Abstract
The aim of this study was to determine the relationship between the prevalence of Porphyromonas gingivalis, its fimA genotypes, Aggregatibacter actinomycetemcomitans, Tannerella forsythia, and Treponema denticola and the evolution of periodontal health. In a longitudinal prospective study, samples of subgingival plaque were taken from 114 patients (37 with chronic periodontitis, 17 with gingivitis, and 60 periodontally healthy) in the course of a full periodontal examination. PCR was employed to determine the presence of the periodontopathogenic bacteria. Four years later, a second examination and sample collection were performed in 90 of these patients (20 with chronic periodontitis, 12 with gingivitis, and 58 periodontally healthy). T. forsythia, P. gingivalis, and T. denticola are the most prevalent bacteria in patients with chronic periodontitis (78.4%, 62.2 y 56.8%, respectively). The P. gingivalis bacterium and its fimA genotypes I, II, and IV showed the highest correlation between the baseline and follow-up assessments. P. gingivalis fimA genotype II and T. forsythia were associated to a significant degree with unfavourable periodontal evolution. Of the variables studied, P. gingivalis fimA genotype II and T. forsythia increase the risk of an unfavourable evolution of periodontal status.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chronic periodontitis is an infectious disease resulting in inflammation within the supporting tissues of the teeth, progressive attachment loss, and bone loss [1]. The composition of subgingival plaque is complex and has been the subject of numerous studies, as the presence of certain bacteria is associated with worse periodontal status, greater pocket depth, and higher bleeding indices [2]. The oral cavity is colonized by a wide range of microorganisms [3]. Some are commensal bacteria that live in harmony with their hosts and do them no harm. Others, known as periodontopathogenic bacteria, are associated with periodontitis. Aggregatibacter actinomycetemcomitans has been associated with aggressive periodontitis, while Porphyromonas gingivalis, A. Actinomycetemcomitans, Tannerella forsythia, Treponema denticola, and Eikenella corrodens have been associated with chronic periodontitis [4].
In 1998, Socransky et al. described a ‘red complex’, formed by a combination of Bacteroides forsythus (now known as T. forsythia), P. gingivalis, and T. denticola, which exhibit a clearer relationship with the clinical signs of chronic periodontitis, specifically pocket depth and bleeding on probing. Subsequently, many studies have been carried out to detect the presence of these three bacteria in different populations, the association between them, and the synergic mechanisms that increase their pathogenicity [5–7].
The virulence of P. gingivalis, a gram-negative anaerobic bacterium, is attributed to its various surface components, such as fimbriae, lipopolysaccharides, and proteases. This surface makes it possible for the bacterium to interact with the external medium and facilitates its growth, nutrient acquisition, colonization, and formation of a biofilm that protects it against the host’s defences [8, 9]. Amano et al. concluded that P. gingivalis can be classified into five genotypes according to genomic differences in the fimA gene which codes fimbrillin, a protein of the major fimbriae [10, 11]. Subsequently, Nakagawa et al. discovered a new variant of the fimA gene, which they named Ib, because it bore a great resemblance to genotype I [12]. P. gingivalis is frequently found in patients with chronic periodontitis, but has also been observed, although to a lesser extent, in periodontally healthy patients [13]. In recent years, studies have been conducted to evaluate the relationship between the different P. gingivalis genotypes and periodontal pathogenesis. Genotype II has been observed to be more prevalent in periodontal patients and to be associated with more aggressive forms of the disease. Some authors attribute this relationship to its possessing greater adhesiveness and invasiveness [14].
The aetiopathogeny of periodontitis is the result of a complex interaction between the oral biofilm and the host’s immune response. New laboratory techniques have given a better understanding of this biofilm and its role in the progress of chronic periodontitis. The aim of this study was to assess the relationship between the presence of periodontal pathogenic bacteria (P. gingivalis and its six fimA genotypes, A. actinomycetemcomitans, T. denticola, T. forsythia, and the ‘red complex’) and periodontal status, whether periodontal bacteria prevalence differed significantly between adult patients in different periodontal status groups and the evolution of periodontal status 4 years later.
