Abstract
Abscisic acid (ABA) response element (ABRE)-binding factors (ABFs) are basic region/leucine zipper motif (bZIP) transcription factors that regulate the expression of ABA-induced genes containing ABRE in their promoters. The amino acid sequence of the wheat bZIP protein, TaABI5, showed high homology to that of Arabidopsis ABA insensitive 5 (ABI5). TaABI5 was classified into the clade of ABI5s in Arabidopsis and rice, unlike TRAB1 of rice, Wabi5 of wheat, and HvABI5 of barley in the bZIP Group A family, by a phylogenetic analysis. TaABI5 was strongly expressed in seeds during the late ripening and maturing stages; however, its expression level markedly decreased after germination. An in situ hybridization analysis showed that TaABI5 mRNA accumulated in seed embryos, particularly the scutellum. In a transient assay using wheat aleurone cells, TaABI5 activated the promoter of Em containing ABRE, which is an embryogenesis abundant protein gene, indicating that TaABI5 acts as a transcription factor in wheat seeds. Furthermore, the seeds of transgenic Arabidopsis lines introduced with 35S:TaABI5 exhibited high sensitivity to ABA and the inhibition of germination. The seed dormancy of the transgenic Arabidopsis lines was stronger than that of Col. These results support TaABI5 playing an important role in mature seeds, particularly before seed germination, and acting as a functional ortholog to Arabidopsis ABI5.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In plants, basic region/leucine zipper motif (bZIP) transcription factors regulate the gene expression processes of pathogen defense, stress and hormone signaling, development, and seed maturation. The Arabidopsis and rice genomes contain 75 and 89 distinct members of the bZIP family (E et al. 2014; Jakoby et al. 2002). They have a basic region that binds DNA and a leucine zipper dimerization motif and have been classified into ten groups based on their structure and function in Arabidopsis (Jakoby et al. 2002). Some bZIP transcription factors assigned to Group A have roles in abscisic acid (ABA) or stress signaling (Choi et al. 2000; Finkelstein and Lynch 2000; Lopez-Molina et al. 2001; Uno et al. 2000). These bZIPs, named ABA response element (ABRE)-binding factors (ABFs) and ABA-responsive element-binding proteins (AREBs), bind to ABRE-containing promoters (Choi et al. 2000; Hobo et al. 1999a; Uno et al. 2000) and activate the expression of genes induced by ABA. Previous studies reported that Arabidopsis ABA insensitive 5 (ABI5) and rice TRAB1 were expressed in dry seeds and induced by abiotic stresses such as cold, drought, and high salinity (Finkelstein and Lynch 2000; Hobo et al. 1999b). The bZIP proteins of other groups have been shown to regulate the expression of genes encoding, for example, seed storage proteins (Onate et al. 1999; Onodera et al. 2001; Vicente-Carbajosa et al. 1997, 1998), pathogenesis-related (PR) proteins, ultraviolet and light-responsive proteins in blue light signal transduction (Kircher et al. 1998; Schindler et al. 1992; Weisshaar et al. 1991), and ent-kaurene oxidase in the gibberellin (GA) biosynthesis pathway (Fukazawa et al. 2000). In addition to the bZIPs classified into these groups, a large number of bZIP proteins are specifically expressed in defined tissues and are transcriptionally activated in plants after stress treatments (e.g. cold, drought, anaerobiosis, and wounding) (Kusano et al. 1995; Martinez-Garcia et al. 1998; Strathmann et al. 2001).
Pre-harvest sprouting (PHS) is one of the most serious issues affecting cereal cultivation because it reduces the quality of crops. In some countries including Japan, high humidity and low temperature during the late ripening and harvesting of wheat seeds damages grains due to PHS. Therefore, the mechanisms underlying seed dormancy and germination need to be elucidated in more detail. Seed dormancy and germination are controlled by a number of factors including the phytohormones, ABA and GA, as well as temperature and humidity during seed development and harvesting. ABA plays important roles in regulating processes, such as the synthesis of seed storage proteins, the promotion of seed desiccation tolerance and dormancy, and the inhibition of germination during reproductive growth (Cutler et al. 2010). As described above, the bZIP proteins of Group A are important key factors in ABA signal transduction. Among them, Arabidopsis ABI5 and OsABI5 of rice are mainly expressed in seeds, particularly dry seeds, and activate the promoters of the genes encoding late embryogenesis abundant proteins, such as Em, in Arabidopsis and rice (Carles et al. 2002; Himmelbach et al. 2003; Nakamura et al. 2001; Schultz et al. 1998; Vasil et al. 1995). In Arabidopsis, abi5 has been isolated as a mutant exhibiting ABA insensitivity in germination. A genetic analysis revealed that Arabidopsis ABI5 is essential for the induction of ABA-dependent growth arrest, which occurs after the breakage of seed dormancy, but prior to autotrophic growth (Lopez-Molina et al. 2001, 2002). ABA insensitive 3/VIVIPAROUS 1 (ABI3/VP1) is another important transcription factor containing a B3 domain for the regulation of ABA-induced gene expression and its loss-of-function causes precocious germination (McCarty et al. 1989; Suzuki et al. 1997, 2014). The Arabidopsis ABI5 protein interacts physically with ABI3 in order to enhance DNA-binding activity to the ABRE (Nakamura et al. 2001). In Arabidopsis, the phosphorylation of ABI5 by SnRK2-type kinase (SnRK2.2 and SnRK2.3) was also shown to be necessary for activating the transcription of ABA-dependent genes (Finkelstein and Lynch 2000; Liu et al. 2010; Lopez-Molina et al. 2001). In wheat, the mRNA of a bZIP, TaABF1, was shown to be expressed in mature seeds and its product was phosphorylated by PKABA1, which is a wheat SnRK2-type kinase that is involved in the ABA signaling pathway (Johnson et al. 2002).
Wheat expresses some ABA-related bZIP transcription factors, such as TaABF1 and a wheat ortholog of HvABI5, Wabi5 (Johnson et al. 2002; Kobayashi et al. 2008). Regarding Wabi5, Wabi5 was shown to be strongly expressed in wheat seedlings treated by low temperature, drought, and exogenous ABA, and Wabi5 functioned as a transcriptional regulator of the Cor/Lea genes in multiple abiotic stress responses (Kobayashi et al. 2008). On the other hand, as described above, TaABF1 was isolated as a bZIP protein specifically bound by PKABA1, which was expressed in mature seeds (Johnson et al. 2002). The deduced amino acid sequence of TaABF1 showed high homology to that of TmABF, which co-localized with one QTL for seed dormancy on chromosome 3Am of diploid wheat mapping lines (Nakamura et al. 2007). Therefore, TaABF1 may have a role in controlling seed germination. In the present study, we attempted to elucidate the role of a ABA-related bZIP transcription factor, TaABI5 cloned from hexaploid wheat in seed dormancy by analyzing mRNA accumulation patterns, the activation of an ABA-induced gene promoter, and transgenic Arabidopsis overexpressing TaABI5.
