Abstract
Nematodes such as Caenorhabditis elegans offer a very effective and tractable system to probe the underlying mechanisms of diverse sensory behaviors. Numerous platforms exist for quantifying nematode behavior and often require separate dependencies or software. Here I describe a novel and simple tool called NemaCount that provides a versatile solution for the quantification of nematode chemotaxis behavior. The ease of installation and user-friendly interface makes NemaCount a practical tool for measuring diverse behaviors and image features of nematodes such as C. elegans. The main advantage of NemaCount is that it operates from within a modern browser such as Google Chrome or Apple Safari. Any features that change in total number, size, shape, or angular distance between control and experimental preparations are suited to NemaCount for image analysis, while commonly used chemotaxis assays can be quantified, and statistically analyzed using a suite of functions from within NemaCount. NemaCount also offers image filtering options that allow the user to improve object detection and measurements. NemaCount was validated by examining nematode chemotaxis behavior; angular distances of locomotory tracks in C. elegans; and body lengths of Heterorhabditis bacteriophora nematodes. Apart from a modern browser, no additional software is required to operate NemaCount, making NemaCount a cheap, simple option for the analysis of nematode images and chemotaxis behavior.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nematodes, like Caenorhabditis elegans, are very widely used for the study of incredibly diverse questions in basic biology ranging from aging (Murphy et al. 2003) to neural behavior (Bargmann et al. 1993) to cell death (Yuan et al. 1993) to magnetotaxis (Vidal-Gadea et al. 2015). In each of these cases, software can be very helpful in measuring and statistically analyzing diverse behaviors and phenotypes (Husson et al. 2012; Kwon et al. 2013; Ramot et al. 2008; Swierczek et al. 2011; Wang and Wang 2013; Yang et al. 2011). Here I present a user-friendly and simple tool called NemaCount, which can be unpacked onto a MAC or PC system in seconds and used immediately to functionally analyze images and behaviors of nematodes from within a modern browser such as Google Chrome or Apple Safari (Fig. 1). All scripts, sample images, frameworks, and libraries are downloaded with NemaCount, and so the user does not need to install any dependencies or additional software apart from a browser. NemaCount is also available as a Google Chrome browser extension from the Chrome Web Store. NemaCount can be used to quantify any features that change in total number, size, shape, or angular distance between control and experimental samples. Linear or freeform distances can easily be measured using NemaCount to quantify animal lengths or a region of interest such as a cell body. The user can also enumerate independent color-coded objects using NemaCount, which can be very helpful in scoring behavioral assays, cell counts, or any other objects that may vary between control and experimental samples. NemaCount offers a variety of functions that will calculate chemotaxis indices for diverse behavioral assays including chemotaxis to volatile odorants (Bargmann et al. 1993), avoidance assays from volatile odorants (Troemel et al. 1997), population-based linear thermotaxis (Goodman et al. 2014; Ito et al. 2006), chemotaxis to soluble compounds (Hart 2006), aversion assays from soluble compounds (Hart 2006; Ward 1973), and also quadrant-based chemotaxis assays (Beets et al. 2012; Wicks et al. 2000). Users can also apply image filters to the locally loaded image; these image filters include a grayscale filter that converts an image to black and white, a white removal option, an invert option that inverts the image coloring, and also a sharpen option that enhances the image quality. Using NemaCount users can input chemotaxis indices from behavioral assays to calculate t test values and p values using Student’s t test density distributions. Although numerous other software solutions exist for quantifying nematode behavior (Cronin et al. 2005; Husson et al. 2012; Kwon et al. 2013; Ramot et al. 2008; Swierczek et al. 2011; Wang and Wang 2013), the utility of NemaCount is that it specifically facilitates the quantification and analysis of nematode chemotaxis from within a browser. Furthermore, the ease of installation and lack of any dependencies required to set up the platform makes NemaCount a cheap, simple option for the analysis of nematode images and chemotaxis behavior.
