Abstract
Background
Trim44 is an important member of the tripartite motif-containing protein (TRIM) family. Recent research reported that Trim44 might play an important role in tumorigenesis, although its role in non-small cell lung cancer (NSCLC) and the related mechanisms is not yet known.
Methods
In this study we analyzed 30 pairs of NSCLC tumors and the matched adjacent normal tissue to define the relationship between Trim44 and NSCLC tumors. The function of Trim44 in cell migration and invasion was determined by overexpression of Trim44 in normal bronchial epithelial cell line 16HE or knockdown of Trim44 in A549 cells, respectively. Whether Trim44-mediated NF-κB signaling activation was involved in Trim44-mediated promotion of lung cancer was tested by q-PCR analysis and cell migration and invasion assay using PDTC, an inhibitor of NF-κB.
Results
We found that Trim44 was upregulated in NSCLC tumors (14/30 cases; 46.7 %). Furthermore, Trim44 was upregulated in many NSCLC cell lines, especially in A549 and H441. Moreover, Trim44 significantly enhanced cell migration and invasion ability, which was related to increased CXCR6 and matrix metalloproteinase 9 (MMP9). Knockdown of Trim44 in A549 cells by siRNA showed a diminished effect in cell migration and invasion. Further investigation revealed that blocking the NF-κB signaling pathway using PDTC, an inhibitor of NF-κB, reversed the expression of CXCR6 and MMP9, and alleviated the promotion of migration and invasion mediated by Trim44.
Conclusions
Our data suggest that Trim44 promotes NSCLC development through activation of NF-κB signaling via upregulating CXCL16 and MMP9 expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is one of the most common malignant tumors and causes 1.4 million deaths per year worldwide. Non-small cell lung cancer (NSCLC) is a subtype of lung cancer and accounts for approximately 85 % of all lung cancers [1]. Although surgical techniques, chemotherapy and radiotherapy have improved greatly over the past decade, the overall prognosis for patients remains extremely poor and the 5-year survival rate is <15 % [1, 2]. It is clear that metastasis is the main reason for failure of NSCLC treatment; however, the exact causes of NSCLC metastasis remain unclear. Therefore, it is important to determine the mechanisms underlying NSCLC metastasis and to develop more effective therapeutic strategies.
The tripartite motif (Trim) family of proteins is characterized by a RING domain, one or two B boxes, and a coiled-coil domain. It has been demonstrated that many Trim family proteins are involved in many important biological processes, such as cell proliferation, differentiation, apoptosis, etc. [3]. Many studies have also shown that Trim family members play critical roles in regulating innate immune reactions [4–7] and show antiviral functions [8–10]. Trim44 is a new member of the Trim family which has been cloned from a mouse brain cDNA library in 2001 [11]. Although there has been some research on Trim44, the function of Trim44 is not well studied. A recent research reported that Trim44 interacts with virus-induced signaling adaptor (VISA) and promotes VISA-mediated antiviral responses [12]. A few studies have revealed that Trim44 is involved in many malignant tumors. Overexpression of Trim44 has been reported in esophageal and junctional adenocarcinoma, and could be used as a clinically relevant prognostic marker [13]. Other research found that the TRIM44 gene is amplified in head and neck cancer [14]. Overexpression of TRIM44 was thought to contribute to malignant outcome in gastric carcinoma [15]. All these studies indicate that Trim44 plays an important role in tumor development; however, the role of Trim44 in NSCLC development and the related mechanisms are still unknown. In this study we investigated the expression of Trim44 in NSCLC and found that Trim44 mRNA and protein were both upregulated in NSCLC tissues, which indicated that Trim44 may play an important role during NSCLC development. Overexpression of Trim44 in normal bronchial epithelial cell line 16HBE resulted in increased cell migration and invasion. Knockdown of Trim44 in A549 cells by siRNA showed a reduced effect on cell migration and invasion. Meanwhile, overexpression of Trim44 induced CXCR6 and MMP9, although no obvious effect was shown on MMP2. Further research revealed that exogenous expression of Trim44 enhanced NF-κB signaling pathways. Inhibition of NF-κB using PDTC (an inhibitor of NF-κB) reversed Trim44-induced CXCR6 mRNA and MMP9 mRNA, especially CXCR6. Blocking the NF-κB signaling pathway also alleviates the promotion of Trim44-mediated migration and invasion. Our findings demonstrated that Trim44 enhances the ability of NSCLC cell migration and invasion through activating the NF-κB signaling pathway resulting in increased CXCR6 and MMP9. Our study suggests that Trim44 might be a potential therapeutic target to manage NSCLC metastasis.