Materials and methods
Plant materials
The highly dormant cultivar Zenkouji-komugi (Zen, Accession 20815), weakly dormant cultivar Chinese Spring (CS, Accession 20054), and Minamino-komugi (Accession 21416) of hexaploid wheat (Triticum aestivum L.) were grown in fields in Kurashiki, Japan. Seeds were harvested at different developmental stages ranging from days after pollination (DAP) 0 to DAP 60. In in situ hybridization, seeds of the cultivar Norin-61 (Accession 21313) were grown at 13 °C in a greenhouse until maturation. These varieties were provided from Genetic Resources Center, the National Agriculture and Food Research Organization (NARO), Tsukuba, Japan (https://www.gene.affrc.go.jp/about.php). A variety of diploid wheat, T. monococcum L. variety KT3-5 (Accession KT003-005) was also grown in fields in Kurashiki and was used for transient expression analyses. This variety was provided from Kihara Institute for Biological Research, Yokohama City University, Japan (https://shigen.nig.ac.jp/wheat/komugi/strains/queryFormNbrp.jsp).
We used the wild-type low-dormancy Arabidopsis accession Columbia (Col, https://plant.rtc.riken.jp/resource/accession/accession_list.html) as a host plant for transformation. In order to germinate Arabidopsis, we pretreated seeds at 4 °C in darkness (i.e., cold conditions) for 5 days, and subsequently sowed them in pots containing compost soil (Jiffy Mix; Sakata, Yokohama, Japan). Plants were grown under continuous white light in a controlled-environment growth room at 24 °C.
RNA extraction and analysis of gene expression by RT-PCR
In the expression analysis of each seed developmental stage, seeds at DAP 0 and DAP 10 and embryos from seeds at DAP 20–60 were used for RNA extraction. In the RNA extraction of imbibed seeds, matured seeds, which were maintained at RT for 2 years, of Minamino-komugi were incubated on filter paper soaked with water at 24 °C during 0–72 h and the embryos were collected. Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) after seeds had been ground with a mortar and pestle. First-strand cDNA synthesis was performed using the PrimeScript™ RT regent Kit (TaKaRa Bio Inc., Japan). qRT–PCR using SYBR premix Ex Taq (TaKaRa Bio Inc., Japan) was performed using a Light cycler (Roche Diagnostics, Tokyo, Japan). Results from triplicate independent biological samples are shown, and error bars represent the SD. TaABI5 gene-specific primers used for qRT-PCR were shown in Table S1.
In situ hybridization analysis
Mature wheat seeds were fixed with Tissue Fixative (Genostaff Co., Ltd., Tokyo, Japan), embedded in paraffin, and sectioned at a thickness of 4 µm. A digoxigenin-labeled RNA probe from the N-terminal regions of TaABI51, 654 bp, with the primer set, ISHTaABI5-F1 (5′–catcggagatgagcaaggac–3′) and ISHTaABI5-R1 (5′–gtgccatcgggtacatcatt–3′), was used. Hybridization was performed according to the procedure of Genostaff Co., Ltd.. Coloring reactions were performed with NBT/BCIP solution (Roche Diagnostics) overnight and seeds were then washed with PBS. Sections were counterstained with Kernechtrot stain solution (Muto Pure Chemicals Co., Ltd., Tokyo, Japan), dehydrated, and mounted with Malinol (Muto Pure Chemicals Co., Ltd.).
Expression analysis for identifying the subcellular localization of the GFP-TaABI5 fusion protein
The pEGAD vector was used in the transient expression analysis of TaABI5 (Cutler et al. 2000). The Hind III-Cla I PCR fragment containing the TaABI5 coding region was cloned into the Hind III-Xba I site of pEGAD. Onion (Allium cepa) bulb scales were cut into small squares and placed on moistened filter paper. One microgram each of the 35S:GFP-TaABI5 plasmid was delivered into onion epidermal cells using gold particle bombardment. Gold particles (1.6 µm) were coated with DNA according to the manufacturer’s directions. Particles were bombarded into onion epidermal cells using the Biolistic PDS-1000/He system (Bio-Rad Laboratories, Inc., Tokyo, Japan) with 1100 psi rupture disks that were operated under vacuum conditions. After bombardment, cells were incubated at 24 °C for 24 h. Images were captured with a Biozero BZ-8000 fluorescence microscope (Keyence, Japan).
Transient expression analysis in wheat aleurone cells
The Act:ABI5 effector construct used in the transient assays was synthesized by cloning the rice Actin1 promoter (McElroy et al. 1990) and fragments containing the TaABI5-coding regions (Nakamura et al. 2007) into the pUC19 vector. The rice Actin1 promoter and 5' untranslated leader sequence (including intron I) of pDM302 (Cao et al. 1992) were cut out as a Hind III fragment and cloned into the Hind III site of pTH-2 (Niwa et al. 1999). The PCR fragment containing the TaABI5 coding region was cloned into the pBluescript SK+ (Agilent, Tokyo, Japan) T-vector. The entire insert was cut out as a Sal I–Not I fragment and cloned into the Sal I–Not I site of the pTH-2-fused Actin1 promoter. Act:TaVp1 and Act:GAmyb were described by Utsugi et al. (2006, 2008).
The aleurone tissues of mature wheat, KT3-5, were prepared for particle bombardment according to the procedure described by Utsugi et al. (2006). Twenty pieces of aleurone tissues were placed on each plate and bombarded by a particle gun using 1.6 µm gold particles coated with plasmid DNAs as described in the manual of the particle delivery system (Bio-Rad biolistic PDS-1000/He particle delivery system, Bio-Rad). In an experiment to examine the effects of TaABI5 on the Em promoter, 0.5 µg of an effector construct, either Act:TaABI5 or Act:TaVp1, and 1 µg of the reporter construct, Em:GUS (pBM113Kp) (Marcotte et al. 1988) were precipitated individually or as a mixture onto 0.75 mg of gold particles. Ubi1:Luciferase (LUC) was added to each sample at a ratio of 1:1 to the reporter plasmid as an internal control for the normalization of GUS expression. After being bombarded, aleurone tissues were incubated on filter paper soaked with water (control) or 10 µM ABA containing 50 U mL− 1 nystatin and 150 µg mL− 1 cefotaxime at 24 °C for 48 h. The preparation of extracts and GUS assays were conducted according to the procedure described by Lanahan et al. (1992). In the quantitative assay using 4-methyl-umbelliferyl β-D-glucuronide (MUG, Merck KGaA, Darmstadt, Germany), the tissue extract for each treatment was prepared from ten pieces by the method of Utsugi et al. (2006). A 100 µL aliquot of each extract from aleurone tissues or 1 µM 4-methylumbelliferone (4-MU, Merck KGaA, Darmstadt, Germany) as a control were added to 400 µL of lysis buffer containing 1 mM MUG and incubated at 37 °C. The reaction was terminated by the addition of 400 µL of 0.2 M Na2CO3, and fluorescence was measured with excitation at 365 nm and emission at 455 nm using a spectrofluorophotometer (RF-5300PC, Shimadzu Co., Japan). The GUS activity was calculated against the standard curve of 1 pmole 4-MU per 1 h. The GUS activity was normalized against the LUC activity of the internal control that was measured using 50 µL of each extract by a luciferase assay system (Promega Co., Madison, USA). Normalized GUS activity was expressed by multiplying the ratio of GUS activity/LUC activity by 1000. Each experiment was replicated at least eight times.
