracing of the liver boundaries. However, this is time consuming and to save time, methods that are based on various algorithms (local filtering, snake and balloon methods, region-growing, etc.) have been applied to segment either fully- or semi-automatically the liver volume using computed tomography (CT) images. Although many scientific groups focus their research on this problem, still the automatic liver segmentation is unresolved and open for further investigations. There are two primary factors that influence the results and the failure of the developed approaches. The first factor is that the absorption characteristics of the liver are very close to these of the neighbour organs like the kidneys, the spleen, the pancreas, etc. This results in X-ray images with unclear liver boundaries in respect to the neighbouring structures and therefore low contrast and blurry edges. The second factor is the huge variability of livers’ shape and size among the patients. This causes difficulties in setting the parameters of segmentation algorithms, which are based on priority knowledge about shape and size of the liver and therefore fail to produce correct results when non-standard shapes are present [4, 5]. Both factors complicate the design of robust segmentation procedure.
Amongst the methods used in liver segmentation, much popularity have received those ones based on grey value evaluation. The simplest one is the simple thresholding. In this approach, the image is partitioned into pixels of two or more values through comparison of pixel values with a value that is predefined. However, this method may not be practical and effective in the case of liver, because the liver intensity differs according to the patient slice and the CT machine. An improved approach includes the application of adaptive thresholding through computing the histogram of the CT image and searching for pixels with intensity that are within a certain margin within the intensity range of the liver region to accommodate any variation in the liver region pixels [6]. Similar method includes segmentation of liver from CT images by a supervised thresholding followed by k-means clustering [7–10]. Lots of iterative morphological filtering operations and k-means clustering are performed. Both approaches are followed by morphological processing to preserve the liver structure and remove the fragments of other organs using the conditions like area and location of liver. Basic weakness of these methods is that the liver grey level estimation does not take into account the high variability of this estimated value observed both in the same and in other CT volumes. The use of texture features extracted from different areas of the CT image like fractal dimensions and entropy may be useful for further liver volume refining. Another technique that demonstrated good segmentation results and performs well with respect to the noise in the images is the region growing algorithm [11]. The basic algorithm and its modifications have been found useful in many image processing applications in the medical domain [12–18]. Several more modifications are needed for the case of the liver in order to face some shortcomings for liver segmentation. One such example is the 3D region growing technique implemented by Campadelli et al. [19] that takes into account both the edge and the grey value information. However, this technique was tested on abdominal contrast-enhanced CT images and reduced image size (256 × 256 pixels) in order to expedite the computational time. In case of patients with kidney failure or diabetes, the use of contrast material would provoke complications. Although new contrast agents are developed, the most safety liver inspection for these patients is by the use of CT without contrast. For these cases, further refinement of region-growing methods and tests are needed.
At present, estimation of the liver volume at the Department of Surgery at the University Hospital of Varna is accomplished by an expert radiologist. The procedure requires the radiologist to manually segment the liver areas on each slice from the acquired CT scan and taking into account the number of pixels in the selected region and the pixel dimensions to calculate the liver volume. This usually takes up to 1 h. However, radiologists cannot replace the surgeon. In many cases, the surgeon needs to perform evaluation of the residual volume himself and therefore to make a prognosis for the liver function after the surgery. Presently, the surgeons assess both the volume and the residual liver function subjectively that may result in inaccuracy and misleading decisions. A dedicated software tool that offers liver volume extraction combined with a virtual interactive surgery and concrete prognosis in terms of residual liver volume, thus providing objective evaluation, would be of great assistance to the surgeons.
The aim of this work is to develop and test a software application dedicated to facilitate the work of the surgeons in our University Hospital. Initially, we developed a methodology for evaluation of the liver volume from CT images. Consequently, the methodology has been encapsulated within a software tool consisting of three modules for (a) accurate liver segmentation based on images from patients’ abdominal CT scans, (b) tracing of the tumour and evaluation of the remaining liver volume and estimation of the liver function and (c) a module for 3D liver rendering. Accurate liver segmentation is obtained with a seeded regional growing algorithm. Several homogeneity criteria, based on intensity voxels values, defined similarly to those proposed by Ogiela and Hachaj [14] for the case of segmentation of the carotid artery bifurcation region, are used to smooth the liver segmentation and prevent excessive growth into neighbouring organs of similar densities like the gallbladder, bile, the stomach, etc. Due to the large amount of CT slices, a prerequisite from the surgeons was the automatic calculation of the liver volume. Therefore, the user involvement is minimal and limited to the selection of the CT slice where the liver appears largest. The segmented images are stacked in a volume that is further rendered in three dimensions.