NemaCount user interface. An image of a chemotaxis assay plate containing C. elegans animals is loaded into NemaCount by clicking the Choose File button and functions selected using the various menus. Animals are counted using the Count function and color coded for animals at the odor (blue) or control (red) or middle (green). NemaCount calculates the chemotaxis index and returns an alert box containing the value which is shown on top of the image. NemaCount will also perform significance comparisons between datasets and return t test values and p values as shown in the alert pop-up box at the bottom
Materials and methods
Animal maintenance
Caenorhabditis elegans strain N2 (wild type) and CX2016 odr-1(n1936) X were maintained at 20 °C on NGM plates seeded with E. coli strain OP50 following standard protocols (Brenner 1974). MT947 unc-31(n422) IV was maintained at 15 °C and assayed at 25 °C. All C. elegans strains were obtained from the Caenorhabditis Genetics Center (CGC). Heterorhabditis bacteriophora strain TTO1 animals (a gift from Dr. John Hawdon at George Washington University) were cultured in vivo at 25 °C in Galleria mellonella (wax moth) larvae using standard protocols (White 1927; Woodring and Kaya 1998). Larvae were infected by dropping a liquid suspension using a dropper containing infective juveniles. The infected larvae were placed on water traps (White 1927) to collect the infective-stage nematodes. The water traps were constructed and nematodes harvested as described previously (O’Halloran and Burnell 2003; O’Leary et al. 1998). To obtain parasitic juvenile stages of H. bacteriophora, the G. mellonella cadavers were harvested 6–8 days post-infection by cutting them open with a scalpel inside a 10-cm Petri dish containing M9 buffer. The nematodes were then washed twice with ddH20. To obtain adult H. bacteriophora nematodes, the G. mellonella cadavers were dissected 5 days post-infection in a 10-cm Petri dish containing M9 buffer and female adults were picked using an aspirator.
Chemotaxis assays
Chemotaxis was performed as described previously (Bargmann et al. 1993). The odor benzaldehyde was diluted using 1 μl benzaldehyde (Sigma) in 200 μl EtOH. The animals were washed from plates using modified S-basal (50 mL 1 M K3PO4 buffer, pH ~6.0, 20 mL 5 M NaClaq and brought to 1 l with ddH2O) three times. Animals were then placed on chemotaxis assay plates (10 ml of 1.6 % agar in 5 mM potassium phosphate (pH 6.0), 1 mM CaCl2, and 1 mM MgSO4) at the origin (bottom of plate—see Fig. 1). One microliter of 1 M sodium azide was placed at the points where the experimental and control compounds are to be placed, and allowed to dry for several minutes before the experimental and control compounds were applied. The animals were allowed to move for 2 h, and then, the number of animals at the odor, control, and middle of the plate counted. All assays were repeated three times on separate days.