Generation of transgenic Arabidopsis plants
The full-length PCR products of TaABI5 were inserted into a binary vector modified from the vector pBI101, in which the expression of the inserted genes was driven by a constitutive 35S promoter. After validating the sequences of the transgenic constructs, we subsequently transformed them into Arabidopsis ecotype Col using the floral-dip method (Clough and Bent 1998). We also generated transgenic Arabidopsis, into which only the transformation vector was introduced. T1 seeds subjected to the cold pretreatment (described above) were selected on 0.8% agar plates containing MS salts supplemented with 75 µg mL− 1 kanamycin (Nacalai Tesque, Kyoto, Japan) and 10 µg mL− 1 hygromycin B (Merck KGaA, Darmstadt, Germany). Resistant T1 seedlings were then transferred into compost soil. To obtain plants homozygous for the transgenes, we randomly selected ten independent plants from the T3 progenies of a resistant T1 plant and tested them for the presence or absence of the transgenes using the PCR assay. We considered the presence of the transgenes in all of the selected T3 plants to be the criterion for considering the plants to be homozygous for the gene.
Seed germination assay of transgenic Arabidopsis plants
In Arabidopsis, the level of seed dormancy gradually decreases after seeds ripen (Alonso-Blanco et al. 2003). Thus, in our germination assay, we evaluated the germination of seeds close to the same maturity stage (DAP 21). In the germination assay, we plated 20 freshly harvested seeds from each plant on 0.8% agar in 9 cm plates and immediately transferred these plates to the growth room. We visually confirmed that all of the seeds sown on agar were mature and undamaged and were, thus, appropriate for the assay. We defined germination as occurring when the radicle was longer than 1 mm. To evaluate the level of dormancy, we counted the number of germinated seeds daily for 14 days after planting and calculated germination percentages.
Results
Phylogenetic analysis of the bZIP group A family
All genes in this phylogenetic analysis shown in Fig. 1 were classified as members of group A (Jacoby et al. 2002). The deduced amino acid sequence of TaABI5 (AB238932) used was nearly identical to those of TaABF1 (AF519804, 99%) and TaABFa (AF519803, 92%). These cDNA sequences were searched on the IWGSC RefSeq database (http://plants.ensembl.org/Triticum_aestivum/Info/Index). The results obtained indicated that both cDNA sequences of TaABI5 and TaABFa showed high homology to a wheat genomic sequence of an annotated gene, TraesCS3D02G364900 (99.4 and 100%, respectively), which was located on chromosome 3D (Table S2). The deduced amino acid sequences were almost identical to that of CS by the search (BLASTP); however, the two cDNAs, TaABI5 and TaABFa, were isolated from cultivars, Minamino-komugi and Brevor, respectively, while the RefSeq was a database of genomic DNA sequences of CS. Thus, it is no wonder that some SNPs are present among their DNA sequences cloned from different cultivars. TaABF1 (TaABFb) showed 100% identity to TraesCS3A02G371800, which was located on chromosome 3A, by the search (BLASTP); therefore, TaABF1 was a homoeologous gene to TaABI5 in the hexaploid wheat genome. TaABI5 also showed high homology to HvABF1 of barley (DQ786408, Schoonheim et al. 2007), OsABI5 of rice (OsABI5 isoform1, EF199630, Zou et al. 2007), and Arabidopsis ABI5 (NP001324734, Finkelstein and Lynch 2000) (88, 63, and 43%, respectively), as shown as clade 1 in Fig. 1. These bZIP proteins, which belong to the TaABI5 subfamily (clade 1), have been reported to be strongly expressed specifically in the dry seeds of Arabidopsis (Finkelstein and Lynch 2000) and rice (Os01g64000 in http://signal.salk.edu/cgi-bin/RiceGE). On the other hand, the amino acid sequences of Wabi5 of wheat (AB193553, Kobayashi et al. 2004), HvABI5 of barley (AY156992, Casaretto and Ho 2003), and TRAB1 of rice (AB023288, Hobo et al. 1999b) showed low homology to TaABI5 (35, 34, and 34%, respectively), as shown as clade 2 in Fig. 1. These results suggest that there are at least two phylogenetically distinct groups in the bZIP group A family (clades 1 and 2 in Fig. 1) as described by Zhou et al. (2017).
Phylogenetic tree of amino acid sequences of plant bZIP-type proteins (Group A). A phylogenetic analysis was performed by Phylogeny.fr (http://www.phylogeny.fr/) (Dereeper et al., 2008) and the resulting tree in the Newick format was rendered using Genetyx-tree (GENETYX CORPORATION, Tokyo) with four barley, three wheat, three rice, six Arabidopsis ABFs, Kfl00015_0390, Phypa235299, and Selmo406659. Clades 1, 2, and 3 (See the Results section) are indicated by red, green, and gray ellipses, respectively. Accession numbers for the corresponding amino acid sequences are: TaABI5 (AB238932); TaABF1 (AF519804); Wabi5 (AB193553); HvABF1 (DQ786408); HvABF2 (DQ786409); HvABF3 (DQ786410); HvABI5 (AY156992); OsABI5 isoform1 (EF199630.1); OsTRAB1 (AB023288); OsABF1 (XP015612904); AtABI5 (NP001324734); AtAREB1 (AB017160); AtAREB2 (AB017161); AtAREB3(AB017162); AtABF1 (NC003070.9); AtABF3 (NM001036708).