Methods
Liver Anatomy and Radiological Appearance
The liver in the human body is the second largest organ after the skin, and it is located in the right upper quadrant of the abdominal cavity. The liver weight ranges from 1,200 to 1,400 g in the adult woman and from 1,400 to 1,500 g in the adult man [20]. In its majority, the liver is composed of two large sections called right and left lobes. These sections are separated by the falciform ligament on the anterior surface of the liver. The right lobe is approximately six times larger than the left lobe. Main role of the liver is to filter the blood coming from the digestive tract, before passing it to the rest of the body. The most reliable method used for liver examination is the CT. It is performed with and without a contrast dye. Figure 1a, b depicts two tomographic slice images that are obtained with and without contrast enhancement. In case of contrast used, the internal structures are better highlighted.
As seen in Fig. 1a, the image of the liver and the gallbladder are merged. There is no boundary that discriminates the two organs due to similar X-ray absorption characteristics. Table 1 lists the elemental composition and densities of the liver and organs that are close and interact with it [21]. The comparison shows how close in terms of chemical composition and density, hence in absorption of X-rays are the liver and the organs that are close to it.
Determining the Residual Volume and Estimation of the Functionality After Liver Resection
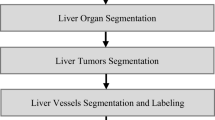
The proposed methodology for evaluation of the residual volume and estimation of the functionality of the liver after resection consists of three major steps: (a) total liver volume extraction, (b) virtual tumour resection and evaluation of the residual volume and functionality and (c) volume rendering of the results.
Liver Volume Extraction
The main stages in the proposed algorithm for extraction and estimation of the liver volume are depicted in Fig. 2a. The input data for the algorithm is the outlined by the surgeon liver boundary performed on a tomographic slice image where the liver appears either the largest or amongst the largests. The selection of this axial slice is done also by the surgeon amongst the images in the patient CT set. The intensity values in all images are converted to floating-point values of double precision and normalised in the range from 0 to 1 for the purposes of the algorithm. A 3 × 3 Gaussian low-pass filter with a Gaussian spread parameter σ = 0.2 (experimentally derived) is applied on all initial CT sets before starting the liver segmentation to remove the image noise in the original images. The homogeneity criteria used in the algorithm are summarised in Fig. 2b and explained below.
To start the segmentation of the liver parenchyma by using seeded region-growing algorithm, several seed points are initially determined within the liver area already segmented in the previous slice. The location of the seed points is sampled randomly, and only points that are located within the area are accepted as seed points for the region-growing algorithm for the current CT slice (denoted as k) of the patient CT set under processing. Specifically, a point is accepted to be a seed point if its intensity value is within an intensity window calculated as I mean ± σ Imean, where σImean is set equal to 0.3*I mean (defined experimentally). I meanis the mean intensity of the pixels in the segmented liver area within the previous CT slice. The initial value of I mean is calculated from the slice selected by the surgeon as:
where I(i, j) is the intensity value of the pixel (i, j) in the previous CT slice, K(i, j) is a mask that has values 1 and 0, and reflects if the pixel belongs or not to the liver area.
An intensity range is calculated for each slice from the given patient CT set. The region-growing algorithm is applied as many times as the seed points are. The region is iteratively grown by comparing and eventually involving the unallocated neighbouring pixels to the region. Specifically, the difference between the intensity value of a given neighbouring pixel and the region’s mean is used as a homogeneity criterion of region-growing algorithm. Further on, any pixel in a slice is considered a voxel in the liver volume. After a voxel is added to the region, its eight neighbours are also processed. This process stops when the intensity difference between region mean and new pixel becomes larger than a certain threshold, set again experimentally to 0.05.
This first and rough segmentation is completed by applying a subsequent morphological image processing. This includes morphological opening with a disc-shaped structuring element having a diameter of 5 pixels, aiming to reduce the noise and remove any isolated regions. Once the whole liver is segmented and available in 3D, the total liver volume, V liver, is calculated simply by multiplying the number of all voxels from the selected liver by their volume. The weight of the liver (W liver) can be obtained by multiplying the calculated volume and the density of the liver parenchyma (ρliver), given in Table 1.
where SliceImage is the current m × n binary 2D CT image with values 0, indicating the background, and 1, indicating the segmented liver area.