Materials and methods
In this longitudinal prospective cohort study, two oral examinations and determinations of periodontal pathogenic bacteria were conducted 4 years apart. The baseline assessment was made in 2009 and the follow-up assessment in 2013.
Baseline assessment
Inclusion and exclusion criteria and study size
The inclusion criteria were patients aged between 25 and 50 years with at least 20 teeth present in the mouth, excluding third molars.
Pregnant women and any with drug-induced gingival hyperplasia, or who had taken antibiotics in the previous six months, or who were taking anti-inflammatory medication to treat a chronic condition were excluded from the study, as were patients with HIV infection, Type I or II diabetes mellitus, coronary heart disease, rheumatoid conditions, lupus erythmatosus, Behçet’s syndrome, Crohn’s disease, herpetic gingivostomatitis, pemphigous, or oral pemphigoid [15].
A consecutive sample of 140 patients of the Dentistry Clinic at the University of Valencia (Spain) who met the inclusion and exclusion criteria was selected for the study. The patients were given the necessary information and took part voluntarily after giving their informed consent. The study design and protocol were both approved by the Research in Humans Ethics Committee of the University of Valencia’s Experimental Research Ethics Committee.
The sample size was calculated on the basis of data from the study by Zhao et al. [13], which estimated the presence of P. gingivalis as around 82% in patients with chronic periodontitis and 22% in periodontally healthy patients. With a confidence level of 1−α = 95% for a statistical power of 85%, a two-tailed test, and an expected loss rate of 25%, the estimated sample size is a minimum of 14 individuals in each study group.
Clinical examinations and sample collection
A full examination of the entire mouth of each patient was conducted, employing a WHO periodontal probe (PCP11.5B, Hu Friedy, Chicago, IL, USA), and the following were recorded at 6 sites on each tooth [16]: bacterial plaque, bleeding, pocket depth, and loss of periodontal attachment. The plaque index was calculated by dividing the number of surfaces with plaque by the total number of surfaces and multiplying the result by 100 to express the result as a percentage [17]. The bleeding index was calculated in the same way.
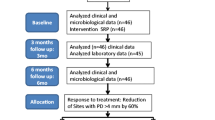
The patients were classified into three groups: Group I was periodontally healthy patients, nowhere presenting a pocket depth greater than 3 mm or loss of periodontal attachment greater than 1 mm [13], Group II was patients with gingivitis, who met the conditions for Group I but presented a Gingival Bleeding Index of over 30% [18], and Group III was patients with chronic periodontitis, presenting at least 4 areas with a probing depth of 5 mm or greater and loss of periodontal attachment of 2 mm or more [16]. Of the 140 patients selected for the study and examined, a total of 114 were included in the study: 60 were assigned to the no disease group, 17 to the gingivitis group, and 37 to the chronic periodontitis group. The other 26 were excluded, as they did not meet the requirements for classification into one of these three groups.
The patients were also classified as ‘non-smokers’, ‘ex-smokers’, or ‘current smokers’. Those who had stopped smoking at least 6 months before the study began were considered ‘ex-smokers’. Those who had stopped smoking less than 6 months previously were included in the ‘current smokers’ group. In the case of patients who smoked, the number of cigarettes per day was also recorded.
The subgingival plaque samples were obtained from the deepest pocket at Ramfjord teeth in the patients in the chronic periodontitis group and from the mesiolabial area of a Ramfjord molar in the no disease group and the gingivitis group. The supragingival plaque was first removed with a sterile Gracey curette, employed with care to avoid bleeding. Three sterile paper points (size 25, Dentsply Maillefer, Ballaigues, Switzerland) were inserted as deeply as possible into the gingival groove, left for 15 s, removed, and placed in a sterile Eppendorf tube [13]. The bacterial DNA was extracted immediately using the Wizard SV DNA Purification System (Promega Cat. # A2360) in accordance with the manufacturer’s instructions. Following extraction, the DNA samples were stored at −20 °C until determination took place.