To clarify the relationship between the appearance of seed plants and differentiation within the group A family, a phylogenetical analysis was performed on the group A family together with orthologs from non-seed plants, lycophyte Selaginella moellendorffii, bryophyte Physcomitrella patens, and charophyte green algae Klebsormidium flaccidum. An ortholog search was conducted with the amino acid sequence of Arabidopsis ABI5 as the initial query using the Arabidopsis genome database and databases of these non-seed plants with genomes that have been sequenced: S. moellendorffii, P. patens, and K. flaccidum. The following ortholog pairs were identified: AtABF1 (NC003070, Sharma et al. 2011) of Arabidopsis and Selm147835 of S. moellendorffii, AtABF1 and Phypa235299 of P. patens, and AtAREB3 (AB017162, Uno et al. 2000) and kfl00015_039 of K. flaccidum. A phylogenetic tree of the group A family including these pairs was shown in Fig. 1. The previous two groups expected from homology to TaABI5 were shown to be clearly distinct in the resulting tree (clade 1 including ABI5s of Arabidopsis and rice, clade 2 including rice TRAB1). Another clade (clade 3) with relatively higher mutation rates was found (Fig. 1). Among the three clades above, Selm147835 and Phypa235299 depart from the same node. This result indicates that the differentiation of clade 1, the genes of which are expressed in dry seeds, such as TaABI5, from other clades occurred as early as the appearance of seed plants.
TaABI5 expression during seed development and dormancy in wheat
In order to investigate the role of TaABI5 in the control of seed germination in wheat, we examined the germination percentage of the dormant wheat cultivar, Zenkouji komugi (Zen) and the low dormant cultivar, Chinese Spring (CS). As shown in Fig. S1, seed weights of CS and Zen increased up to DAP 30 and 35, respectively, and gradually decreased after that. The water contents of both seeds decreased from DAP 30 to 40, and it was finally approximately 10% in fully matured seeds (DAP60). The results of the germination tests on the seeds of Zen and CS were shown in Fig. 2. In CS, the germination percentage of seeds at DAP 20 to 25 was 13%, and this gradually increased and reached 100% after DAP 55 (Fig. 2a). However, the seeds of Zen hardly germinated until DAP 55 and slightly germinated at DAP 60 (20%) even though they had completely acquired the ability to germinate (all half-grain seeds to break dormancy germinated after DAP 45, data not shown). The germination percentage of Zen seeds was significantly different from that of CS (P < 0.01).
Germination percentages and TaABI5 expression levels during the ripening of CS and Zen seeds. a Seed germination test of the dormant wheat cultivar Zen and low dormant wheat cultivar CS. Seeds harvested every 5 days during days after pollination (DAP) 20–60 were used for the germination test. Germination percentages for 14 days were shown. bTaABI5 expression in the ripening seeds of Zen and CS. qRT-PCR was performed by using RNA samples prepared from ovaries collected just after pollination (DAP 0), seeds at DAP10, and embryos at DAP 20, 30, 40, and 50 from CS and Zen. A total RNA of 0.1 µg from each tissue was reverse-transcribed and amplified by PCR. cDNA from wheat Actin1 was also amplified as a positive control for PCR. Each relative mRNA level of TaABI5 was normalized to the value of Actin1 as 1. The asterisk indicates that the level is significantly different from that of CS (P < 0.01). Error bars represent the SE (n = 3)
To examine the temporal expression pattern of TaABI5 during seed ripening, qRT-PCR was performed using RNA samples prepared from ovaries collected just after pollination (DAP 0), seeds at DAP 10, and embryos at DAP 20, 30, 40, and 50 from CS and Zen. As shown in Fig. 2b, TaABI5 mRNA was weakly expressed at DAP 0, but was more strongly expressed in seeds at DAP 10, and expression levels increased in embryos from DAP 20 to 50 in both CS and Zen. TaABI5 expression levels in Zen embryos peaked at DAP 40 and remained high at DAP 50, and these levels were significantly different from those of CS (P < 0.01) (Fig. 2b). Therefore, a marked difference was observed in expression levels between the two cultivars with different dormancy strengths.
To confirm the relationship between the accumulation of TaABI5 mRNA and germination, another qRT-PCR was performed using RNA extracted from the embryos of imbibed seeds. The non-dormant seeds of CS also germinated within only 1 day after imbibition. Whereas the non-dormant seeds of Zen did not germinate so quickly, and they needed at least 3 days to germinate after imbibition. On the other hand, Minamino-komugi is a dormant cultivar and the transcripts of TaABI5 were expressed in the mature seeds (Fig. 3). The matured seeds were harvested at DAP 60 and were maintained at RT for 2 years. These non-dormant seeds started germinating at 24 h after imbibition, and all seeds stably germinated until 36 h after imbibition (Fig. S2). Therefore, the seeds were used to analyze the expression pattern of TaABI5 in imbibed seeds in more detail. The results obtained showed that TaABI5 mRNA levels gradually increased in imbibed seeds and peaked at 24 h, but markedly decreased 36 h after the emergence of radicles and were not detected after 72 h (Fig. 3).
TaABI5 expression in Minamino-komugi seeds imbibed for 0, 12, 24, 36, 48, and 72 h. DAP 60 seeds after-ripened for 2 years from Minamino-komugi were imbibed for 0, 12, 24, 36, 48, and 72 h, and each RNA sample was prepared for qRT-PCR. A total RNA of 0.1 µg from each tissue was reverse-transcribed and amplified by PCR. cDNA from wheat Actin1 was also amplified as a positive control for PCR. Relative mRNA levels were normalized to the value of Actin1 as 1. Error bars represent the SE (n = 3)
Accumulation of TaABI5 mRNA in wheat seed tissues
TaABI5 mRNA accumulates in maturing seed grains, but not in seedlings treated with ABA and exposed to stresses, such as cold, NaCl, and drought (Johnson et al. 2002). In an attempt to demonstrate the expression pattern of TaABI5 in wheat seed tissues in detail, an in situ hybridization experiment was performed on the mature seeds of Norin-61 using TaABI5 RNAs as a probe. As shown in Fig. 4, TaABI5 mRNA accumulated in the embryo, particularly the scutellum.
Activation of the Em promoter by TaABI5
In order to confirm the intercellular localization of TaABI5, GFP-TaABI5 driven by the CaMV 35S promoter was introduced into onion epidermal cells by particle bombardment. As shown in Fig. S3, GFP was detected in the nuclei of cells into which the 35S:GFP-TaABI5 construct was introduced.
We conducted an efficient transient assay on aleurone tissues from the seeds of the diploid wheat line, KT3-5, using the reporter construct Em:GUS containing a GUS gene driven by the Em promoter (Fig. 5a) to clarify whether the TaABI5 product transactivates the promoter of Em, one of the ABA-responding genes in wheat. As an effector construct, we used Act:TaABI5 expressing TaABI5 driven by the rice Actin1 promoter (Fig. 5a). To investigate the effects of TaABI5 on the activation of the Em promoter, we also used the construct Act:TaVp1 expressing TaVp-B1 in wheat, which is another effector to the Em promoter (Utsugi et al. 2008). Aleurone tissues were co-bombarded with the Em:GUS reporter construct or a mixture of this reporter construct and one or two of the effector constructs, Act:TaVp1 and/or Act:TaABI5. The bombarded tissues were incubated on filter paper soaked with water or 10 µM ABA solution.