Virtual Tumour Resection
A virtual liver resection that involves virtual removal of the tumour volume by the surgeon is performed iteratively by using another dedicated software module. From this resection, the virtually removed volume can be calculated, similarly to the volume of the whole liver. To define the relative residual liver volume (RLV) associated with the development of postoperative hepatic dysfunction and infections, we used the relation of residual to total functional liver volume:
Finally, a volume-rendering module is used to visualise the segmented liver volume and pathologies. The liver segmentation and visualisation script were developed in Matlab (MathWorks, Inc.), while the virtual resection was developed under C++.
Patients
Forty-one sets of abdominal CT images were obtained at the University Hospital of Varna, Bulgaria. The CT images were of size 512 × 512, 16-bit grey level resolution. The total number of slices per patient varied from 60 to 180. From these, the number of slices with the presence of the liver varied from 45 to 60. The images were acquired by using a standard acquisition protocol that provides slices with pixel size of 0.87 mm in each direction and slice thickness of 3 mm. The CT images were obtained using SOMATOM Definition Flash (Siemens Healthcare), composed of a pair of sources and detectors that move around the patient. Data were extracted from the initial DICOM images and stored in a TIFF image format to facilitate the further applying and monitoring of the image processing techniques. The dataset statistics is summarized in Table 2.
Evaluation of the Accuracy of the Proposed Algorithm
The evaluation of the algorithm was accomplished (i) subjectively, by visual inspection from the surgeons, and (ii) more objectively, by calculating the relative difference RDliver_volume between the liver volume V liver_manually obtained after a manual segmentation and the volume V liver_algorithm obtained according to the suggested semi-automatic approach, for each dataset:
The averaged relative difference is computed as
where N is the number of datasets used in the study (41 in our investigation), was considered as an estimator of the overall relative error of the suggested semi-automatic approach and a criterion for the feasibility of its application.
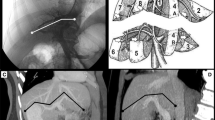
An evaluation was carried out by comparing calculated volumes from automatic liver segmentation obtained by applying the algorithm outlined in Fig. 2 with volumes that were selected and calculated by two surgeons from all the imaging sets after manual selection. Both surgeons were highly trained specialists in surgical treatment of various liver cancers with an expertise of more than 15 years in this domain. Each surgeon performed liver segmentation on different datasets, i.e. one used the first 20 patients’ datasets, while the second surgeon used the rest 21 patients’ datasets. Therefore, we treated the volume they selected as a gold standard for the comparison. For this purpose, CT images from patient sets were loaded in another dedicated software application developed for this study and the experts manually traced the appearance of the liver tissue on each slice. Figure 3 shows a screenshot from the interface used by the surgeons to select manually the tumour and the liver outlines. Subsequently, the tomographic slice images with the outlined liver boundaries were processed to produce a stack of slices which contain only the segmented liver and thus the total liver volume. This program code actually was applying thresholding and morphological closing that resulted into binary slices with segmented liver regions.
Results and Discussion
Performance of the Proposed Computational Tool
The manual approach took on average 25 min per patient scan to outline the liver borders on all CT slices and 30 min in total to calculate the total liver volume. The comparison between all 41 volumes (after a manual procedure and according to the suggested semi-automatic approach) showed an average relative difference of 3.50 ± 2.6 %. Largest error was observed for the case of CT images obtained without a contrast agent due to the negligible difference in contrast between the liver and neighbouring regions. Some results that include the worst and the best cases are summarised in Table 3.
Figure 4 reveals the comparison of the performance of the proposed algorithm (Fig. 4 lower row) versus the manual selection accomplished by one of the surgeons (Fig. 4 upper row). The parallel made in Fig. 4 concerns three consecutive CT slices from dataset 1 (Table 3). The liver segmentation was performed on a set of 156 patients’ abdomen CT slices with planar dimensions of 512 in each direction acquired without contrast agent. The liver of the patient was healthy, without any pathologies. As shown in the slice segmentation results (lower row in Fig. 4a–c), the method was successfully applied to outline the shape of the liver in different slices as well as to calculate successfully the liver volume.
Liver segmentation: a–c three subsequent CT slices with outlined liver boundaries; d liver volume visualisation. First row, regions selected by the surgeon. Second row, automatically outlined regions by applying the algorithm in Fig. 2
In general, the algorithm has been successfully applied to various sets of CT data extracted from multiphase liver CT scans that involved precontrast, arterial phase, portal venous phase and delayed phase. Discussions with the surgeons showed that the present 3.50 % average deviation of the semi-automatic liver evaluation from the manual approach is acceptable. Besides, they agree that the manual outlining is not always done to a maximum level of image details, which can explain that difference. The computational time for applying the algorithm was about 3 min (using medium computational power PC), while the experts required on average 25 min to trace the volume and calculate its value. In addition, the surgeon involvement is minimal.
