Abstract
Rhizosphere acidification is a general response to Pi deficiency, especially in dicotyledonous plants. However, the signaling pathway underlying this process is still unclear. Here, we demonstrate that miR156 is induced in the shoots and roots of wild type Arabidopsis plants during Pi starvation. The rhizosphere acidification capacity was increased in 35S:MIR156 (miR156 overexpression) plants, but was completely inhibited in 35S:MIM156 (target mimicry) plants. Both 35S:MIR156 and 35S:MIM156 plants showed altered proton efflux and H+-ATPase activity. In addition, significant up-regulation of H+-ATPase activity in 35S:MIR156 roots coupled with increased citric acid and malic acid exudates was observed. qRT-PCR results showed that most H+-ATPase and PPCK gene transcript levels were decreased in 35S:MIM156 plants, which may account for the decreased H+-ATPase activity in 35S:MIM156 plants. MiR156 also affect the root architecture system. Collectively, our results suggest that miR156 regulates the process of rhizosphere acidification in plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phosphorus (P) is a macronutrient that plays a vital role in many physiological processes, such as energy transfer, signal transduction, the synthesis of biological molecules, photosynthesis, and respiration. Plant roots acquire P in the form of phosphate (Pi) from the soil (Vance et al. 2003). Although Pi is abundant in the soil, most of it is in the form of precipitates with cations and is immobile, limiting Pi uptake (Raghothama 1999). The excretion of organic acids and protons from the roots is one of the strategies plants use to cope with Pi deficiency. Root exudation acidifies the soil to release insoluble P, which helps plants to take up and use P (Vance et al. 2003; Zhang et al. 2004). The induction of rhizosphere acidification by Pi deficiency has been reported in many species. White lupin (Lupinus albus L.) develops so-called proteoid roots to acidify the rhizosphere soil under Pi deficient conditions (Li et al. 1997). During this process, the release of organic acids (mainly citric acid) is coupled to proton efflux (Neumann and Römheld 1999; Yan et al. 2002). Furthermore, it was found that there is a close link between the burst of citrate exudation and plasma H+-ATPase-catalyzed proton efflux (Tomasi et al. 2009; Zhu et al. 2005). Salicylic acid and citramalic acid were exuded by sugar beet roots to solubilize P in response to Pi deficiency (Khorassani et al. 2011). The roots of rape (Brassica napus L.) plants excreted malic and citric acids under Pi deficiency, resulting in rhizosphere acidification and solubilization of rock phosphate (Hoffland et al. 1989). Acid soluble Ca phosphates were used by chickpea and tomato in response to Pi deficiency (Dinkelaker 1990; Hinsinger and Gilkes 1997).
The rhizosphere acidification phenomenon was explored very early, but studies on the molecular mechanism of acidification are still at the initial stage. A phosphoenol pyruvate carboxylase gene (LaPEPC3) was reported to be related to citrate exudation in the cluster roots of white lupin (Cheng et al. 2014; Penaloza et al. 2005). In Arabidopsis, phosphorylation of AtPPC1 by AtPPCK1 and AtPPCK1 contributed to the metabolic adaptations in response to Pi starvation (Gregory et al. 2009; Plaxton and Tran 2011). The root plasma membrane H+-ATPase was involved in the modulation of Pi uptake in soybean under Pi starvation (Shen et al. 2006). The plasma membrane H+-ATPase gene OsA8 influenced the uptake and translocation of Pi in rice (Chang et al. 2009). H+-pyrophosphatase AVP1 (AVP, Arabidopsis vacuolar pyrophosphatase) mediated rhizosphere acidification and root proliferation in Arabidopsis (Yang et al. 2007).
In recent years, the role of plant microRNAs in responses to environmental stress has generated widespread interest. A number of significant achievements have already been made in determining the function of the key molecules miR399 and miR827 in the regulation of phosphorus homeostasis (Bari et al. 2006; Fujii et al. 2005; Huang et al. 2013; Lin et al. 2008). miR399 is upregulated under Pi deficiency and directs the cleavage of PHOSPHATE2 (PHO2) (Aung et al. 2006; Bari et al. 2006; Chiou et al. 2006; Fujii et al. 2005). miR827 has been functionally characterized in both Arabidopsis and rice with regard to its roles in Pi starvation signaling (Lin et al. 2010; Wang et al. 2012).The target of miR827 in Arabidopsis, NITROGEN LIMITATION ADAPTATION (NLA) and PHO2 direct the degradation of plasma membrane-localized PHT1s in different subcellular compartments (Lin et al. 2013; Liu et al. 2014). In addition, deep sequencing of Arabidopsis small RNAs was conducted to reveal miRNAs that were differentially expressed in response to phosphate (Pi) deficiency, such as miR156, miR169, miR395 and miR2111 (Hsieh et al. 2009). However, the molecular basis of these Pi responsive miRNAs remains to be explored.