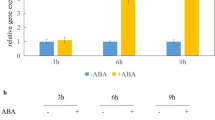
Effects of transiently expressed TaABI5 on Em gene expression in wheat aleurone cells. a Structures of the effector and reporter constructs. Em:GUS (pBM113Kp) is a reporter construct with a wheat Em promoter fused to the GUS and NOS reporter cassettes. GUS the β-glucuronidase coding sequence, NOS the nopaline synthase terminator sequence. Act:TaVP1, Act:TaABI5, and Act:TmGAmyb are effector constructs with TaVp-B1, TaABI5, and TmGAmyb coding sequences, respectively, inserted between the rice Actin1 promoter and NOS terminator. b Effects of the effectors Act:TaVP1 and Act:TaABI5 on the activation of the Em promoter. Aleurone tissues were bombarded with Em:GUS alone or co-bombarded with a mixture containing Em:GUS and either or both Act:TaVP1 and Act:TaABI5. Experiments were repeated at least eight times. The mean values of GUS activities in tissues incubated on filter paper soaked with ABA solution and in water are presented by black (ABA) and open (water) columns, respectively. See “Materials and methods” for the unit of GUS activity (pmole 4-MU h− 1 LUC1000− 1). The error bars indicate the standard errors of the means. + and – below the graph indicate the presence and absence of each construct, respectively. Act:TmGAmyb was used as a positive control for the assay. Asterisks mean each activity is significantly different from GUS activity in Em:GUS-introduced tissues in the absence of ABA (P < 0.01).
As shown in Fig. 5b, ABA increased the expression of GUS in Em:GUS-introduced tissues (4.2-fold increase). GUS activity was also enhanced by the TaVp1 effector in the absence of ABA (3.1-fold that of Em:GUS-introduced tissues), and its activity in the presence of ABA was almost the same as that of Em:GUS-introduced tissues. In tissues co-bombarded with Em:GUS and Act:TaABI5, GUS activities were significantly higher in the absence of ABA (5.1-fold) than Em:GUS-introduced tissues and were similar to those of Em:GUS-introduced tissues treated with ABA. GUS activity in the presence of ABA was markedly higher than that of Em:GUS-introduced tissues (7.6-fold increase), indicating that TaABI5 functions as an activator of the expression of Em. In addition, in tissues co-bombarded with Em:GUS and both Act:TaVp1 and Act:TaABI5 effectors, GUS activity was similar to those achieved through co-bombardment with Em:GUS and Act:TaABI5, suggesting that TaABI5 activates the Em promoter more strongly than the TaVp1 effector. As a negative control for effectors, GUS activity in tissues co-bombarded with Em:GUS and Act:GAmyb was shown (Fig. 5b). This pattern was similar to that in tissues bombarded with Em:GUS alone.
ABA sensitivity of transgenic Arabidopsis overexpressing TaABI5
In order to confirm the effects of dormancy by TaABI5, we introduced TaABI5 driven by the CaMV 35S promoter to Arabidopsis and analyzed seed germination by the transgenic plants. Three TaABI5-overexpressing transgenic lines, #2, #4, and #5, were obtained and the T4 homozygote seeds harvested at DAP 21 were used in subsequent germination tests and expression analyses. Harvests and experiments were repeated four times for each transgenic line.
The mRNA accumulation of introduced TaABI5 was confirmed in the seeds of transgenic lines #2, #4, and #5 (Fig. 6a). To examine ABA sensitivity in seed germination, 20 seeds of Col, E74-1 (abi5-7 in the Col background, Nambara et al. 2002), and the overexpressed lines were sown on MS medium containing 0, 0.3, 0.5, and 1 µM ABA, and were then incubated at 24 °C after a cold treatment at 4 °C for 4 days in order to break dormancy (Fig. 6). Germinated seeds were counted after 7 days. As shown in Fig. 6, the germination percentages of the three overexpressing lines on each ABA-containing medium were lower than that of WT, except for that of #2 seeds treated with 0.3 µM ABA, which was 55%. The germination percentages of #4 and #5 seeds on 0.3 µM ABA-containing medium were low at 20 and 5%, respectively. The germination percentages of #2, #4 and #5 seeds on 0.5 µM ABA-containing medium were low at 1, 0, and 3%, respectively. Transgenic plants grown on MS medium containing 0 or 0.3 µM ABA were shown in Fig. 6c. These results suggest that transgenic Arabidopsis lines were hypersensitive to ABA for seed germination by the ectopic overexpression of TaABI5.
ABA sensitivity of WT and T4 seeds of TaABI5-overexpressing transformants in Arabidopsis. a RT-PCR used Col, WS, the abi5 mutant (E74-1/Col background), and TaABI5-overexpressing transgenic lines #2, #4, and #5. TaABI5-specific primers (upper) and Actin primers for an internal control (bottom) were used. b ABA sensitivities of Col, TaABI5-overexpressing transgenic lines #2, #4, and #5, and the abi5 mutant (E74-1/Col background). Each T4 homozygote seed was harvested at DAP 21 and germination was tested. Seeds of WT, E74-1, and the 3 transgenic lines were sown on 0, 0.3, 0.5, and 1 µM ABA-containing MS medium, transferred to 24 °C after the cold treatment for 4 days, and germination percentages were then calculated 7 days after sowing. These experiments were repeated three times. Germination percentages were indicated by the symbols open circle, filled square, filled triangle, filled dimaond, and × for Col, transgenic lines #2, #4, #5, and E74-1, respectively. Bars indicate standard errors. c Plants of WT (Col) and TaABI5-overexpressing transgenic lines grown on MS or 0.3 µM ABA plates for 3 weeks. Asterisks (green) mean each germination percentage is significantly different from Col (P < 0.05) and E74-1 (P < 0.01) on 0.3 µM ABA-containing medium, and asterisks (blue) mean each is significantly different from Col (P < 0.05) and E74-1 (P < 0.01) on 0.5 µM ABA-containing medium.