Behavioral quantification in NemaCount
NemaCount is written using JavaScript and uses the fabric.js library and jQuery framework. The user interface was written using HTML5 and CSS. NemaCount has been tested and validated on both PC (Windows 7) and MAC (OS X) using Google Chrome (v 48.0.2564.116) and Apple Safari (v 8.0.2) browsers. NemaCount provides a facility to calculate behavioral indices for a variety of commonly tested behaviors in C. elegans (see Fig. 2 for overview of each assay). Function that calls to calculate the chemotaxis index (Fig. 2a) implements a formula described previously (Bargmann et al. 1993), where the total number of animals at the control point (B) is subtracted from the total number of animals at the odor spot (A) and then divided by the total number of animals (N):
NemaCount returns indices for various behavioral assays. These assays include: a odortaxis to attractive odors; b odor-avoidance from repellent odors; c thermotaxis for linear population assays; d soluble compound attractant assays; e aversion assays; and f quadrant assays. For each assay plate, the cartoon images of nematodes refer to the point of origin for the animals with red spots indicating the point source for each compound and blue spots indicating the control spots
The function that calculates the avoidance index (Fig. 2b) implements the formula described previously (Troemel et al. 1997) where the chemotaxis assay plate is divided into six sections and the following formula used to calculate the avoidance index:
where N is the total number of animals across all six panels, A is the panel on which the odor is applied, and F is the panel on which the control odor is applied. The thermotaxis index for linear population assays (Fig. 2c) is calculated as described previously (Goodman et al. 2014; Ito et al. 2006):
where N x is the number of animals in each panel, which is then multiplied by the corresponding value x (i.e., +4 to −4) for that panel, and N is the total number of animals. To calculate the chemotaxis index for water-soluble attractive compounds (Fig. 2d), the formula described by (Hart 2006) was implemented to calculate the chemotaxis index:
where A + B is the total number of animals, A is the number of animals at the soluble compound, and B is the number of animals at the control. The function that calculates the avoidance index (Fig. 2e) implements the formula below where the assay plate is divided into two sections (A and B) separated by a barrier (Cu2+) and the following formula used to calculate the avoidance index (Hart 2006; Ward 1973):
where A is the total number of animals in the section with the odor compound and B is the total number of animals in the other half. Soluble compounds can also be assayed using assay plates that are divided into four quadrants (Fig. 2f) with the water-soluble compound placed into two quadrants (A and A′) and controls or other compounds placed in the other two quadrants (B and B′). The chemotaxis index is calculated as described by (Beets et al. 2012; Wicks et al. 2000) and shown below:
where N is the total number of animals across all quadrants. For all formulae, NemaCount pushes measurements into an array and calculates the average, median, variance, standard deviation, and standard error across all measurements. These statistics can be retrieved by clicking the Count Stats button, Line Stats button, or Free Draw Stats button. The chemotaxis indices can also be typed into the data array textboxes as comma separated values to calculate t test values where the null hypothesis is that there is no significant difference between each array data set using the following formula:
where \(\overline{x1}\) is the mean of the first array; \(\overline{x2}\) is the mean of the second array; \(s_{1}^{2}\) is the standard deviation of array 1; \(s_{2}^{2}\) is the standard deviation of array 2; and N 1 and N 2 are the number of elements in array 1 and 2, respectively. p values are calculated using Student’s t test density distributions by calculating the area under the curve interval (a,b) as follows:
with data type = ‘1’ for paired data; ‘2’ for two-sample equal variance; or ‘3’ for two-sample unequal variance.
Image analysis in NemaCount
NemaCount will load jpeg or png files for visualization within a browser. Sample images contained within the sample images directory that is part of the download were taken with an iPhone6 of a chemotaxis assay to a volatile odorant using C. elegans and also of an adult wild-type C. elegans animal. Images of behavioral assay plates can be captured using a smartphone and loaded directly into NemaCount as a jpeg, thereby facilitating the digital archiving of behavior and making the analysis of behaviors very fast and easy. The user can download the project from GitHub (https://github.com/dohalloran/NemaCount) and double click the file named: run_NemaCount.html in a browser to launch the software. Alternatively, the user can install NemaCount as a Google Chrome extension from the Chrome Web Store (Fig. 3). The extension option is only available on Chrome browsers and is freely available here: https://chrome.google.com/webstore/detail/nemacount/lbdnkeodmclalnjhbhppkjpjkbianfmk.
NemaCount Google Chrome Extension. NemaCount is also freely available as a Google Chrome Extension from the Chrome Web Store. The user only needs to click the add to Chrome button, and the extension is loaded into the browser. The user can click the microscope icon to generate the NemaCount HTML page and load and analyze images directly
Smartphone chemotaxis imaging station
An imaging station was developed so that researchers could go from image acquisition directly to image analysis in NemaCount by using a very low-cost implementation. The imaging station relies on a smartphone camera which is modified with a magnification lens (available here: http://www.aixiz.com/store/product_info.php/products_id/374 or here: http://www.microphonelens.com/products/micro-phone-lens). The modified smartphone is mounted on a flexible silicone-PVC rod that allows the user to adjust the working distance between lens and sample. A Perspex plate with a basic LED light source underneath provides a stable stage for the sample as well as sample illumination.