miR156 and its targets the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) family play critical roles in diverse developmental processes such as phase transition, flowering time, leaf development and anthocyanin biosynthesis (Bergonzi et al. 2013; Gou et al. 2011; Usami et al. 2009; Wang et al. 2008, 2009; Wu et al. 2009; Zhou et al. 2013). Recently, miR156 has also been reported to participate in multiple stress responses such as recurring heat stress and salt stress (Cui et al. 2014; Stief et al. 2014). However, whether miR156 is involved in rhizosphere acidification under Pi deficiency was still unknown. Therefore, the objective of the present study was to investigate the influence of miR156 on the process of root acidification. Our results may serve as an important basis for detailed studies of miR156 that regulate rhizosphere acidification in plants.
Materials and methods
Plant growth and stress treatments
Dry seeds of Arabidopsis thaliana were surface sterilized using 0.1 % HgCl2 for 5 min, washed 5–7 times using sterilized water and placed on MS medium plates (containing MS salts, 3 % sucrose and 0.6 % agar, pH 5.8) at 4 °C for 3 day. The plates were placed in growth chambers (day/night cycle of 16/8 h at 22 °C; light intensity of 90–120 mmol m−2 s−1; relative humidity of 80 %) for 1–2 weeks. Low Pi stress treatment of plants was made by modification of MS medium such that the Pi concentration in LP medium was 12.5 µM. The agar of LP medium was replaced by agarose (Promega, Madison, WI, USA) to avoid contamination with phosphorous. For nutrition solution treatment, the LP solution was made without Pi.
Quantitative RT-PCR
Total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). After DNase I treatment, cDNA was synthesized from 0.5 μg total RNA using Moloney murine leukemia virus reverse transcriptase (Invitrogen) with the oligo(dT) primer. Mature miR156 expression levels were determined following the methods of Xing et al. (2010) and Yang et al. (2009). Total RNA was extracted using a miRNA extraction kit (Kang Wei, Beijing, China). After DNase I treatment, 200 ng RNA was used for first-strand cDNA synthesis. SYBR Green was used in subsequent qRT-PCR. All qRT-PCR experiments were performed with two or three biological replicates. All specific primers used are listed in Table S2. Relative expression levels were normalized to ACTIN2 or 18S rRNA.
Histochemical analysis of GUS activity
Plants expressing the PromiR156a:GUS and PromiR156d:GUS transgenes in the wild-type background were used for histological analysis. Seedlings of transgenic plants grown in MS or low Pi (LP, 12.5 μM) medium were incubated at 37 °C for at least 4 h in staining buffer (100 mM phosphate buffer pH 7.0, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1 % Triton X-100, 0.1 % X-gluc) in the dark. Chlorophyll was removed with an ethanol series consisting of 50, 70 and 100 % ethanol washes at room temperature for 30 min each. The GUS activity was determined according to Jefferson et al. (1987).
Detection of rhizosphere acidification
After cultivation for 9 day on MS medium, seedlings were placed on MS medium or LP medium that contained 0.75 % (w/v) agar (or agarose) and 0.006 % (w/v) bromocresol purple (pH 5.8). Rhizosphere acidification was visualized after incubation for 8 h. The pH calibration was performed according to the color of medium containing bromocresol purple with different pH (from 5.2 to 6.8).
Quantitative measurements were performed as previously described (Santi and Schmidt 2009) with some modifications. Three hundred microliters of MS or low Pi (LP) solution (0.5 % sucrose, 0.006 % bromocresol purple, pH 5.8) was added into 96-well plates. The 9-day-old seedlings were transferred to the plates and three seedlings were pooled as one sample; three samples were set per treatment. The plates were placed in growth chambers for 2–3 h. A 100 μL solution of each sample was removed to another 96-well plate. Rhizosphere acidification capacity was accessed by reading the adsorption at 590 nm (A 590). To determine the pH values of the rhizosphere, the adsorption was converted to pH according to the pH calibration. The pH calibration was performed according to the color of MS solution containing bromocresol purple with different pH (from 5.3 to 6.0).
Assay of cytoplasmic pH in root epidermal cells
The assay was performed as described previously (Gao et al. 2004; Moseyko and Feldman 2001) with some modifications. 35S:MIR156 and 35S:MIM156 plants containing pH sensitive GFP were created by crossing with N9561 lines obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/StockInfo?NASCid=9561). Nine-day-old seedlings were transferred from 0.6 MS medium to MS solution for 1 day, and then transferred to MS solution with or without Pi for 2 h. The root hairs of seedlings were used to monitor intracellular pH. GFP fluorescence was monitored using a FV 1000 confocal microscope (exciters, 405 and 488 nm; emitter, 525 nm). Fluorescence images were taken every 3 s. The F405 to F488 ratio was used as a measure of pH. pH levels were pseudo-color-coded according to the calibrated 410/470 nm ratio image of the same root hair.