Polymerase chain reaction (PCR)
The PCR method was used to detect the four bacteria: P. gingivalis, A. actinomycetemcomitans, T. denticola, and T. forsythia [19, 20]. The method described by Zhao et al. [13] was used to analyse the fimA genotypes of P. gingivalis. To genotype the fimA gene, the specific primers for each subtype described by Amano et al. [11] and Nakagawa et al. [12] were used. Table 1 shows the species-specific primers used for PCR in this study.
The PCR reaction was performed with 100 ng of the bacterial DNA extracted in the previous step, 200 μM of each of the dNTPs, 3 mM MgCl2, 50 pmol of each primer, and 0.5 U of AmpliTaq Gold (9800 Fast ThermalCycler of Applied Biosystems®). The PCR conditions were an initial taq polymerase activation step at 95 °C for 10 min, followed by 40 denaturing cycles at 94 °C for 30 s, annealing at 58 °C for 30 s, elongation at 72 °C for 30 s, and finally, a single final elongation step at 72 °C for 7 min.
The PCR products were separated by electrophoresis in 1.5% agarose gels and stained with ethidium bromide, then viewed under ultraviolet light. All the samples were analysed in duplicate using two different PCR reactions for each primer pair. A no-template control (PCR reactions without a DNA template) for each primer pair was included in each group of PCR reactions to check for foreign DNA contamination and for non-specific amplification (primer-dimer formation). Samples in which types I and II were both found were amplified further using specific primers for type Ib. The resulting amplicons were digested with RsaI, and cast in 1.5% agarose gels stained with ethidium bromide and viewed under ultraviolet light. Samples in which 2 fragments appeared were considered type Ib [12].
Negative controls were included for each primer set in each round of PCR reactions. Positive controls were included to verify the negative amplification results. For Porphyromonas, DNA isolated from cell lines (P. gingivalis ATCC33277 and P. gingivalis W83) was used. For the other bacterial species, DNA isolated from the patients’ samples was used and those samples that yielded PCR amplification products were included in each round of amplification.
Follow-up assessment
Of the initial 114 patients included in the study, 90 could be re-examined at the follow-up assessment 4 years later. Two patients were not re-examined, because they did not meet the inclusion criteria established for the chronic conditions at the baseline assessment. Informed consent was again obtained, and a new examination record was filled in registering whether the patient had received any periodontal treatment since the initial examination.
The patients were again classified into one of the three groups (no disease, gingivitis, or chronic periodontitis) according to the clinical signs observed, as in the baseline assessment, to determine whether their periodontal evolution had been favourable or unfavourable. The subgingival plaque samples were obtained at the same point of the gingival sulcus in the no disease and gingivitis groups and in the same periodontal pocket in the chronic periodontitis group. The classification criteria and sample collection and processing followed the same protocol, as described above.
Study variables
-
Bacteria0/Genotype0/Red Complex0: The presence of bacteria (A. actinomycetemcomitans, T. denticola, P. gingivalis, or T. forsythia), a P. gingivalis fimA genotype (I–V or Ib), or Socransky’s ‘red complex’ in the baseline assessment.
-
Bacteria1/Genotype1/Red Complex1: The presence of bacteria (A. actinomycetemcomitans, T. denticola, P. gingivalis, or T. forsythia), a P. gingivalis fimA genotype (I–V or Ib), or Socransky’s ‘red complex’ in the follow-up assessment 4 years later.
-
Periodontal treatment during the follow-up period (none/sporadic scaling/constant periodontal maintenance).
-
Stable or favourable periodontal evolution: patients whose periodontal condition in the follow-up assessment 4 years later was the same or better than in the baseline assessment.
-
Unfavourable periodontal evolution: patients who had passed from the no disease to the gingivitis or periodontitis group between the baseline and follow-up assessments, or who were already affected initially by chronic periodontitis and whose pocket depth had increased during the follow-up period.
Statistical analyses
The mean values of the quantitative variables and the prevalence rates for the dichotomous or ordinal variables were obtained. The 95% confidence intervals were calculated. In the two-tailed statistical analysis, the relationships between the dichotomous variables were evaluated by the Chi-square test and the linear trend test. Student’s t test and ANOVA were used to compare means and the Pearson coefficient to assess correlations. The significance level was set at p < 0.05 or p < 0.01. A multivariate binary logistic regression analysis was performed. Computations were carried out by the SPSS 19.0 statistical analysis software (SPSS Inc., Chicago, IL USA).