Results
The NemaCount user interface is very intuitive and does not require any programming knowledge from the end user (Fig. 1). The user can download the project from GitHub and open the file named run_NemaCount.html in a browser or install NemaCount as a browser extension for Google Chrome (see Fig. 3 for Chrome extension). All scripts, frameworks, and libraries come with the download, and so no additional dependencies are necessary. The user can click the user guide button on the interface page to get a step-by-step overview of all features. Once an image file is loaded, the user can apply various imaging filters prior to analysis by clicking the grayscale, remove white, invert, or sharpen buttons. Next, the user must then click one of the following functions: (1) Count; (2) Measure Line; (3) Measure Angle; or (4) Free Draw, to begin quantifying or measuring objects within the image file (Fig. 1).
The first function, Count, allows the user to enumerate objects using unique color and size options; the size and color of the dot counts can be altered by changing the Color and Line Width options (see Fig. 1). To facilitate the analysis of behavioral assays of nematodes, NemaCount will calculate chemotaxis indices, avoidance indices, or thermotaxis indices from behavioral assays that are enumerated using the Count function in NemaCount. To calculate the index, the individual counts for each assay can be entered into the appropriate pop-up box that appears after clicking the Chemotaxis Index button (for either volatile or water-soluble compounds), the Avoidance Index (for either volatile or water-soluble compounds) button, Quadrant Chemotaxis Index button for quadrant-based chemotaxis assays, or the Thermotaxis Index button. The formulae for the various assays are described above in the methods section. Figure 1 demonstrates how the Count function can be used to score the number of animals in a chemotaxis assay at the odor point, the control point, and the rest of the plate. These values can then be used to calculate a chemotaxis index which is shown in the pop-up box (Fig. 1). Basic statistics for counts are returned by clicking the Count Stats button (see Fig. 3: statistics include the average, median, variance, standard deviation, and standard error).
The second function is Measure Line and allows the user to measure any number of linear distances within the image file. In order to obtain accurate measurements, the user must complete the options contained within the orange rectangle (see Fig. 1) that are necessary to compute precise distances. These metrics include the dimensions of the original image in pixels, the pixel size for the camera on which the image was taken, whether binning was performed, whether C-mount was used, and the objective and lens magnifications. The line width and color can be set by the user to measure distinct objects and features at varying resolution.
The third function is Measure Angle, which generates a protractor object within the image file that can be used to measure angles of objects or their relative positioning within the image file. Any number of protractor objects can be generated with varying colors set by the user. The tips of the protractors can be moved using the mouse to measure angular distances. The mouse position is detected using JavaScript event listeners once the user clicks on the end of the protractor tip. The user can also move the protractor around the image by clicking the apex of the protractor and moving the mouse to the required position.
The fourth function is Free Draw, which enables the user to draw directly onto the image and measure the length of freely drawn objects. As per the Measure Line function, the line width and color used in the Free Draw function can be set by the user. The Free Draw feature can be used to trace nematode tracks on agar to measure distances travelled, or to capture and measure a region of interest (ROI) for area comparisons, which can be analyzed further using other functions within NemaCount. Basic statistics for measurements can be obtained by clicking the Line Stats, or Free Draw Stats buttons (see Fig. 1). In each case, the average, median, variance, standard deviation, and standard error of the dataset of measurements are returned.