Materials and methods
Human lung tumor tissues and normal lung tissues
Between January 2011 and July 2012, 30 pairs of NSCLC specimens and matched normal lung tissues were collected from Shanghai Chest Hospital, Shanghai, China. All NSCLC and compared normal lung tissues samples were obtained at the time of surgery, snap frozen and stored at −80 °C until use. All the cases were evaluated to confirm the pathological diagnosis. Written informed consent was obtained from all patients or their guardians in accordance with the ethical committee standards. All human samples were used in accordance with the policies of the institutional review board at the Shanghai Chest Hospital of Jiao Tong University, China.
q-PCR
Total RNA samples from NSCLC tissues and A549 cells were extracted using TRIzol reagent (Invitrogen). Reverse transcription of RNA (1 mg) was performed using the Reverse Transcriptase M-MLV (TaKaRa, Otsu, Japan). qRT-PCR was performed using Fast Start DNA Master SYBR Green I reagent (Roche Diagnostics) on a 7900HT Fast Real-Time PCR System (Applied Biosystems). GAPDH was used as an internal control. The primer sequences used for q-PCR were as follow:
CXCR6
Forward primer: 5′-GGCCTATGCAGGCATCCAT-3′
Reverse primer: 5′-CCCAGTAGGCTCTTGCACATG-3′
MMP9
Forward primer: 5′-CGCCAGTCCACCCTTGTG-3′
Reverse primer: 5′-CAGCTGCCTGTCGGTGAGA-3′
MMP2
Forward primer: 5′-CGTCTGTCCCAGGATGACATC-3′
Reverse primer: 5′-ATGTCAGGAGAGGCCCCATA-3′
GAPDH [12]
Forward primer: 5′-TGGGCTACACTGAGCACCAG-3′
Reverse primer: 5′-GGGTGTCGCTGTTGAAGTCA-3′
Plasmid, cell culture and transfection
Human Trim44 was amplified by PCR using cDNA from A549 cells and subcloned into a pcDNA3-HA vector (Invitrogen) to construct pcDNA-HA-trim44 plasmid. The primer sequences used were as follow:
Forward primer: 5′-TGGCCACAGCTCATGTGACT-3′
Reverse primer: 5′-CATCAACCTATCCATGTGGGATT-3′
All human lung carcinoma cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10 % fetal bovine serum (FBS), 100 U/mL penicillin and 100 μg⁄mL streptomycin. The cells were maintained at 37 °C in a humidified atmosphere of 5 % CO2. For transfection, 16HBE cells were placed onto a 6-well plate (1 × 104 per well), and 24 h later pcDNA-HA-Trim44 or control vector was transfected into the cells using Lipofectamine 2000 Transfection Reagent (Invitrogen) according to the manufacturer’s protocol. Cells were collected 24–48 h post-transfection and subjected to further investigation.
Wound-healing assay
For wound-healing assay, 16HBE cells (1 × 104 per well) were transfected with pcDNA-HA-Trim44 or control vector as indicated. Cells were collected 24 h post-transfection and seeded onto 6-well plates and incubated at 37 °C. The following day, cells in the individual wells were wounded by dragging a rubber policeman through the confluent monolayers. The detached cells were removed by washing with PBS, and the cells were then incubated with DMEM medium containing 10 % FBS at 37 °C. Photographs were taken at 0, 24 and 48 h, respectively. Three independent experiments were performed.