Results
The distribution of the sample by mean age, gender, or smoking did not differ significantly between the no disease, gingivitis, and chronic periodontitis groups. The data and the inter-group statistical differences found among the variables—number of cigarettes per day, plaque index, bleeding index, probing depth, and level of attachment loss—are shown in Table 2.
Percentage of individuals with periodontal bacteria at the beginning of the study, by group (baseline assessment)
Table 3 shows the prevalence of the different bacteria found in the subgingival plaque. The three periodontal status groups presented significant differences in rates of P. gingivalis, A. actinomycetemcomitans, T. denticola, T. forsythia, and the ‘red complex’, which increased significantly as periodontal condition worsened and reached their highest point in the chronic periodontitis patients. The percentages of individuals with T. forsythia (78.4%), P. gingivalis (62.2%), and T. denticola (56.8%) in the subgingival plaque samples were higher than those with A. actinomycetemcomitans (27%) in the chronic periodontitis group. The ‘red complex’ appeared in 35.1% of the individuals taking part in the study.
On studying the distribution of the four bacteria under study and the ‘red complex’ by non-smoker, ex-smoker or ‘current smoker’, regardless of periodontal status, the only bacterium showing statistically significant differences between smokers and non-smokers or ex-smokers was T. denticola (data not shown).
Changes in periodontal status and in the percentage of individuals with periodontal bacteria 4 years later, at the end of this study (follow-up assessment)
Of the 114 persons initially examined, 90 remained for the follow-up assessment 4 years later, signifying a loss of 21.1% of the sample. At follow-up, most of the patients remained in their initial groups. On the Landis and Koch scale, the agreement between the baseline status and follow-up status was considered high (kappa statistic = 0.722). The clinical status of 10 patients from the initial no disease group had worsened (8 had developed gingivitis and 2 periodontitis). Of the patients initially placed in the gingivitis group, 2 were moved to the no disease group and another 2 to the periodontitis group. All the patients initially placed in the periodontitis group remained there. Of the 90 individuals examined at follow-up, only 6 had received constant periodontal maintenance (periodical scaling and root planning with chlorhexidine mouthrinses before treatment), 40 had received sporadic scaling, and the remaining 44 had not received any type of treatment.
Variations in the composition of the patients’ subgingival plaque were recorded. Table 4 shows the correlations for each bacterium, the ‘red complex’, and P. gingivalis fimA genotypes at the beginning (baseline assessment) and end of the study (follow-up assessment 4 years later). Those with the greatest correlation between baseline and follow-up were genotypes I, II, and IV, the bacterium P. gingivalis, and the ‘red complex’.
Bacteria associated with unfavourable periodontal evolution
Table 5 shows the prevalence of P. gingivalis bacteria and fimA genotypes in the baseline assessment of the patients with a stable or favourable periodontal evolution and those with an unfavourable periodontal evolution. The bacteria with statistically significant differences were P. gingivalis and its fimA genotype II, T. forsythia, and the ‘red complex’. This table also shows the incidence ratios of unfavourable periodontal evolution due to the presence of the different bacteria. The highest are those associated with the presence of A. actinomycetemcomitans and the ‘red complex’.
A multivariate binary logistic regression analysis was performed with ‘unfavourable periodontal evolution’ as the dependent variable and all the measurements of bacteria or P. gingivalis fimA genotype presence in the baseline assessment as the independent variables, together with age, gender, the number of cigarettes smoked by the patients who were smokers, and any periodontal treatment received during the 4-year follow-up period. The final model contained significant independent variables: P. gingivalis fimA genotype II (p = 0.017; Expβ = 3.84) and T. forsythia (p = 0.008; Expβ = 4.16). The other variables entered did not prove significant. The omnibus test of the model was significant, with p = 0.001 and a Nagelkerke’s R 2 value of 0.212. The Hosmer and Lemeshow goodness of fit test found p = 0.901.