Once the user has calculated a series of behavioral indices or image measurements, these measurements can then be statistically analyzed for control and experimental samples by entering values into one of two arrays located within the red box (see Fig. 1). The user can also specify the data type: ‘1’ for paired data; ‘2’ for two-sample data with equal variance; or ‘3’ for two-sample data with unequal variance. The user then clicks the calculate significance button and an alert box will appear containing the various statistics including t test values and p values determined from Student’s t test distributions (see Fig. 1 for example of alert box).
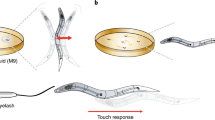
To validate NemaCount, the Count function was used to compare chemotaxis behavior between wild-type and odr-1(n1936) mutant animals to the AWC sensed odor benzaldehyde (Fig. 4a). Significant differences were observed between odr-1(n1936) mutants and wild-type animals as previously observed (L’Etoile and Bargmann 2000). The Measure Line function was also used to sample the mean lengths of wild-type Heterorhabditis bacteriophora infective juveniles (IJs) and adults (Fig. 4b). Significant differences in body lengths of IJs and adults of H. bacteriophora were observed. The Measure Angle function was used to measure a sample body angle at the vertices indicated in Fig. 4c for wild-type and unc-31(n422) mutant (Avery et al. 1993) animals using locomotion tracks on agar plates (Fig. 4d). Multiple protractors were generated and manipulated by moving the cursor with the mouse to adjust the protractor tips. When comparing body angles made by wild-type and unc-31(n422) mutant animals, significant differences were not observed (Fig. 4d).
Sample quantification using NemaCount. a Quantifying behavior using the Count function to enumerate wild-type and odr-1(n1936) mutant animals from three independent chemotaxis assays. b Measuring body length of Heterorhabditis bacteriophora infective juveniles (IJs) and adults using the Measure Line function. Three animals are examined for each stage. c Waveform locomotion tracks of wild-type C. elegans and unc-31(n422) mutant animals. d Body angles calculated at vertices highlighted in (c) for three wild-type and three unc-31(n422) mutant animals using the Measure Angle functions. In all cases, error bars represent the SEM. p values calculated using the Student’s t test
Discussion
Using NemaCount, users can quickly and accurately measure nematode chemotaxis behavior and features in a browser. Although NemaCount does not offer the automated features of other worm tracker software (Husson et al. 2012; Kwon et al. 2013; Ramot et al. 2008; Swierczek et al. 2011; Wang and Wang 2013) or ImageJ plugins, the ease of installation and simple interface makes NemaCount a versatile solution for both quantifying and statistically analyzing various nematode behaviors and image features. Furthermore, only NemaCount offers a suite of functions aimed at measuring and analyzing diverse nematode behaviors including chemotaxis, odortaxis, avoidance, and thermotaxis assays.
The goal in developing NemaCount was to provide a simple and low-cost option for analyzing nematode behavior that could especially be used by researchers new to C. elegans research, such as undergraduate students. To further achieve this goal, I coupled NemaCount to a low-cost and simple imaging station (Fig. 5a). The imaging station uses a smartphone camera with a modified magnification lens (Fig. 5b) to obtain images of nematodes and nematode chemotaxis assays. Detailed images can be obtained using this approach (Fig. 5c) and then simply uploaded to NemaCount for quantification and analysis. The smartphone imaging station uses a flexible rod for mounting the smartphone which allows the user to easily attain the correct working distance. The sample is placed on a Perspex plate that contains an LED light housing underneath in order to illuminate the sample. The entire station costs ~ $20 to prepare. By coupling NemaCount to a budget imaging station, it provides a simple platform that allows multiple researchers new to C. elegans to work together in small spaces to both image and analyze nematodes and behavior. The use of smartphones as microscopes has been used previously in multiple areas of research (Bogoch et al. 2013; Pirnstill and Cote 2015; Skandarajah et al. 2014; Switz et al. 2014), and using other approaches can enhance the field of view and achieve greater magnification.