Cold stratification on seed germination in transgenic Arabidopsis overexpressing TaABI5
Three overexpressed lines were examined to assess the effects of cold stratification. All germination tests were performed using freshly harvested seeds. Twenty seeds of Col and each overexpressing line were sown on MS medium, transferred to 24 °C after being treated at 4 °C for 0–4 days, and germinated seeds were counted for 14 days (Fig. 7a–c). Col seeds not exposed to the cold treatment started to germinate 4 days after sowing and the germination percentage reached 92% by 14 days (Fig. 7a). The seeds of transgenic lines #2 and #5 started to germinate 8 and 5 days after sowing, and germination percentages were 70 and 56%, respectively, after 14 days (Fig. 7a). However, the germination percentage of transgenic line #4 not exposed to the cold treatment was only 3% 14 days after sowing (Fig. 7a). When the seeds were cold-treated for 1 day after sowing, all Col seeds germinated 4 days after being transferred to 24 °C, and seed germination by transgenic lines #2 and #5 was slightly delayed and reached 100% after 5 days (Fig. 7b). On the other hand, the germination percentage of line #4 after the cold treatment for 1 day was 78% after 5 days and 80% after 14 days. Four days after the cold treatment, all Col seeds and the three transgenic lines germinated 3 days after being transferred, indicating that the dormant state of each line was completely broken by the cold treatment for 4 days (Fig. 7c). These results suggest that the freshly harvested seeds of TaABI5-overexpressing lines needed longer cold stratification processing than those of Col to break dormancy.
Effects of cold stratification and after-ripening on seed germination in Col and T4 of TaABI5-overexpressing transgenic Arabidopsis lines. a–c Effects of cold stratification and seed germination on Col and TaABI5-overexpressing transformants. The CaMV35S promoter:TaABI5 construct was introduced into Arabidopsis. Freshly harvested seeds of Col and transgenic lines were sown onto MS medium and treated at 4 °C for 0 days (no cold treatment) (a), 1 day (b), and 4 days (c) and were then transferred to 24 °C. d–f Effects of after-ripening on the germination of Col and TaABI5-overexpressing transformants. Seeds incubated at RT for 1 week (d), 3 weeks (e), and 5 weeks (f). All experiments were repeated three times. Germination percentages were indicated by the symbols open circle, filled square, filled triangle and filled dimaond for Col, transgenic lines #2, #4, and #5, respectively. Asterisks (* or **) mean each germination percentage is significantly different from Col (P < 0.05 or P < 0.01, respectively) after 14 days.
After-ripening effects on harvested seeds of TaABI5-overexpressing Arabidopsis
In order to analyze the seed dormancy of post-harvested transgenic lines, the germination percentages of seeds incubated at room temperature for 1, 3, and 5 weeks after the harvesting of Col and three transgenic lines were compared with those of freshly harvested seeds. When seeds were incubated at 24 °C for 1 week after harvesting, each germination curve of Col and the transgenic lines (Fig. 7d) was very similar to that of freshly harvested seeds (Fig. 7a). The seeds of Col started to germinate 4 days after sowing and the germination percentage was 98% after 14 days (Fig. 7d). The seeds of transgenic lines #2 and #5 started to germinate 5 and 4 days after sowing, and germination percentages were 75 and 72%, respectively, on day 14 (Fig. 7d). The germination rate of transgenic line #4 was very low with a percentage of only 5% 14 days after sowing (Fig. 7d). Col seeds incubated for 3 weeks at 24 °C started to germinate after 4 days and the germination percentage reached 100% by day 8 (Fig. 7e). The seeds of transgenic lines #2 and #5 incubated for 3 weeks started to germinate 5 days after sowing and the germination percentage of both lines reached almost 100% after 14 days (Fig. 7e). However, the seeds of transgenic line #4 hardly germinated after being incubated for 3 weeks (10% 14 days after sowing) (Fig. 7e). After a 5-week incubation at 24 °C, 67% of Col seeds germinated after 3 days and the germination percentage reached 100% at 7 days (Fig. 7f). The seeds of transgenic lines #2, #4, and #5 incubated for 5 weeks started to germinate after 4 days (4.9, 5.0, and 5.0%, respectively, they were significantly different from Col (P < 0.01)) and the germination percentages of lines #2 and #5 rapidly increased and reached almost 100% after 11 days, whereas that of line #4 slowly increased and reached only 35% (Fig. 7f). These results showed that the germination of Col and three transgenic lines gradually increased after ripening; however, the germination percentages of transgenic lines were always lower than that of Col. The germination curves of transgenic lines #2 and #5 after ripening for 3 weeks were similar to that of Col after ripening for 1 week, suggesting that the dormancy of these transgenic lines became stronger or longer than that of Col after harvesting (seed ripening).
Discussion
The clade including TaABI5 may be characterized by seed-specific expression
Figure 1 shows that the bZIP group A family of seed plants was divided into three subgroups by a phylogenetic analysis. TaABI5 was shown to be specifically expressed in seeds, and Arabidopsis ABI5, belonging to the same clade (clade1), were reported to be expressed in seeds including dry seeds (Bensmihen et al. 2002; Finkelstein et al. 2000). On the other hand, TRAB1 of rice, Wabi5 of wheat, and HvABI5 of barley, which belong to a different clade (clade 2) from TaABI5, were reported to have different expression patterns to that of TaABI5 (Hobo et al. 1999b; Kobayashi et al. 2008; Mezer et al. 2014). Although phylogenetic analyses are not directly related to gene expression, these results suggest that the expression patterns of genes belonging to a phylogenetic group are similar to each other because of the inheritance of their promoter and UTR regions from a common ancestor gene. Therefore, genes belonging to clade 1 may be mainly expressed in seeds. The tree shown in Fig. 1 provides no insight into the time line of the differentiation of group A because Selm147835 of S. moellendorffii and Phypa235299 of P. patens from non-seed plants and three clades from seed plants departed from the same node. However, the probability that clade 1 consists of genes mainly expressed in seeds is high because orthologs from non-seed plants and their ortholog (AtABF1; clade 2, as shown by the ortholog search of the phylogenetic analysis) in the Arabidopsis genome are out of clade 1. As described above, group A of seed plants differentiates into three subgroups; two (clades 1 and 2) have been differently characterized, while the other one (clade 3) remains unclear. Commonly named genes are irregularly found among these clades, and very similar genes are assigned different names. This may be confusing for researchers, and, hence, the renaming of genes according to a set of rules is needed.