Cell migration and invasion assays
For invasion assay, Matrigel-coated Transwell cell culture chambers (8 μm pore size; Coster, Boston, MA, USA) were introduced. Briefly, 5 × 105 16HBE cells transfected with pcDNA-HA-Trim44 or control vector or A549 cells transfected with Trim44 siRNA or control siRNA were seeded onto the upper chamber of the Transwell (BD Falcon Labware, Bedford, MA, USA) with serum-free DMEM medium and the lower chamber with DMEM medium containing 10 % FBS. After incubation at 37 °C for 48 h, the cells in the upper chamber were removed by wiping with a cotton swab. The cells attached to the reverse phase of the membrane were fixed in 4 % paraformaldehyde at room temperature for 30 min, and stained with 4,6-diamidino-2-phenylindole (DAPI) (C1002; Beyotime Institude of Biotechnology, China).The cells on the lower surface of the filters were then observed under a microscope, and the cells in 10 randomly selected fields were counted. The experiment was carried out four times to reduce the possible effects of biological variability. For migration assay, a chemotaxis chamber (Corning Inc., Corning, NY, USA) was used. 16HBE cells transfected with pcDNA-HA-Trim44 or control vector or A549 cells transfected with Trim44 siRNA or control siRNA were washed with serum-free medium and resuspended in 100 μl of serum-free medium and added to the upper chambers. The lower chambers were then filled with 0.6 ml of complete DMEM cell culture with or without 100 ng/ml CXCL16. To maintain the chambers at 37 °C for 24 h, the filters were fixed with 4 % formaldehyde in PBS and were stained with DAPI. Cells in the lower surface of the filter which penetrated through the filter were counted under a microscope.
Loss-of-function by siRNA
siRNA targeting Trim44 and control siRNA were purchased from Dharmacon (Lafayette, CO, USA). Each siRNA (10 nmol/L) was transfected into A549 cells using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. The knockdown of a target gene was confirmed by Western blot analysis.
Histology
Lung tissue was fixed in formalin and embedded in paraffin. Sections were stained with anti-Trim44 antibody. Light microscopic examination was performed.
Protein extraction and Western blotting
Tissues or cells were lysed on ice using RIPA buffer (0.05 M Tris–HCl, pH 7.4, 0.15 M NaCl, 0.25 % deoxycholic acid, 1 % NP-40, 1 mM EDTA, 0.5 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, and 10 μg/ml aprotinin). The lysates were centrifuged at 12,000 rpm for 10 min at 4 °C. The supernatants were transferred to a fresh EP tube and stored at −80 °C until use. The protein concentration was measured using the BCA™ Protein Assay Kit (Thermo Scientific Pierce, Sunnyvale, CA, USA). A 30–50 μg protein sample was separated by 10 % SDS–polyacrylamide gel electrophoresis and transferred onto a PVDF membrane. The membranes were then blocked with block buffer (5 % skim milk in TBST) for 1 h at room temperature. Immunoblotting was performed using antibodies against Trim44 (1:2,000, Proteintech Group), P-65 (1:2,000, Cell Signaling Technology), p-P65 (1:2,000, Cell Signaling Technology), MMP9 (1:1,000, Santa Cruz Biotechnology), MMP2 (1:1,000; Santa Cruz Biotechnology), and a monoclonal mouse anti-β-actin (1:10,000; Cell Signaling Technology). The membranes were then washed three times with TBST, followed by incubation with indicated secondary antibody (horseradish peroxidase-conjugated goat anti-rabbit secondary antibody or horseradish peroxidase-conjugated donkey anti-mouse secondary antibody, 1:2,000, Santa Cruz Biotechnology). The membranes were then washed three times with TBST and visualized using an enhanced chemiluminescence kit (ECL; Pierce Biotechnology). β-actin was used as an internal control.