Measurement of H+-ATPase activity
Fourteen-day-old Arabidopsis seedlings were transferred to MS or LP solution (0.5 % sucrose, pH 5.8) for 1 day. After the seedling roots were isolated, the plasma membranes were prepared as described previously (Larsson et al. 1987) and the protein was quantified according to Bradford (1976). The H+-ATPase activity was determined as described previously (Ames 1966; Yan et al. 2002).
Collection of root exudates and analysis of carboxylates
Three hundred microliters of MS or LP solution (0.5 % sucrose, pH 5.8) were added into 96-well plates. Ten-day-old seedlings were transferred to the plates and three seedlings were pooled as one sample. The plates were placed in growth chambers for 1 day and the solution was collected. The roots were carefully rinsed twice with deionized water, dried and weighed. The root exudate solution was filtered through a 0.2 μm filter and analyzed by HPLC with a 300 × 7.8 mm column (Aminex HPX-87H); the mobile phase was 10 mmol/L H2SO4 with a flow rate of 1 mL min−1 at 55 °C.
Results
miR156 is induced during Pi starvation
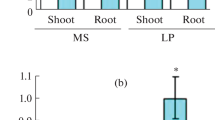
Eight loci (MIR156A to H) that encode pri-miR156 have been reported in A. thaliana and miR156 may be involved in Pi deficiency (Hsieh et al. 2009; Xing et al. 2010; Yang et al. 2013). To investigate the function of miR156 in Pi deficiency, the expression of mature miR156 in Arabidopsis was analyzed. The seeds of wild-type (WT) seedlings were sowed on MS medium for 7 day and then transferred to Pi sufficient or LP medium for 5 day. Quantitative real time (qRT) PCR results indicated that miR156 was induced significantly in both roots and shoots during Pi starvation (Fig. 1a). To further study the expression pattern of miR156, we observed the GUS staining of PromiR156a:GUS and PromiR156d:GUS plants. As shown in Fig. 1b, when grown in low Pi, the PromiR156a:GUS seedlings displayed stronger GUS activity in their shoots. Additionally, marginally stronger GUS activity was detected in the root tips of PromiR156d:GUS seedlings. Consistent with the above qRT-PCR results, GUS activity in PromiR156a:GUS and PromiR156d:GUS plants also increased in response to Pi deficiency (Fig. 1c). However, the induction level of PromiR156a:GUS and PromiR156d:GUS plants were much lower compared with qRT-PCR results. Thus, the expression of MIR156A-H were examined (Fig. S1a). The results show that MIR156B and MIR156E play dominant roles within the miR156 gene family in response to Pi starvation. Although MIR156A and D were also induced, their expression levels were lower than those of MIR156B and MIR156E.
miR156 is induced in response to phophate starvation. a qRT-PCR analysis of miR156 expression. Wild-type seedlings were grown on MS medium for 7 day and transferred to low Pi (LP) medium for 6 h. b GUS staining of PromiR156a:GUS and PromiR156d:GUS transgenic lines. Tissues were stained 7 day after transfer to MS or LP medium. Bar 1 mm. c The assay of GUS activity in PromiR156a:GUS and PromiR156d:GUS plants in response to normal states and Pi deficiency. Error bars indicate the ±SD of three biological replicates
The only target family of miR156 is SQUAMOSA PROMOTER BINDING LIKE (SPL) transcription factors (Rhoades et al. 2002). To confirm whether SPLs was repressed in Pi starvation, the expression of SPLs transcript in wild type seedlings after transferring from MS to LP medium were also examined by qRT-PCR. As shown in Fig. S2, the expression of SPL genes was generally lower at low Pi. In addition, SPL3 mRNA level is the most significant repression of expression at low Pi conditions.
The rhizosphere acidification capacity was changed in miR156 transgenic plants under Pi deficiency
Rhizosphere acidification is one of the strategies by which plants promote the release of undissolved Pi from soil minerals. We tested whether miR156 was involved in the process of rhizosphere acidification. The transgenic plants 35S:MIR156 (miR156 overexpression; Schwab et al. 2005) and 35S:MIM156 (Franco-Zorrilla et al. 2007) were analyzed. It is noteworthy that 35S:MIM156 is an artificial target mimics which contains a motif with sequence complementarity to miR156, but the pairing is interrupted by a mismatched loop. It sequesters miR156, thereby resulting in increased SPL mRNA accumulation and activity. The expression of MIR156B was induced dramatically in 35S:MIR156 plants (Fig. S1b), which is consistent with the previous report (Schwab et al. 2005). The expression of MIR156B was also repressed significantly in 35S:MIM156 plants.