TaABI5 is a positive regulator of dormancy in wheat seeds
In the present study, we clarified that TaABI5 mRNA highly accumulated in late ripening seeds, particularly in the scutellum and surrounding embryo of mature seeds (Figs. 2b, 4). The scutellum plays an important role in the secretion and transport of materials, such as starch hydrolysis enzymes, signal molecules, and water (Rathjen et al. 2009), suggesting that the transcriptional regulation of some enzymes by TaABI5 controls germination by transporting materials through the scutellum. Furthermore, the mRNA of wheat MOTHER OF FT AND TFL1 (TaMFT), the expression of which was up-regulated in the seeds of plants grown at a low temperature after physiological maturity and the precocious germination of which was suppressed in immature embryos overexpressing TaMFT, also accumulated in the scutellum and embryos of wheat seeds. The expression pattern was spatially and temporally similar to that of TaABI5 (Nakamura et al. 2011). TaABI5 and TaMFT may interact with each other and regulate gene expression in wheat seeds, similar to ABI5 and MFT in Arabidopsis (Xi et al. 2010). In Arabidopsis seeds, Penfield et al. (2006) reported that ABI5:GUS was expressed in the embryo and micropylar region of the endosperm, suggesting that this region controls seed germination by physically holding the root emergency process. Although the structure of wheat seeds is different from that of Arabidopsis, TaABI5 mRNA may also accumulate around the embryo containing the plumule and radicle in order to regulate emergence for germination. In rice, Zou et al. (2007) demonstrated that two alternative splicing products from OsABI5, OsABI5-1 and OsABI5-2, have different expression patterns, specificities to bind to G-box in ABRE, and transactivation activities; however, they have completely identical functional domains. The results of an analysis of TaABI5 transcripts revealed that some types of isoforms, which are different from TaABI5 in the C-terminal sequence following the leucine zipper, were included in the transcripts of wheat dry seeds (data not shown). However, this was hard to decipher due to the difficulties associated with distinguishing missplicings from homeologs derived from the hexaploid wheat genome. Although we checked TaABI5 transcripts in developmental seeds (DAP 10 and DAP 30) and seedlings, we failed to detect alternatively spliced products of TaABI5 in different tissues or at different developmental stages, unlike OsABI5 (data not shown).
ABI5 interacts with ABI3/VP1 in seeds. They directly bind to G-box (–CACGTG–) in ABRE of the promoter of ABA-regulated genes, such as Em, which encodes one of the LEA proteins. ABI5 enhances its own DNA-binding activities by interacting with ABI3/VP1 and activating the transcription of ABA-regulated genes (Carles et al. 2002). In the present study (Fig. 5b), when TaVp1 or TaABI5 was introduced with Em promoter:GUS to wheat aleurone cells, GUS activity in these cells was higher than that of cells into which TaVp1 was introduced. This result suggests that TaABI5 directly and strongly binds to the Em promoter region and activates it. As shown in Fig. 5b, TaVp1 also activated the Em promoter in aleurone cells, suggesting that TaVp1 binds directly to the promoter of Em or that it may interact with endogenous TaABI5 or other bZIPs that bind directly to the Em promoter. Nevertheless, these results suggest that TaABI5 effectively activates the expression of ABA-induced genes by interacting with VP1, and regulates the ABA signal in wheat seeds.
We demonstrated that TaABI5 activated the transcription of the Em promoter containing ABRE using a transient assay with wheat aleurone cells. Furthermore, Arabidopsis transgenic lines overexpressing TaABI5 exhibited hyper-sensitivity to ABA and decreased germination rates (Fig. 6). These results suggest that TaABI5 is a positive regulator that activates ABA-induced genes containing ABRE in the promoter, and the accumulation of TaABI5 suppresses seed germination. The contribution of TaABI5 to seed dormancy has also been supported by a QTL analysis. Nakamura et al. (2007) reported that TaABI5 is a positional candidate gene for seed dormancy because one QTL for seed dormancy was co-located with TmABF on chromosome 3Am in four QTLs of diploid wheat mapping lines. As described previously, TaABI5 is specifically expressed in seeds and belongs to the same clade as Arabidopsis ABI5 in the phylogenetic tree, and its expression increases sensitivity to ABA and seed dormancy, suggesting that it is functionally an ortholog of Arabidopsis ABI5.
Furthermore, as another target gene of ABI5, a tonoplast intrinsic protein from barley (HvTIP3;1) was reported by Lee et al. (2015). The HvTIP3;1 promoter contained three cis-acting elements that respond to ABA, and HvTIP3;1 prevented the coalescence of protein storage vacuoles, which is a vacuolar structural change for mobilization during cereal grain germination, in aleurone cells. However, because they also showed that the HvTIP3;1 promoter did not increase at all in their ABA responsiveness by knock-down of HvABI5 or HvVP1 in transient expression assays (Lee et al. 2015), ABA signaling factors other than HvABI5 may need the response for HvTIP3;1 by ABA. At least, it was reported that HvTIP3;1 is highly expressed in seeds and is involved in the control of intracellular water transportation in a previous study (Utsugi et al. 2015). These results indicate that TaABI5 controls seed activation and inactivation through its regulated gene products, suggesting that it plays an important mechanistic role in seed dormancy regulated by ABA.
References
Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-De Vries H, Koorneef M (2003) Analysis of_natural_allelic_variation_at_seed_dormancy_loci_of Arabidopsis thaliana. Genetics 164:711–729
Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, Giraudat J, Parcy F (2002) The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14:1391–1403
Cao J, Duan X, McElroy D, Wu R (1992) Regeneration of herbicide resistant transgenic rice plants following microprojectile-mediated transformation of suspension culture cells. Plant Cell Rep 11:586–591
Carles C, Bies-Etheve N, Aspart L, Léon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30:373–383
Casaretto J, Ho TH (2003) The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. Plant Cell 15:271–284
Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA responsive element binding factors. J Biol Chem 275:1723–1730
Clough SJ, Bent AF (1998) Floral dip: a simplified method_for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant_J 16:735–743
Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97:3718–3723
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61:651–579
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O Dereeper (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469 (Web Server issue)
E ZG, Zhang YP, Zhou JH, Wang L (2014) Roles of the bZIP gene family in rice. Genet Mol Res 13:3025–3036
Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12:599–609
Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y (2000) REPRESSION OF SHOOT GROWTH, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12:901–915
Himmelbach A, Yang Y, Grill E (2003) Relay and control of abscisic acid signaling. Curr Opin Plant Biol 6:470–479
Hobo T, Asada M, Kowyama Y, Hattori T (1999a) ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J 19:679–689
Hobo T, Kowyama Y, Hattori T (1999b) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. PNAS 96:15348–15353
Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111
Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK (2002) The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol 130:837–846
Kircher S, Ledger S, Hayashi H, Weisshaar B, Schäfer E, Frohnmeyer H (1998) CPRF4, a novel plant bZIP protein of the CPRF family: comparative analysis of light dependent expression, post-transcriptional regulation, nuclear import and heterodimerisation. Mol Gen Genet 257:595–605
Kobayashi F, Maeta E, Terashima A, Takumi S (2008) Positive role of a wheat HvABI5 ortholog in abiotic stress response of seedlings. Physiol Plant 134:74–86
Kobayashi F, Takumi S, Nakamura C (2004) Regulation of cold-responsive Cor/Lea genes and their transcription factors by the major freezing tolerance locus Fr-1 in wheat. Recent Res Dev Plant Sci 2:249–266
Kusano T, Berberich T, Harada M, Suzuki N, Sugawara K (1995) Amaize DNA-binding factor with a bZIP motif is induced by low temperature. Mol Gen Genet 248:507–517
Lanahan MB, Ho THD, Rogers SW, Rogers JC (1992) A gibberellin response complex in cereal [alpha]-amylase gene promoters. Plant Cell 4:203–211
Lee SE, Yim HK, Lim MN, Yoon IS, Kim JH, Hwang YS (2015) Abscisic acid prevents the coalescence of protein storage vacuoles by upregulating expression of a tonoplast intrinsic protein gene in barley aleurone. J Exp Bot 66:1191–1203
Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98:4782–4787
Liu H, Stone SL (2010) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22:2630–2641
Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32:317–328
Martinez-Garcia JF et al (1998) Two bZIP proteins from Antirrhinum flowers preferentially bind a hybrid C-box/G-box motif and help to define a new subfamily of bZIP transcription factors. Plant J 13:498–505
Marcotte WR, Bayley C Jr, Quatrano RS (1988) Regulation of a wheat promoter by abscidic acid in rice protoplast. Nature 335:454–457
McCarty DR, Carson CB, Stinard PS, Robertson DS (1989) Molecular analysis of viviparous-1: an abscisic acid-insensitive mutant of maize. Plant Cell 1:523–532
McElroy D, Zhang W, Cao J, Wu R (1990) Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2:163–171
Mezer M, Turska-Taraska A, Kaczmarek Z, Glowacka K, Swarcewicz B, Rorat T (2014) Differential physiological and molecular response of barley genotypes to water deficit. Plant Physiol Biochem 80:234–248
Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, Utsugi S, Ogawa T, Handa H, Ishida H, Mori M, Kawaura K, Ogihara Y, Miura H (2011) A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23:3215–3229
Nakamura S, Komatsuda T, Miura H (2007) Mapping diploid wheat homologues of Arabidopsis seed ABA signaling genes and QTLs for seed dormancy. Theor Appl Genet 114:1129–1139
Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26:627–635
Nambara E, Suzuki M, Abrams S, McCarty DR, Kamiya Y, McCourt P (2002) A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics 161:1247–1255
Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H (1999) Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J 18:455–463
Oñate L, Vicente-Carbajosa J, Lara P, Díaz I, Carbonero P (1999) Barley BLZ2, a seed-specific bZIP protein that interacts with BLZ1 in vivo and activates transcription from the GCN4-like motif of B-hordein promoters in barley endosperm. J Biol Chem 274:9175–9182
Onodera Y, Suzuki A, Wu CY, Washida H, Takaiwa F (2001) A rice functional transcriptional activator, RISBZ1, responsible for endosperm-specific expression of storage protein genes through GCN4 motif. J Biol Chem 276:14139–14152
Penfield S, Li Y, Gilday AD, Graham S, Graham IA (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18:1887–1899
Rathjen JR, Strounina EV, Mares DJ (2009) Water movement into dormant and non-dormant wheat (Triticum aestivum L.) grains. J Exp Bot 60:1619–1631
Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR (1992) Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J 11:1261–1273
Schoonheim PJ, Sinnige MP, Casaretto JA, Veiga H, Bunney TD, Quatrano RS, de Boer AH (2007) 14-3-3 adaptor proteins are intermediates in ABA signal transduction during barley seed germination. Plant J 49:289–301
Schultz TF, Medina J, Hill A, Quatrano RS (1998) 14-3-3 proteins are part of an abscisic acid-VIVIPAROUS1 (VP1) response complex in the Em promoter and interact with VP1 and EmBP1. Plant Cell 10:837–847
Sharma PD, Singh N, Ahuja PS, Reddy TV (2011) Abscisic acid response element binding factor 1 is required for establishment of Arabidopsis seedlings during winter. Mol Biol Rep 38:5147–5159
Strathmann A, Kuhlmann M, Heinekamp T, Dröge-Laser W (2001) BZI-1 specifically heterodimerises with the tobacco bZIP transcription factors BZI-2, BZI-3/TBZF and BZI-4, and is functionally involved in flower development. Plant J 28:1–15
Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97:11632–11637
Suzuki M, Kao CY, McCarty DR (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9:799–807
Suzuki M, Wu S, Li Q, McCarty DR (2014) Distinct functions of COAR and B3 domains of maize VP1 in induction of ectopic gene expression and plant developmental phenotypes in Arabidopsis. Plant Mol Biol 85:179–191
Utsugi S, Maekawa M, Noda K (2006) An efficient transient gene expression system using aleurones of diploid wheat seeds. Plant Biotechnol 23:413–417
Utsugi S, Nakamura S, Noda K, Maekawa M (2008) Structural and functional properties of Viviparous1 genes in dormant wheat. Genes Genet Syst 83:153–166
Utsugi S, Shibasaka M, Maekawa M, Katsuhara M (2015) Control of the water transport activity of barley HvTIP3;1 specifically expressed in seeds. Plant Cell Physiol 56:1831–1840
Vasil V, Marcotte WR Jr, Rosenkrans L, Cocciolone SM, Vasil IK, Quatrano RS, McCarty DR (1995) Overlap of Viviparous1 (VP1) and abscisic acid response elements in the Em promoter: G-box elements are sufficient but not necessary for VP1 transactivation. Plant Cell 7:1511–1518
Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ (1997) A maize zincfinger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci USA 94:7685–7690
Vicente-Carbajosa J, Oñate L, Lara P, Diaz I, Carbonero P (1998) Barley BLZ1: a bZIP transcriptional activator that interacts with endosperm-specific gene promoters. Plant J 13:629–640
Weisshaar B, Armstrong GA, Block A, da Costa e Silva O, Hahlbrock K (1991) Light-inducible and constitutively expressed DNA-binding proteins recognizing a plant promoter element with functional relevance in light responsiveness. EMBO J 10:1777–1786
Xi W, Liu C, Hou X, Yu H (2010) MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22:1733–1748
Zhou K, Yang J, Wang ZX, Wang JR (2017) Sequence analysis and expression profiles of TaABI5, a pre-harvest sprouting resistance gene in wheat. Genes Genom 39:161–171
Zou M, Guan Y, Ren H, Zhang F, Chen F (2007) Characterization of alternative splicing products of bZIP transcription factors OsABI5. Biochem Biophys Res Commun 360:307–313
Acknowledgements
We would like to thank Dr. Quatorano for kindly providing the pBM113Kp construct and Dr. Nambara for providing E74-1 (abi5-7). We are grateful to Dr. Nakazono for providing the wheat material Norin-61.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material
Rights and permissions
About this article
Cite this article
Utsugi, S., Ashikawa, I., Nakamura, S. et al. TaABI5, a wheat homolog of Arabidopsis thaliana ABA insensitive 5, controls seed germination. J Plant Res 133, 245–256 (2020). https://doi.org/10.1007/s10265-020-01166-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-020-01166-3