Discussion
The composition of the subgingival plaque and, more specifically, the presence of particular periodontal pathogenic bacteria are related to clinical signs which define the patient’s periodontal status and its evolution. The three study groups presented significant differences in T. forsythia, P. gingivalis, and T. denticola prevalence. These differences also existed in the case of A. actinomycetemcomitans, but this bacterium was only found in 27% of the chronic periodontitis patients. The same differences were found in P. gingivalis fimA genotypes II, III, and Ib. FimA genotype V was not found in any of the patients. On studying 90 of the same patients 4 years later, collecting the subgingival plaque sample from the same area as before, genotypes I, II, and IV, the P. gingivalis bacterium, and the ‘red complex’ were the pathogens with the highest correlation between the baseline and follow-up assessments. The bacteria with significant differences between patients whose periodontal status had remained steady or improved and those in whom it had worsened were P. gingivalis and its fimA genotype II, T. forsythia, and the ‘red complex’. The results showed that P. gingivalis and T. forsythia were the most prevalent bacteria in patients with an unfavourable evolution. However, the incidence ratio shows that A. actinomycetemcomitans presents the highest relative risk (4.63) despite being the least prevalent of the bacteria studied.
The aim of most longitudinal studies has been to follow the evolution of cases who had been given a particular treatment compared to a control group, or to compare two different treatments [21, 22]. In the present case, the treatment received during the follow-up interval did not influence the results (p = 0.237). It should be noted that this study had a lengthy follow-up interval, 4 years, in keeping with the evolution of chronic periodontitis. One salient result is that 2 patients who were included in group II at the beginning of the study were placed in group I in the follow-up assessment, because their gingival bleeding index had dropped below 30%. This confirms that gingivitis is a reversible condition. The main limitation in this type of study is the loss of a proportion of the sample for a variety of reasons, such as a change of address or of the contact telephone number. It must be acknowledged that the loss rate of 21.1% in this case constitutes a limitation for the evaluation of the results.
It has already been reported by Simonson et al. that T. denticola can serve as a prognostic marker for periodontal disease recurrence [23]. In 2012, Nomura et al. published a longitudinal study with the aim of ascertaining which salivary biomarkers and which bacteria could be used to predict the progression of chronic periodontitis. The bacteria they studied were P. gingivalis, Prevotella intermedia, and T. forsythia, and their follow-up period was 18 months. The bacteria that proved to be significant predictors of the progression of chronic periodontitis were P. gingivalis and P. intermedia [24]. As regard longitudinal studies of the presence of P. gingivalis fimA genotypes, Fujise et al. [25] assessed the influence of the different genotypes in patients with chronic periodontitis who received scaling and root planing treatment. They found that genotype I was a predictor of bleeding on treatment.
The fimA genotypes of P. gingivalis and the bacteria A. actinomycetemcomitans, T. forsythia, and T. denticola have all been widely investigated, with numerous cross-sectional studies in different age groups and ethnic groups, and have been related to the presence of systemic pathologies, such as diabetes mellitus or tobacco use. These studies show that P. gingivalis very frequently appears in the subgingival plaque of patients with chronic periodontitis, ranging from 29.6 to 97.5% prevalence [10, 13, 18, 26–29]. While P. gingivalis is considered one of the bacteria most closely associated with chronic periodontitis, it is also present in the subgingival plaque of periodontally healthy patients, although in a smaller percentage, ranging from 1.5 to 57.8% depending on the study [11, 13, 18, 27–30]. The present study detected P. gingivalis in 62.2% of the patients with chronic periodontitis and 23.3% of the healthy individuals.
As regard the distribution of P. gingivalis fimA genotypes, one fact that every study repeats is the greater prevalence of fimA genotype II in patients with chronic periodontitis. In the first studies in which the 6 genotypes were known, it was already being said that type II had the strongest association with periodontitis, while type Ib, the last to be identified, was also shown to be associated with progression of the disease, although to a lesser degree [12]. Nevertheless, variations in the distribution of the genotypes have been found, attributed to ethnic and geographical differences [13, 18]. Indeed, certain studies show marked variations in the composition of the subgingival plaque of chronic periodontitis patients from different countries with the same age, pocket depth, gender, and exposure to tobacco use [17, 31, 32].