Nine-day-old 35S:MIR156, 35S:MIM156 and WT seedlings were transferred to MS or low Pi (12.5 μM) medium that contained 0.006 % (w/v) bromocresol purple. Bromocresol purple was chosen as the pH indicator; its color changed from yellow to purple when the pH increased. After 8 h, WT plants (the control) acidified the low Pi medium. At the same time, the rhizosphere acidification capacity was increased in 35S:MIR156 plants, but was completely inhibited in 35S:MIM156 plants (Fig. 2a). To detect whether plasma membrane H+ pumping was involved in the process of root acidification, the H+-ATPase inhibitor vanadate (1 mM) was added into the medium. After 8 h of cultivation, the color of the rhizosphere did not change for any of the P-deficient plants, indicating that root acidification was completely inhibited by 1 mM vanadate (Fig. 2b).
Acidification capacity are enhanced or decreased in 35S:MIR156 and 35S:MIM156 transgenic plants. a Seedlings grown for 9 day were transferred to MS and low Pi (LP) medium with the addition of 0.006 % (w/v) bromocresol purple for 1 day. Four biological replicates were analyzed, with similar results. Bar 1 cm. b 1 mM vanadate was added in the corresponding medium for study of the inhibitory effect of vanadate. Four biological replicates were analyzed, with similar results. Bar 1 cm. c For the determination of pH values of the rhizosphere, 9-day-old plants were incubated in the test solution for 24 h. Error bars show standard deviation of 12 plants analysed in two independent experiments. Asterisks represent significant differences from the wild type (P ≤ 0.05 from a Student’s t test)
To further quantify the pH variation of plants in the rhizosphere, 9-day-old seedlings were transferred to 96-well plates containing MS or LP standard nutrient solution (0.5 % sucrose, 0.006 % bromocresol purple, pH 5.8). After 3 h incubation in the test solution, the absorption at 590 nm (A 590) was read, and then converted into pH according to a pH curve. The results showed that the rhizosphere pH in MS solution was 5.57 ± 0.01 for the WT, 5.46 ± 0.02 for the 35S:MIR156 plants and 5.69 ± 0.01 for the 35S:MIM156 plants. In the LP solution, the rhizosphere pH was 5.32 ± 0.03 for the WT, 5.2 ± 0.04 for the 35S:MIR156 plants and 5.43 ± 0.03 for the 35S:MIM156 plants (Fig. 2c).
The above results showed that rhizosphere acidification by the plants was enhanced under low Pi stress and the pH decreased obviously. Compared with the WT, the rhizosphere acidification capacity of the 35S:MIR156 plants was enhanced whereas that of the 35S:MIM156 plants was decreased.
miR156 modulates the cytosolic pH of root hair cells in response to Pi deficiency
Plant roots acidify the rhizosphere soil and increase proton export in response to P deficiency (Yan et al. 2002). However, how the intracellular pH changes is not clear. 35S:MIR156 and 35S:MIM156 plants containing ratiometric pH-sensitive GFP were obtained by crossing with the Pt-GFP line N9561 (Moseyko and Feldman 2001). Nine-day-old seedlings were transferred to MS solution for 1 day. Then, the seedling roots were used to monitor cytosolic pH by incubating them in low Pi solution for 2 h. We monitored GFP fluorescence using a confocal microscope. The 405/488 nm ratio image was converted to intracellular pH according to a pH standard curve. As shown in Fig. 3b, the cytosolic pH of the WT was increased from 6.98 ± 0.17 to 7.31 ± 0.24 after low Pi treatment, while the pH of 35S:MIR156 plants changed from 6.82 ± 0.24 to 8.21 ± 0.18, which was higher than the WT. The pH of 35S:MIM156 plants increased marginally from 7.01 ± 0.09 to 7.12 ± 0.17, which was marginally lower than the WT after low Pi treatment (Fig. 3a).
Cytoplasmic pH in root epidermal cells of 35S:MIR156 and 35S:MIM156 transgenic plants. a 405/488 ratio fluorescence images of different processing in root epidermal cell of 35S:MIR156, 35S:MIM156 transgenic plants and wild type plants. Bar 10 µm. b cytoplasmic pH in root epidermal cell of 35S:MIR156, 35S:MIM156 transgenic plants and wild type plants. Error bars represent mean ± SD of three biological replicates with two technical replicates each. Asterisks represent significant differences from the wild type (P ≤ 0.05 from a Student’s t test)
Next, the root cells were treated with a H+-ATPase inhibitor (1 mM vanadate) for 15 min. We observed that the ratio image became extremely weak, indicating that the pH in root epidermal cells decreased significantly (Fig. 3b).