Luciferase assays
Luciferase assays were performed as described previously [16]. Briefly, A549 cells were transfected with the reporter and the indicated expression plasmids, together with Renilla luciferase plasmids as an internal control. 24 h post-transfection, the cells were stimulated with or without LPS for an additional 12 h. The cells were then lysed and analyzed using a dual luciferase reporter assay system (Promega).
Statistical analysis
Each experiment was repeated at least three times. All data were presented as mean ± SEM. Student’s t test (two-tailed) was used to compare two groups. Results were considered statistically significant when P < 0.05.
Results
High expression of Trim44 in human lung cancer cells
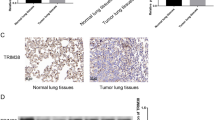
To examine the potential role of Trim44 in NSCLC development, we first analyzed the expression of Trim44 mRNA in 30 pairs of NSCLC specimens and matched normal lung tissues using q-PCR. As showed in Fig. 1a, Trim44 mRNA was significantly up-regulated in NSCLC samples compared with normal lung tissue samples. Similar results were also obtained when we examined the expression of TRIM4 protein using Western blot analysis (Fig. 1b). Histopathological analysis of lung sections from normal tissues and tumor tissues supported the upregulation of Trim44 in tumor tissue (Fig. 1c). Western blot analysis and q-PCR analysis were also carried out to test what level of Trim44 protein is expressed in four lung cancer cell lines such as A549, H441, H460 and SPC-A1. Normal bronchial epithelial cell line (16HBE) was used as the control cell line. Results showed that overexpression of Trim44 was observed in A549 and H441 cells, suggesting this gene plays a role in these cell lines (Fig. 1d, e). These results suggested that Trim44 might be involved in NSCLC tumor progression.
Trim44 is highly expressed in human lung cancer cells. a mRNA levels of Trim44 were detected by q-PCR in both human normal lung and NSCLC tissue samples. b Protein levels of Trim44 were detected by Western blot analysis in both normal lung and NSCLC tissue samples. Quantitative data are presented in the low panel. c Trim44 immunohistochemical staining on tissue sections from human lung adenocarcinoma and normal lungs. Sections were stained with anti-Trim44 antibody. d Western blot analysis of protein levels of Trim44 in several lung cancer cell lines. e q-PCR analysis of mRNA levels of Trim44 in several lung cancer cell lines. All data shown are from a single experiment that is representative of two independent experiments performed. Bars show mean ± SD. *P < 0.05, **P < 0.01, Student’s t test
Trim44 promotes cell migration of NSCLC cells by induction of CXCR6
To find the potential role of Trim44 in NSCLC development, 16HBE cells were adopted. Results of the cell wound healing assay showed that after overexpression of Trim44, the wound healing rate was dramatically increased compared with the control group, especially at 48 h (Fig. 2a, b). The results indicated that Trim44 promoted motility of 16HBE cells. Chemokine CXCL16 is a ligand of CXCR6. It was reported that expression of CXCR6/CXCL16 was higher in NSCLC tissues than in normal lung tissues and CXCR6/CXCL16 was involved in metastasis of human lung cancer. We next examined whether CXCR6 was responsible for the role of Trim44 in promoting NSCLC progression. As shown in Fig. 2c, Trim44 obviously elevated the level of CXCR6 mRNA. Next, we performed Transwell assay to confirm the effect of Trim44 on migration of 16HBE cells. Consistent with the result of cell wound healing, 16HBE cells overexpressed with Trim44 dramatically promoted cell migration to CXCL16 (Fig. 2d, e). These results suggested that induction of CXCR6 might contribute to the promotion role of Trim44 in inducing migration of 16HBE cells.