In the same way as P. gingivalis is present in both healthy subjects and in patients with chronic periodontitis, so are T forsythia, T denticola, and A. actinomycetemcomitans. In all cases, the prevalence of these bacteria is lower in healthy individuals than in patients with chronic periodontitis. It should be mentioned that of the four bacteria studied, A. actinomycetemcomitans is the one that appears least frequently in both healthy and periodontal patients (around 30%), even in studies conducted in patients with aggressive periodontitis [4]. Wara-aswapati et al. [7] found a high frequency of the three ‘red complex’ bacteria in the moderate to severe periodontitis patients group, whereas A. actinomycetemcomitans was only present in 35% of the patients.
On studying the influence of tobacco on the composition of subgingival bacterial plaque, the present study found no differences in the distribution of P. gingivalis fimA genotypes when current smokers were compared with ex-smokers and non-smokers. Some previous studies have found no differences in the quantities of P. gingivalis obtained from the subgingival plaque samples of smokers and non-smokers [7]. As regard tobacco use and the bacteria studied, the present study did find significant differences in T. denticola prevalence between smokers, on the one hand, and ex-smokers and non-smokers, on the other hand. Haffajee and Socransky [31] found that certain bacteria, such as P. gingivalis and T. denticola, were more prevalent in smokers than in ex-smokers or non-smokers.
Of all the variables studied, those that proved significant in the multivariate logistic regression analysis were P. gingivalis fimA genotype II and T. forsythia, although owing to the complexity of the aetiopathogeny of chronic periodontitis, further longitudinal studies are needed to determine which variables influence the evolution of periodontitis for the worse to develop different patient follow-up protocols depending on the risk profile.
References
(1999) 1999 International workshop for a classification of periodontal diseases and conditions. Papers. Oak Brook, Illinois, October 30–November 2, 1999. Ann Periodontol 4:1–112.
Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44.
Zaura E, Nicu EA, Krom BP, Keijser BJ. Acquiring and maintaining a normal oral microbiome: current perspective. Front Cell Infect Microbiol. 2014;4:85. doi:10.3389/fcimb.2014.00085.eCollection.
Feng Z, Weinberg A. Role of bacteria in health and disease of periodontal tissues. Periodontol 2000. 2006;40:50–76.
Suzuki N, Yoneda M, Hirofuji T. Mixed red-complex bacterial infection in periodontitis. Int J Dent. 2013;. doi:10.1155/2013/587279.
Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–90.
Wara-aswapati N, Pitiphat W, Chanchaimongkon L, Taweechaisupapong S, Boch JA, Ishikawa I. Red bacterial complex is associated with the severity of chronic periodontitis in a Thai population. Oral Dis. 2009;15:354–9.
Yoshimura F, Murakami Y, Nishikawa K, Hasegawa Y, Kawaminami S. Surface components of Porphyromonas gingivalis. J Periodontal Res. 2009;44:1–12.
Lamont RJ, Jenkinson HF. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol Immunol. 2000;15:341–9.
Amano A, Nakagawa I, Kataoka K, Morisaki I, Hamada S. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J Clin Microbiol. 1999;37:1426–30.
Amano A, Kuboniwa M, Nakagawa I, Akiyama S, Morisaki I, Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res. 2000;79:1664–8.
Nakagawa I, Amano A, Ohara-Nemoto Y, Endoh N, Morisaki I, Kimura S, et al. Identification of a new variant of fimA gene of Porphyromonas gingivalis and its distribution in adults and disabled populations with periodontitis. J Periodontal Res. 2002;37:425–32.
Zhao L, Wu YF, Meng S, Yang H, OuYang YL, Zhou XD. Prevalence of fimA genotypes of Porphyromonas gingivalis and periodontal health status in Chinese adults. J Periodontal Res. 2007;42:511–7.
Amano A, Nakagawa I, Okahashi N, Hamada N. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J Periodontal Res. 2004;39:136–42.
Schulz S, Machulla HK, Altermann W, Klapproth J, Zimmermann U, Glaser C, et al. Genetic markers of tumour necrosis factor alpha in aggressive and chronic periodontitis. J Clin Periodontol. 2008;35:493–500.