These results suggested that proton export by root cells was induced in response to Pi deficiency, which caused cytoplasmic alkalization. The intracellular pH of 35S:MIR156 plants was remarkably changed under Pi starvation. On the contrary, the pH of 35S:MIM156 plants did not change significantly. When the H+-ATPase activity was inhibited, the cytosolic pH of all of the transgenic plants and WT plants was decreased (Fig. 3b), which indicates that H+-ATPase is involved in the process of Pi deficiency response.
The H+-ATPase activity of 35S:MIR156 plants was enhanced during Pi starvation
To gain a deeper insight into the relationship between rhizosphere acidification and H+-ATPase, the plasma membrane ATPases of WT, 35S:MIR156 and 35S:MIM156 roots were assayed. Fourteen-day-old seedlings were treated with MS or LP solution for 1 day. The roots were collected, the total membrane protein was extracted and the H+-ATPase activity was determined. The plasma membrane H+-ATPase activity of WT and 35S:MIR156 plants was increased dramatically. Moreover, the activity of 35S:MIR156 plants was higher than the WT. By contrast, the H+-ATPase activity of 35S:MIM156 plants decreased significantly (Fig. 4). Consistent with a previous report (Yan et al. 2002), plasma membrane H+-ATPase was closely related with rhizosphere acidification. The increased H+-ATPase activity of 35S:MIR156 plants may be one of the reasons for their higher rhizosphere acidification capacity. Likewise, the decreased H+-ATPase activity of 35S:MIM156 plants after LP treatment may inhibit their rhizosphere acidification capacity.
Comparison of H+-ATPase activity of plasma membranes derived from different roots of 35S:MIR156, 35S:MIM156 transgenic plants and wild type plants. Plants were grown on MS medium for 14 day and transferred to nutrition solution with or without Pi for 1 day. Bars represent mean ± SD of three biological replicates with two technical replicates each. Asterisks represent significant differences from the wild type (P ≤ 0.05 from a Student’s t test)
Root organic acid exudates of 35S:MIR156 plants were increased in response to Pi deficiency
It is generally accepted that enhanced exudation of organic anions from roots is connected with Pi deficiency (Ryan et al. 2001). To determine whether organic acid exudates were changed in 35S:MIR156 and 35S:MIM156 plants, the root exudates of WT, 35S:MIR156 and 35S:MIM156 plants were tested by HPLC. We mainly assayed citric acid, malic acid and succinic acid from the root exudates of the WT and transgenic plants. These acids were present in root exudates from both MS and LP treatments (Fig. 5). As expected, the concentrations of the three excreted compounds from the WT were all higher with LP than with MS treatment. Under both treatments, the citric acid and malic acid exuded from 35S:MIR156 plants were significantly higher than the WT, whereas the exudates of 35S:MIM156 plants were not significantly different from the WT except that the concentration of citric acid was marginally higher. These results showed that the root organic acid (mainly citric acid and malic acid) exudates of 35S:MIR156 plants were increased in response to Pi deficiency. Maybe this is one of the reasons for the increased rhizosphere acidification of 35S:MIR156 plants.
The assay of organic acid contents in root and root exudates. 35S:MIR156, 35S:MIM156 transgenic plants and wild type plants were grown on MS medium for 21 day and transferred to nutrition solution with or without Pi for 1 day. The root exudates were collected and assayed by HPLC. Bars represent mean ± SD of three biological replicates. Asterisks represent significant differences from the wild type with MS treatment (P ≤ 0.05 from a Student’s t test)
Expression of acidification-related genes
To understand the molecular basis of changes in rhizosphere acidification, we monitored the expression of H+-ATPase and H+-pyrophosphatase genes, such as AHA1, AHA2, AHA7 and AVP1. Seven-day-old seedlings were transferred to LP medium for 1 day, and were then harvested for RNA extraction. The qRT-PCR results showed that transcripts of all four genes were lower in 35S:MIM156 seedlings than in the WT under Pi deficiency (Fig. 6). The transcript level of AHA7 in 35S:MIR156 seedlings was the same as in the WT in response to Pi deficiency, whereas transcripts of AVP1 and the other two genes (AHA2 and AHA7) were increased and decreased marginally respectively compared with the WT. In addition, the expression of PPCK1 and PPCK2, encoding the PEPC protein kinase, was also evaluated. Compared with WT plants, the expression of PPCK1 was decreased in 35S:MIM156 seedlings, while the expression of PPCK2 was also decreased in 35S:MIR156 during Pi deficiency. These results indicated that the overexpression and suppression of miR156 had different effects on the genes involved in rhizosphere acidification.