Trim44 promotes cell migration of A549 cells by induction of CXCR6. a Representative wound healing images of 16HBE cells that were transfected with or without Trim44. b The number of wound healing cells in a. Cells in 10 randomly selected fields were counted. C CXCR6 mRNA expression was tested in 16HBE cells with overexpression of Trim44. d Representative cell field images of migration cells of 16HBE cells transfected with or without Trim44. e The number of cells that migrated in d. Cells in 10 randomly selected fields were counted. All data shown are from a single experiment that is representative of two independent experiments performed. Bars show mean ± SD. *P < 0.05, **P < 0.01, Student’s t test
Trim44 facilitates cell invasion by enhancing MMP9
We then analyzed the effects of Trim44 on the invasion of 16HBE cells. As shown in Fig. 3a and 3b, Trim44 showed significant promotion in cell invasion, suggesting that Trim44 may be involved in the invasion of tumor cells. It is well known that matrix metalloproteinases (MMPs) play a critical role in tumor growth, invasion and metastasis. Of all MMPs, MMP2 and MMP9 are considered to be the most important for basement membrane type IV collagen degradation, especially MMP9. We then explored the effect of Trim44 on the expression of MMPs. The results of q-PCR and Western blot analysis showed that both MMP9 mRNA and protein were significantly increased in 16HBE cells that overexpressed Trim44, while MMP2 mRNA and protein levels showed no obvious changes (Fig. 3c, d). Therefore, it seemed that Trim44 modulated 16HBE cell invasion through enhancing MMP9 but not MMP2 expression.
Trim44 facilitates invasion of human lung cancer cells. a Representative cell field images of Matrigel invasion cells of 16HBE cells transfected with Trim44 or control vector. b The numbers of invaded cells in a. Cells in 10 randomly selected fields were counted. c Trim44 induces MMP9 protein expression. d Trim44 induces MMP9 mRNA expression. All data shown are from a single experiment that is representative of two independent experiments performed. Bars show mean ± SD. *P < 0.05, **P < 0.01, Student’s t test
Suppression of cell migration and invasion by downregulation of TRIM44 expression
To gain further insight into the potential role of Trim44 in lung cancer, we first carried out a Western blot assay to test the knockdown efficiency of Trim44 by using siRNA specific to Trim44 in A549 cells. Results showed that the Trim44 protein level was markedly reduced (Fig. 4a). We then examined the effects of Trim44 knockdown on the migration and invasion of A549 cells. The number of cells that migrated through the uncoated (migration assay) or Matrigel-coated (invasion assay) membrane into the lower chamber were significantly lower in Trim44 siRNA transfected cells than in control siRNA cells, further supporting the notion that Trim44 promoted migration and invasion of A549 cells (Figs. 4b, c, d).
Suppression of cell migration and invasion by downregulation of Trim44 expression. a Western blot analysis of the knockdown efficiency of Trim44 in A549 cells. b, c The effects of Trim44 knockdown on the migration and invasion of A549 cells. d, e The number of A549 cells that migrated and invaded were calculated. Cells in 10 randomly selected fields were counted. All data shown are from a single experiment that is representative of two independent experiments performed. Bars show mean ± SD. *P < 0.05, **P < 0.01, Student’s t test
The NF-κB signaling pathway is involved in Trim44-induced NSCLC migration and invasion
It has been suggested that activation of NF-κB might be involved in chemokine production [17]. Therefore, we performed Western blot analysis to evaluate the effect of Trim44 on NF-κB activation. As shown in Fig. 5a, overexpression of Trim44 in 16HBE may increase phosphorylation of p65, and the additional Trim44 also enhanced the activation of LPS-induced NF-κB. The luciferase assay results revealed that overexpression of Trim44 enhanced LPS-induced NF-κB activity (Fig. 5b), while PDTC (an inhibitor of NF-κB) effectively reversed the effect of Trim44 on activating NF-κB (Fig. 5c). Furthermore, after treatment with PDTC, the mRNA level of CXCR6 induced by Trim44 was inhibited (Fig. 5d). The mRNA level of MMP9 induced by Trim44 was also inhibited while MMP2 was unaffected (Fig. 5e). Our data suggest that NF-κB signaling activation accounts for the upregulation of Trim44-mediated CXCR6 and MMP9.