Herrera D, Contreras A, Gamonal J, Oteo A, Jaramillo A, Silva N, et al. Subgingival microbial profiles in chronic periodontitis patients from Chile, Colombia and Spain. J Clin Periodontol. 2008;35:106–13.
O’Leary TJ, Drake RB, Naylor JE. The plaque control record. J Periodontol. 1972;43:38.
Missailidis CG, Umeda JE, Ota-Tsuzuki C, Anzai D, Mayer MP. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol Immunol. 2004;19:224–9.
Ashimoto A, Chen C, Bakker I, Slots J. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol Immunol. 1996;11:266–73.
Kulekcia G, Leblebicioglub B, Keskina F, Ciftcia S, Badurc S. Oral and dental bacteriology and infection Salivary detection of periodontopathic bacteria in periodontally healthy children. Anaerobe. 2008;14:49–54.
Janatova T, Najmanova L, Neubauerova L, Kyselkova M, Novotna G, Spizek J, et al. Changes in the incidence of periodontal pathogens during long-term monitoring and after application of antibacterial drugs. Folia Microbiol (Praha). 2009;54:429–35.
Jervoe-Storm PM, AlAhdab H, Semaan E, Fimmers R, Jepsen S. Microbiological outcomes of quadrant versus full-mouth root planing as monitored by real-time PCR. J Clin Periodontol. 2007;34:156–63.
Simonson LG, Robinson PJ, Pranger RJ, Cohen ME, Morton HE. Treponema denticola and Porphyromonas gingivalis as prognostic markers following periodontal treatment. J Periodontol. 1992;63:270–3.
Nomura Y, Shimada Y, Hanada N, Numabe Y, Kamoi K, Sato T, et al. Salivary biomarkers for predicting the progression of chronic periodontitis. Arch Oral Biol. 2012;57:413–20.
Fujise O, Miura M, Hamachi T, Maeda K. Involvement of Porphyromonas gingivalis fimA genotype in treatment outcome following non-surgical periodontal therapy. J Periodontol. 2005;76:1661–6.
Beikler T, Peters U, Prajaneh S, Prior K, Ehmke B, Flemmig TF. Prevalence of Porphyromonas gingivalis fimA genotypes in Caucasians. Eur J Oral Sci. 2003;111:390–4.
Miura M, Hamachi T, Fujise O, Maeda K. The prevalence and pathogenic differences of Porphyromonas gingivalis fimA genotypes in patients with aggressive periodontitis. J Periodontal Res. 2005;40:147–52.
Hayashi F, Okada M, Oda Y, Kojima T, Kozai K. Prevalence of Porphyromonas gingivalis fimA genotypes in Japanese children. J Oral Sci. 2012;54:77–83.
Moon JH, Herr Y, Lee HW, Shin SI, Kim C, Amano A, et al. Genotype analysis of Porphyromonas gingivalis fimA in Korean adults using new primers. J Med Microbiol. 2013;62:1290–4.
Yang HW, Huang YF, Chou MY. Occurrence of Porphyromonas gingivalis and Tannerella forsythensis in periodontally diseased and healthy subjects. J Periodontol. 2004;75:1077–83.
Haffajee AD, Socransky SS. Relationship of cigarette smoking to the subgingival microbiota. J Clin Periodontol. 2001;28:377–88.
Lopez NJ, Socransky SS, Da Silva I, Japlit MR, Haffajee AD. Subgingival microbiota of chilean patients with chronic periodontitis. J Periodontol. 2004;75:717–25.
Acknowledgements
The authors wish to thank Mary Georgina Hardinge for translating the manuscript into English.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by the University of Valencia’s project no. UV-INV-AE11-40221.
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study design and protocol were both approved by the Research in Humans Ethics Committee of the University of Valencia’s Experimental Research Ethics Committee.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Puig-Silla, M., Montiel-Company, J.M., Dasí-Fernández, F. et al. Prevalence of periodontal pathogens as predictor of the evolution of periodontal status. Odontology 105, 467–476 (2017). https://doi.org/10.1007/s10266-016-0286-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10266-016-0286-x