Expression patterns of root acidification related genes in mutants and wild type. Seeds of the 35S:MIR156, 35S:MIM156 transgenic plants and wild type plants were grown on MS agar medium for 7 day and then transferred to the MS or LP medium for 1 day. The ACTIN2 transcript level was used for normalization. Bars represent mean ± SD of three biological replicates with two technical replicates each. Asterisks represent significant differences from the wild type (P ≤ 0.05 from a Student’s t test)
As transcription factor family, the core binding motif of SPLs is GTAC consensus sequence (Birkenbihl et al. 2005). An analysis of the promoter regions of the above acidification related genes, indicated that all of them have one or more GTAC sequence on their promoters (Table S1).
MiR156 affect the root system architecture
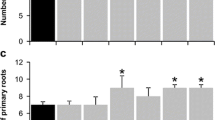
It has been reported that the root system architecture (RSA) affect the process of rhizosphere acidification (Li et al. 1997; Yan et al. 2002). Therefore, it was logical to examine the RSA of 35S:MIR156, 35S:MIM156 and wild-type seedlings under normal and low Pi states (Fig. S3a–c). 35S:MIM156 plants caused decreases in the number and length of lateral roots compared with wild type under normal and low Pi conditions. Besides, the number of lateral roots increased in 35S:MIR156 under LP condition. The number and length of root hairs were also detected under both conditions (Fig. S3d–f). It was found that the length of root hairs increased in miR156 overexpression mutants under both states. Moreover, the number of root hairs of 35S:MIR156 increased only in normal condition. Together, these data mean that miR156 is involved in the lateral root and root hair development process. The involvement of miR156 in RSA development may also contribute to the phenotype of rhizosphere acidification.
Discussion
Rhizosphere acidification is a common response to Pi deficiency, especially in dicotyledonous plants. The roots of plants exude more H+ than HCO3− or OH−, resulting in acidification of the rhizosphere soil (Neumann and Römheld 1999; Raghothama 1999). A previous study screened dozens of rhizosphere acidification deficiency mutants, one of which was the spl1 mutant (Lei et al. 2015). Our research showed that the upstream regulator of SPL genes-miR156 also take part in the process of rhizosphere acidification. In Arabidopsis, the process of rhizosphere acidification occurred in a short time. There were significant color changes around the roots of seedlings grown on LP medium containing 0.006 % (w/v) bromocresol purple for 8 h. We observed that the acidification ability of 35S:MIR156 and 35S:MIM156 seedlings increased and decreased, respectively (Fig. 2a). In addition, the H+-ATPase inhibitor 1 mM vanadate blocked the acidification process completely (Fig. 2b). Yan et al. (2002) reported that plasma membrane H+-ATPase is essential for enhanced rhizosphere acidification by active proteoid roots in white lupin. Combined with the H+-ATPase activity assay results (Fig. 4), we confirmed that plasma membrane H+-ATPase plays a vital role in the process of rhizosphere acidification in Arabidopsis.
To clarify the cytosolic pH changes of root hair cells in response to Pi deficiency, the cytosolic pH was assayed using pH-sensitive GFP fluorescence. The cytosolic pH of 35S:MIR156 plants increased compared with the WT under Pi deficiency (Fig. 3). This phenomenon was in accordance with the increased H+-ATPase activity of 35S:MIR156 plants. The increased H+-ATPase activity may promotes proton efflux, resulting in an increase of cytosolic pH in the root hair cells of 35S:MIR156 plants.
It was reported that citric acid release was closely correlated with H+ release in white lupin (Zhu et al. 2005). In this study, we found significant up-regulation of H+-ATPase activity in 35S:MIR156 roots coupled with high citric acid and malic acid exudation (Fig. 5). These results demonstrate that H+ release and organic acid exudates from roots contribute to rhizosphere acidification in Arabidopsis.
In Arabidopsis, 11 out of 17 SPL genes are targeted by miR156 (Rhoades et al. 2002). Thus, we tested the molecular mechanism by which miR156 is involved in rhizosphere acidification under Pi deficiency. MiR156 was induced and targeted SPL family was repressed in response to Pi starvation (Fig. 1, Figs. S1 and S2). Table S1 also showed that the promoter of these acidification related genes all have GTAC motifs. In addition, we found that AHA1/2/7 and PPCK genes transcript levels were decreased in 35S:MIM156 plants, which may account for the decreased H+-ATPase activity in 35S:MIM156 plants (Fig. 6). Furthermore, the expression levels of these genes in 35S:MIR156 plants were the same as or lower than in WT plants. Only the expression of AVP1 gene in 35S:MIR156 plants increased compared with WT. The AVP1 gene might be the target of SPL gene, which needs further research.