Overexprssion of Trim44 activates the NF-κB signaling pathway. a Overexpression of Trim44 increases phosphorylation of p65. Trim44 or control vector were transfected into 16HBE cells. 24 h post-transfection, cells were stimulated with or without LPS for 12 h. Cells were then collected and subjected to Western blot analysis. b Trim44 positively regulates NF-κB signaling. 16HBE cells were transfected with indicated plasmids. 24 h post-transfection, cells were stimulated with or without LPS for 12 h and then lysed. The luciferase activity of the lysates was analyzed using a dual luciferase reporter assay system. c Inhibition of NF-κB effectively reverses the effect of Trim44 on activating NF-κB. 16HBE cells were transfected with indicated plasmids. After 24 h, cells were treated with or without 50 μM PDTC for 1 h and then stimulated with or without LPS for 12 h. d, e Inhibition of NF-κB effectively restrains the expression of Trim44-induced d CXCR6 mRNA and e MMP9 mRNA. 16HBE cells were transfected with indicated plasmids. After 24 h, cells were treated with or without 50 μM PDTC for 1 h and then stimulated with or without LPS for 12 h. RNA samples were isolated from cells and used for q-PCR analysis. All data shown are from a single experiment that is representative of two independent experiments performed. Bars show mean ± SD. *P < 0.05, **P < 0.01, Student’s t test
Blocking the NF-κB signaling pathway alleviates the promotion of Trim44-mediated migration and invasion
To further verify whether the NF-κB signaling pathway plays a critical role in Trim44-induced cell migration and invasion, we performed Transwell assays. As shown in Fig. 6a–d, after treatment with PDTC, the amount of Trim44-induced cell migration and invasion was significantly reduced, suggesting that the NF-κB signaling pathway is involved in the promotion of Trim44-mediated migration and invasion.
Blocking the NF-κB signaling pathway alleviates the promotion of Trim44-mediated migration and invasion. a, c 16HBE cells were transfected with Trim44 and then treated with or without 50 μM PDTC for 1 h. Migration assays and invasion assays were performed as described in ‘Materials and methods’. b The number of cells that migrated in a. Cells in 10 randomly selected fields were counted. d The number of cells that invaded in b. Cells in 10 randomly selected fields were counted. All data shown are from a single experiment that is representative of two independent experiments performed. Bars show mean ± SD. *P < 0.05, **P < 0.01, Student’s t test
Discussion
In this study we detected the expression of Trim44 in NSCLC tissues, and found that Trim44 was upregulated in NSCLC tissue specimens compared to corresponding normal lung tissues. We then explored the role of Trim44 in NSCLC cells and found that Trim44 significantly promoted cell migration and invasion of these cells, which was related to the promotion of CXCR6 and MMP9 expression by Trim44. Further investigation revealed that Trim44 may function through activating the NF-κB signaling pathway. Blocking the NF-κB signaling pathway using PDTC (an inhibitor of NF-κB) reversed the expression of CXCR6 and MMP9, and alleviated the promotion of Trim44-mediated migration and invasion.