Smartphone chemotaxis imaging station. a A simple and cheap imaging station is implemented to capture images of nematodes. The setup uses a lens to provide magnification to a smartphone camera. The camera is held in place by a flexible mount that can be adjusted to the correct height based on sample size and working distance (yellow circle 1). A clear Perspex stage is used to house the sample being imaged (yellow circle 2), and an LED light source is placed underneath the Perspex stage to provide sample illumination (yellow circle 3). b Image showing the magnification lens which is placed over the built-in smartphone lens. c A sample image of a C. elegans adult animal is taken using a modified iPhone 6
In conclusion, NemaCount is a cheap and simple option for measuring nematode behavior in a browser. Any number of commonly tested behavioral assays is appropriate for analysis within NemaCount. These include all forms of chemotaxis (Bargmann et al. 1993) and thermotaxis assays (Ito et al. 2006; Komatsu et al. 1996; Wang et al. 2013), lethargus (Raizen et al. 2008) [whereby C. elegans form a unique hockey stick shape (Tramm et al. 2014)], egg laying (Schafer and Kenyon 1995), and survival (Murphy et al. 2003). In terms of image analysis, any features that change in total number, size, shape, or angular distance between control and experimental preparations are suited to NemaCount for functional analysis.
References
Avery L, Bargmann CI, Horvitz HR (1993) The Caenorhabditis elegans unc-31 gene affects multiple nervous system-controlled functions. Genetics 134:455–464
Bargmann CI, Hartwieg E, Horvitz HR (1993) Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74:515–527
Beets I, Janssen T, Meelkop E, Temmerman L, Suetens N, Rademakers S, Jansen G, Schoofs L (2012) Vasopressin/oxytocin-related signaling regulates gustatory associative learning in C. elegans. Science 338:543–545. doi:10.1126/science.1226860
Bogoch II, Andrews JR, Speich B, Utzinger J, Ame SM, Ali SM, Keiser J (2013) Mobile phone microscopy for the diagnosis of soil-transmitted helminth infections: a proof-of-concept study. Am J Trop Med Hyg 88:626–629. doi:10.4269/ajtmh.12-0742
Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77:71–94
Cronin CJ, Mendel JE, Mukhtar S, Kim YM, Stirbl RC, Bruck J, Sternberg PW (2005) An automated system for measuring parameters of nematode sinusoidal movement. BMC Genet 6:5
Goodman MB, Klein M, Lasse S, Luo L, Mori I, Samuel A, Sengupta P, Wang D (2014) Thermotaxis navigation behavior. WormBook. doi:10.1895/wormbook.1.168.1
Hart A (2006) Behavior. In: The C. elegans Research Community (ed) WormBook. doi:10.1895/wormbook.1.87.1
Husson SJ, Costa WS, Schmitt C, Gottschalk A (2012) Keeping track of worm trackers. WormBook. doi:10.1895/wormbook.1.156.1
Ito H, Inada H, Mori I (2006) Quantitative analysis of thermotaxis in the nematode Caenorhabditis elegans. J Neurosci Methods 154:45–52
Komatsu H, Mori I, Rhee JS, Akaike N, Ohshima Y (1996) Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron 17:707–718
Kwon N, Pyo J, Lee SJ, Je JH (2013) 3-D worm tracker for freely moving C. elegans. PLoS ONE 8:e57484. doi:10.1371/journal.pone.0057484
L’Etoile ND, Bargmann CI (2000) Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron 25:575–586
Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424:277–283. doi:10.1038/nature01789
O’Halloran DM, Burnell AM (2003) An investigation of chemotaxis in the insect parasitic nematode Heterorhabditis bacteriophora. Parasitology 127:375–385
O’Leary SA, Stack CM, Chubb MA, Burnell AM (1998) The effect of day of emergence from the insect cadaver on the behavior and environmental tolerances of infective juveniles of the entomopathogenic nematode Heterorhabditis megidis (strain UK211). J Parasitol 84:665–672