On the other hand, the lateral roots and root hairs development of two transgenic plants of miR156 are different from wild type (Fig. S3), which suggest that miR156 is involved in regulation of lateral root and root hair development. The increase of root surface area may be attributed to the increased rhizosphere acidification capacity. Thus, further investigation is required to elucidate the mechanisms of interaction between the root development and the process of rhizosphere acidification.
Cui et al. (2014) reported that miR156 integrates environmental signals (such as salt stress) to ensure timely flowering. In this study, miR156 is also involved in the rhizosphere acidification process in response to Pi deficiency. MiR156 may be a key regulator which integrate the environmental signals into developmental process. Taken together, our results demonstrate that miR156 is involved in the rhizosphere acidification process in terms of proton release and carboxylate exudation. miR156 positively regulates the rhizosphere acidification process and contributes to coping with insoluble phosphorus stress. This research opens a whole new research direction in the search for novel miRNA signaling machinery related to rhizosphere acidification and is potentially a substantial advance in understanding the molecular mechanisms of rhizosphere acidification.
References
Ames BN (1966) Assay of inorganic phosphate, total phosphate and phosphatase. Methods Enzymol 18:115–118
Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141:1000–1011
Bari R, Datt Pant B, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141:988–999
Bergonzi S, Albani MC, Loren Ver, van Themaat E, Nordstrom KJ, Wang R, Schneeberger K, Moerland PD, Coupland G (2013) Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340:1094–1097
Birkenbihl RP, Jach G, Saedler H, Huijser P (2005) Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol 352:585–596
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chang C, Hu Y, Sun S, Zhu Y, Ma G, Xu G (2009) Proton pump OsA8 is linked to phosphorus uptake and translocation in rice. J Exp Bot 60:557–565
Cheng L, Tang X, Vance CP, White PJ, Zhang F, Shen J (2014) Interactions between light intensity and phosphorus nutrition affect the phosphate-mining capacity of white lupin (Lupinus albus L.). J Exp Bot 65:2995–3003
Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL (2006) Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant cell 18:412–421
Cui LG, Shan JX, Shi M, Gao JP, Lin HX (2014) The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J 80:1108–1117
Dinkelaker B (1990) Genotypische Unterschiede in der Phosphateffizienz von Kichererbse (Cicer arietinum L.). Ph.D. Thesis, Universität Hohenheim, Stuttgart
Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39:1033–1037
Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15:2038–2043
Gao D, Knight MR, Trewavas AJ, Sattelmacher B, Plieth C (2004) Self-reporting Arabidopsis expressing pH and [Ca2+] indicators unveil ion dynamics in the cytoplasm and in the apoplast under abiotic stress. Plant Physiol 134:898–908
Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23:1512–1522
Gregory AL, Hurley BA, Tran HT, Valentine AJ, She YM, Knowles VL, Plaxton WC (2009) In vivo regulatory phosphorylation of the phosphoenolpyruvate carboxylase AtPPC1 in phosphate-starved Arabidopsis thaliana. Biochem J 420:57–65
Hinsinger P, Gilkes RJ (1997) Dissolution of phosphate rock in the rhizosphere of five plant species grown in an acid, P-fixing mineral substrate. Geoderma 75:231–249
Hoffland E, Findenegg G, Nelemans J (1989) Solubilization of rock phosphate by rape. Plant Soil 113:161–165
Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151:2120–2132
Huang TK, Han CL, Lin SI, Chen YJ, Tsai YC, Chen YR, Chen JW, Lin WY, Chen PM, Liu TY, Chen YS, Sun CM, Chiou TJ (2013) Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant cell 25:4044–4060
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Khorassani R, Hettwer U, Ratzinger A, Steingrobe B, Karlovsky P, Claassen N (2011) Citramalic acid and salicylic acid in sugar beet root exudates solubilize soil phosphorus. BMC Plant Biol 11:121
Larsson C, Widell S, Kjellbom P (1987) Preparation of high-purity plasma-membranes. Meth Enzymol 148:558–568
Lei KJ, Xie JY, Zhu YY, Song CP, An GY (2015) Screening and analysis of rhizosphere acidification deficiency mutants in Arabidopsis thaliana under low phosphorus. Soil Sci Plant Nutr 61:1–8
Li M, Shinano T, Tadano T (1997) Distribution of exudates of lupin roots in the rhizosphere under phosphorus deficient conditions. Soil Sci Plant Nutr 43:237–245
Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ (2008) Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147:732–746