Feasibility Studies
The feasibility of the proposed technique has been demonstrated within several cases of tumour resection.
Case 1: A liver with simulated large pathology.
We used the CT dataset 1 (Table 3) of a healthy 54-year-old woman to simulate metachronous liver metastasis from breast cancer. We considered a significant chemotherapy-associated steatohepatitis after systematic anti-cancer treatment with a liver-limited metastatic disease but significant comorbidity and renal insufficiency. Renal insufficiency precludes intra-venous contrast application. On the CT slices, the surgeon traced virtually the outlines of a metastasis with quite large margins. The reconstruction of the whole volume with the metastasis is shown in Fig. 5. For this case, the removed volume was evaluated as being 40 ml, while the RLV was calculated to be 96 %. In the native CT scan, surgeon could suspect but cannot exactly detect the metastasis shown below. However, with the assistance of image processing this is now possible. This example also demonstrates the feasibility of the tool for the training of students in medical universities.
Case 2: A liver with several metastases.
This is a case of a 59-year-old man, with multiple synchronous colorectal liver metastases which are difficult for detection but seem to be resectable. The input patient data were from an abdomen CT scan obtained without contrast agent, corresponding to dataset 3 from Table 3. The volume of the liver was reconstructed by applying the algorithm, and in consequence, the surgeon performed a virtual resection of the tumour volume. Figure 6a shows three consecutive slices with tumours that were marked for resection, while Fig. 6b reviews the reconstructed liver volume with the metastases. The tumour volume was estimated to be 50 ml, while the evaluated RLV was 94 %.
While detecting all metastases, even the smallest one is essential for building of oncosurgical strategy for radical treatment, and it is crucial for removal the tumour load of human body with R-0 index—no residual tumour after surgery. This patient underwent two-stage hepatectomy. This example also demonstrates the feasibility of this framework for the exercising of last year students in the medical universities.
Case 3: A liver with a giant hepatocellular cancer.
A case of 66-year-old man has been diagnosed with a giant hepatocellular carcinoma due to hepatitis C virus cirrhosis, who is discussed for liver resection— major hepatic surgery with critically low future liver remnant. Input patient data correspond to dataset 4 (Table 3). Figure 7a shows two consecutive slices with tumours that were marked for resection. We applied the described process to extract the whole liver volume, the tumour itself, and evaluate the RLV. The estimated liver volume was 2,150 ml (shown in Fig. 7b), while the removed one was 1,500 ml. As a consequence, the residual liver volume was calculated to 650 ml that gives a RLV of 30 %. The total liver volume with reconstructed giant hepatocellular carcinoma is shown in Fig. 7c.
Computed tomography images with agent contrast showing a two successive slices with giant hepatocellular carcinoma of patient from dataset 4 (Table 3), b the total liver volume reconstructed with the proposed technique from Fig. 2 and c the total liver volume including the giant hepatocellular carcinoma
The RLV of 30 % and the liver function Child-Pugh B puts patient according to Barcelona Clinic Liver Cancer Staging and Treatment Strategy criteria [22] in radically non-resectable group, and he is indicated for palliative procedure or to be discussed on liver transplant board.
The tool has been evaluated positively by the Department of Surgery at the University Hospital of Varna. Until now, the surgeons were evaluating the liver volume and the resection volume mainly subjectively and intuitively. This is primarily due to lack of experience to manage image processing software applications and to handle programming codes for overall calculation of the selected pixels and respectively the volume itself. The tool has been designed not to replace but to facilitate the surgeons; thus, the requirements of minimal involvement, easy to use and fast volume evaluation have been respected. Further, an extensive comparative study is planned with the involvement of this algorithm, existing commercial software applications, as well as other available in-house developed algorithms implemented in other areas of medical imaging (as for example [14]) to segment properly the liver volume from abdominal CT images.
Conclusions
An approach and an application tool for evaluation of the residual function of the liver prior to the intervention of the surgeons are proposed. The approach includes volume determination based on segmentation of liver from computed tomography images, virtual tumour resection and estimation of the residual liver function and volume rendering. The tool is characterised by robust technique for liver segmentation. It allows the surgeons to interactively and objectively evaluate the liver volume and the residual functionality after tumours are removed. In addition to its main purpose as an assisting tool for facilitating the decision making, it appears to be appropriate for educational purposes—as a teaching tool for medical students specializing in this area as well as for virtual training of young surgeon specialists.