Pirnstill CW, Cote GL (2015) Malaria diagnosis using a mobile phone polarized microscope. Sci Rep 5:13368. doi:10.1038/srep13368
Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV, Pack AI (2008) Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451:569–572. doi:10.1038/nature06535
Ramot D, Johnson BE, Berry TL Jr, Carnell L, Goodman MB (2008) The Parallel Worm Tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PLoS ONE 3:e2208. doi:10.1371/journal.pone.0002208
Schafer WR, Kenyon CJ (1995) A calcium-channel homologue required for adaptation to dopamine and serotonin in Caenorhabditis elegans. Nature 375:73–78. doi:10.1038/375073a0
Skandarajah A, Reber CD, Switz NA, Fletcher DA (2014) Quantitative imaging with a mobile phone microscope. PLoS ONE 9:e96906. doi:10.1371/journal.pone.0096906
Swierczek NA, Giles AC, Rankin CH, Kerr RA (2011) High-throughput behavioral analysis in C. elegans. Nat Methods 8:592–598. doi:10.1038/nmeth.1625
Switz NA, D’Ambrosio MV, Fletcher DA (2014) Low-cost mobile phone microscopy with a reversed mobile phone camera lens. PLoS ONE 9:e95330. doi:10.1371/journal.pone.0095330
Tramm N, Oppenheimer N, Nagy S, Efrati E, Biron D (2014) Why do sleeping nematodes adopt a hockey-stick-like posture? PLoS ONE 9:e101162. doi:10.1371/journal.pone.0101162
Troemel ER, Kimmel BE, Bargmann CI (1997) Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell 91:161–169
Vidal-Gadea A, Ward K, Beron C, Ghorashian N, Gokce S, Russell J, Truong N, Parikh A, Gadea O, Ben-Yakar A, Pierce-Shimomura J (2015) Magnetosensitive neurons mediate geomagnetic orientation in Caenorhabditis elegans. Elife. doi:10.7554/eLife.07493.10.7554/eLife.07493
Wang SJ, Wang ZW (2013) Track-a-worm, an open-source system for quantitative assessment of C. elegans locomotory and bending behavior. PLoS ONE 8:e69653. doi:10.1371/journal.pone.0069653
Wang D, O’Halloran D, Goodman MB (2013) GCY-8, PDE-2, and NCS-1 are critical elements of the cGMP-dependent thermotransduction cascade in the AFD neurons responsible for C. elegans thermotaxis. J Gen Physiol 142:437–449. doi:10.1085/jgp.201310959
Ward S (1973) Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc Natl Acad Sci USA 70:817–821
White GF (1927) A method for obtaining infective nematode larvae from cultures. Science 66:302–303
Wicks SR, de Vries CJ, van Luenen HG, Plasterk RH (2000) CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev Biol 221:295–307. doi:10.1006/dbio.2000.9686
Woodring JL, Kaya HK (1998) Steinernematid and heterorhabditid nematodes: a handbook of biology and techniques. Southern Cooperative Series Bulletin Arkansas Agricultural Experiment Station, Fayetteville
Yang JS, Nam HJ, Seo M, Han SK, Choi Y, Nam HG, Lee SJ, Kim S (2011) OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS ONE 6:e23525. doi:10.1371/journal.pone.0023525
Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR (1993) The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 75:641–652
Acknowledgments
I thank members of the O’Halloran laboratory for critical reading of the manuscript.
Funding
The George Washington University (GWU) Columbian College of Arts and Sciences, GWU Office of the Vice-President for Research, and the GWU Department of Biological Sciences. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Author contributions
D.M.O’H conceived the idea for NemaCount, wrote the code, and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Rights and permissions
About this article
Cite this article
O’Halloran, D.M. NemaCount: quantification of nematode chemotaxis behavior in a browser. Invert Neurosci 16, 5 (2016). https://doi.org/10.1007/s10158-016-0188-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10158-016-0188-1