Lin SI, Santi C, Jobet E, Lacut E, El Kholti N, Karlowski WM, Verdeil JL, Breitler JC, Perin C, Ko SS, Guiderdoni E, Chiou TJ, Echeverria M (2010) Complex regulation of two target genes encoding SPX-MFS proteins by rice miR827 in response to phosphate starvation. Plant Cell Physiol 51:2119–2131
Lin WY, Huang TK, Chiou TJ (2013) Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant cell 25:4061–4074
Liu TY, Lin WY, Huang TK, Chiou TJ (2014) MicroRNA-mediated surveillance of phosphate transporters on the move. Trends Plant Sci 19:647–655
Moseyko N, Feldman LJ (2001) Expression of pH-sensitive green fluorescent protein in Arabidopsis thaliana. Plant Cell Environ 24:557–563
Neumann G, Römheld V (1999) Root excretion of carboxylic acids and protons in phosphorus-deficient plants. Plant Soil 211:121–130
Penaloza E, Munoz G, Salvo-Garrido H, Silva H, Corcuera LJ (2005) Phosphate deficiency regulates phosphoenolpyruvate carboxylase expression in proteoid root clusters of white lupin. J Exp Bot 56:145–153
Plaxton WC, Tran HT (2011) Metabolic adaptations of phosphate-starved plants. Plant Physiol 156:1006–1015
Raghothama KG (1999) Phosphate aquisition. Annu Rev Plant Physiol Plant Mol Biol 50:665–693
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110:513–520
Ryan P, Delhaize E, Jones D (2001) Function and mechanism of organic anion exudation from plant roots. Annu Rev Plant Physiol Plant Mol Biol 52:527–560
Santi S, Schmidt W (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183:1072–1084
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Shen H, Chen J, Wang Z, Yang C, Sasaki T, Yamamoto Y, Matsumoto H, Yan X (2006) Root plasma membrane H+-ATPase is involved in the adaptation of soybean to phosphorus starvation. J Exp Bot 57:1353–1362
Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Baurle I (2014) Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26:1792–1807
Tomasi N, Kretzschmar T, Espen L, Weisskopf L, Fuglsang AT, Palmgren MG, Neumann G, Varanini Z, Pinton R, Martinoia E, Cesco S (2009) Plasma membrane H+-ATPase-dependent citrate exudation from cluster roots of phosphate-deficient white lupin. Plant Cell Environ 32:465–475
Usami T, Horiguchi G, Yano S, Tsukaya H (2009) The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136:955–964
Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157:423–447
Wang JW, Schwab R, Czech B, Mica E, Weigel D (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20:1231–1243
Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138:738–749
Wang C, Huang W, Ying Y, Li S, Secco D, Tyerman S, Whelan J, Shou H (2012) Functional characterization of the rice SPX-MFS family reveals a key role of OsSPX-MFS1 in controlling phosphate homeostasis in leaves. New Phytol 196:139–148
Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138:750–759
Xing S, Salinas M, Hohmann S, Berndtgen R, Huijser P (2010) miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22:3935–3950
Yan F, Zhu Y, Muller C, Zorb C, Schubert S (2002) Adaptation of H+-pumping and plasma membrane H+-ATPase activity in proteoid roots of white lupin under phosphate deficiency. Plant Physiol 129:50–63
Yang H, Knapp J, Koirala P, Rajagopal D, Peer WA, Silbart LK, Murphy A, Gaxiola RA (2007) Enhanced phosphorus nutrition in monocots and dicots over-expressing a phosphorus-responsive type I H+- pyrophosphatase. Plant Biotechnol J 5:735–745
Yang H, Schmuke JJ, Flagg LM, Roberts JK, Allen EM, Ivashuta S, Gilbertson LA, Armstrong TA, Christian AT (2009) A novel real-time polymerase chain reaction method for high throughput quantification of small regulatory RNAs. Plant Biotechnol J 7:621–630
Yang L, Xu M, Koo Y, He J, Poethig RS (2013) Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2:e00260
Zhang WH, Ryan PR, Tyerman SD (2004) Citrate-permeable channels in the plasma membrane of cluster roots from white lupin. Plant Physiol 136:3771–3783
Zhou CM, Zhang TQ, Wang X, Yu S, Lian H, Tang H, Feng ZY, Zozomova-Lihova J, Wang JW (2013) Molecular basis of age-dependent vernalization in Cardamine flexuosa. Science 340:1097–1100
Zhu Y, Yan F, Zorb C, Schubert S (2005) A link between citrate and proton release by proteoid roots of white lupin (Lupinus albus L.) grown under phosphorus-deficient conditions? Plant Cell Physiol 46:892–901
Acknowledgments
We thank Professor Jiawei Wang for providing the 35S:MIR156 and 35S:MIM156 seeds. We also thank Dr. Shuping Xing for providing the PromiR156a:GUS and PromiR156d:GUS seeds. This work was supported by the National Natural Science Foundation of China (31371426, 31570263).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lei, K.J., Lin, Y.M. & An, G.Y. miR156 modulates rhizosphere acidification in response to phosphate limitation in Arabidopsis . J Plant Res 129, 275–284 (2016). https://doi.org/10.1007/s10265-015-0778-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-015-0778-8