References
Moreno Elola-Olaso A, Davenport DL, Hundley JC, Daily MF, Gedaly R: Predictors of surgical site infection after liver resection: a multicentre analysis using National Surgical Quality Improvement Program data. HPB (Oxford) 14:136–141, 2012
Schindl MJ, Redhead DN, Fearon KC, Garden OJ, Wigmore SJ: The value of residual liver volume as a predictor of hepatic dysfunction and infection after major liver resection. Gut 54:289–296, 2005
Guglielmi A, Ruzzenente A, Conci S, Valdegamberi A, Iacono C: How much remnant is enough in liver resection? Dig Surg 29:6–17, 2012
Tomoshige S, Oost E, Shimizu A, Watanabe H, Nawano S: A conditional statistical shape model with integrated error estimation of the conditions; Application to liver segmentation in non-contrast CT images. Med Image Anal 18:130–143, 2014
Heimann T, van Ginneken B, Styner MA, Arzhaeva Y, Aurich V, et al: Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans Med Imaging 28:1251–1265, 2009
Kumar S, Moni R: Diagnosis of liver tumor from CT images using countourlet transform. Int J Biomed Eng Technol 7:15, 2011
Liu F, Zhao B, Kijewski PK, Wang L, Schwartz LH: Liver segmentation for CT images using GVF snake. Med Phys 32:3699–3706, 2005
Lim SJ, Jeong YY, Ho YS: Automatic liver segmentation for volume measurement in CT images. J Vis Commun Image Represent 17:860–875, 2006
Goryawala M, Guillen MR, Cabrerizo M, Barreto A, Gulec S, et al: A 3-D liver segmentation method with parallel computing for selective internal radiation therapy. IEEE Trans Inf Technol Biomed 16:62–69, 2012
Goryawala M, Guillen MR, Gulec S, Barot T, Suthar R, et al: An accurate 3d liver segmentation method for selective internal radiation therapy using a modified k-means algorithm and parallel computing. Int J Innov Comput Inf Control 8:6515–6538, 2012
Ballard DH, Brown CM: Computer Vision, Prentice-Hall, 1982
Zanaty EA, Asaad A: Probabilistic region growing method for improving magnetic resonance image segmentation. Connect Sci 25:179–196, 2013
Ogiela MR, Hachaj T: The automatic two - step vessel lumen segmentation algorithm for carotid bifurcation analysis during perfusion examination, In: Smart Innov. Syst. Technol., Lakhmi C., Robert J., Junzo Toyohide, Phillips-Wren G.(eds), Vol 16: 485–493, 2012
Ogiela MR, Hachaj T: Automatic segmentation of the carotid artery bifurcation region with a region-growing approach. J Electronic Imaging 22: 2013
Mohd Saad N, Abu-Bakar SAR, Muda S, Mokji M, Abdullah AR: Fully automated region growing segmentation of brain lesion in diffusion-weighted MRI. IAENG Int J Comput Sci 39:155–164, 2012
Gasca F, Ramrath L, Huettmann G, Schweikard A: Automated segmentation of tissue structures in optical coherence tomography data. J Biomed Opt 14: 2009
Fadzil MHA, Izhar LI, Venkatachalam PA, Karunakar TVN: Extraction and reconstruction of retinal vasculature. J Med Eng Technol 31:435–442, 2007
Liu RM, Yang L, Liu E, Yang J, Li M, Wang F: Automatic extraction of infrared small target based on support vector regression and adaptive region growing algorithm. Opt Eng 46: 2007
Campadelli P, Casiraghi E, Esposito A: Liver segmentation from computed tomography scans: a survey and a new algorithm. Artif Intell Med 45:185–196, 2009
Wolf D: In: Walker HK, Hall WD, Hurst JW Eds. Clinical methods: the history, physical, and laboratory examinations: chapter 94: evaluation of the size, shape, and consistency of the liver, 3rd edition. Butterworths, Boston, 1990
Schneider W, Bortfeld T, Schlegel W: Correlation between CT numbers and tissue parameters needed for Monte Carlo simulations of clinical dose distributions. Phys Med Biol 45:459–478, 2000
Pons F, Varela M, Llovet JM: Staging systems in hepatocellular carcinoma. HPB (Oxford) 7:35–41, 2005
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bliznakova, K., Kolev, N., Buliev, I. et al. Computer Aided Preoperative Evaluation of the Residual Liver Volume Using Computed Tomography Images. J Digit Imaging 28, 231–239 (2015). https://doi.org/10.1007/s10278-014-9737-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-014-9737-5