It has been reported that many Trim family members participate in regulating tumorigenesis and development. Researchers also found that TRIM family proteins are involved in NSCLC development and progression. For example, TRIM28 was reported to have a tumor-suppressing role in the early stages of lung cancer [18]. TRIM29 may participate in the development of squamous cell carcinoma (SC) and was considered to play a reference role in distinguishing between poorly differentiated SC and adenocarcinoma of NSCLC [19]. Trim44 is a member of the TRIM family. A few earlier studies found that Trim44 is implicated with esophageal and junctional adenocarcinoma [13], head and neck cancer [14] and gastric carcinoma [15], suggesting that Trim44 might be a clinically relevant prognostic marker for these tumors. Thus, these findings encouraged us to explore the role of Trim44 in NSCLC. Therefore, we first investigated the expression of Trim44 in NSCLC tissues. As shown in Fig. 1, compared to normal lung tissues, Trim44 mRNA and protein were both upregulated in NSCLC tissues. The results indicated that Trim44 might be involved in NSCLC development. For confirmation, we performed wound healing assay and Transwell assay, which showed that overexpression of Trim44 in 16HBE cells significantly increased the ability for cell migration and invasion. The data demonstrate that Trim44 plays an important role in NSCLC progression.
MMPs are a multigene family; the members play important roles in degrading extracellular matrix during tumor invasion and metastasis [20]. Among MMPs, MMP2 and MMP9 are considered to be the most important in degrading basement membrane type IV collagen [21–23], and are associated with invasive, aggressive or metastatic tumor phenotypes [24–26]. Moreover, MMP9 is identified as a representive epithelial−mesenchymal transition marker [27]. In our study, we found that ectopic expression of Trim44 obviously increased the expression of MMP9 mRNA and MMP9 protein, but showed no significant effect on MMP2. The data indicate that MMP9 is also involved in the role of Trim44-induced NSCLC invasion and metastasis.
It is well known that NF-κB is an important transcription factor which plays a critical role in the development of inflammatory skin diseases and in regulating the transcription of chemokine genes [28, 29]. Beck et al. [17] also reported that the expression and production of chemokines by endothelial cells depend on activation of NF-κB. In the present study, we found that overexpression of Trim44 in 16HBE cells significantly increased the activation of NF-κB, which was consistent with previous reports [12]. The luciferase assay results further demonstrated that Trim44 plays a positive role in regulating NF-κB. To explore whether Trim44 induced CXCR6 and MMP9 through NF-κB, we introduced PDTC, an inhibitor of NF-κB. Treatment with PDTC effectively inhibits the effect of Trim44 on activating NF-κB; however, upregulation of CXCR6 mRNA and MMP9 mRNA by Trim44 was also inhibited. The data reveal that Trim44 induces CXCR6 and MMP9 and this might be through activating NF-κB. Transwell assays were performed to further verify whether the NF-κB signaling pathway plays a critical role in Trim44-induced NSCLC cell migration and invasion. The results showed that the NF-κB signaling pathway is involved in the promotion of Trim-mediated migration and invasion.
In conclusion, this study provides the first evidence that Trim44 facilitates NSCLC migration and invasion. As stated above, our study demonstrated the molecular mechanism by which Trim44 participates in the migration and invasion of NSCLC cells. Our results strongly support Trim44 as a potential target for NSCLC treatment. Suppression of Trim44 expression may be useful for the prevention of chemokine production in Trim44-induced NSCLC cell migration.
References
Jemal A, Bray F, Center MM et al (2011) Global cancer statistics CA. Cancer J Clin 61:69–90
Kamangar F, Dores GM, Anderson WF (2006) Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 24:2137–2150
Carthagena L, Bergamaschi A, Luna JM et al (2009) Human TRIM gene expression in response to interferons. PLoS One 4:e4894
McNab FW, Rajsbaum R, Stoye JP et al (2011) Tripartite-motif proteins and innate immune regulation. Curr Opin Immunol 23:46–56
Ozato K, Shin DM, Chang TH et al (2008) TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol 8:849–860
Jefferies C, Wynne C, Higgs R (2011) Antiviral TRIMs: friend or foe in autoimmune and autoinflammatory disease? Nat Rev Immunol 11:617–625
Li Q, Yan J, Mao AP et al (2011) Tripartite motif 8 (TRIM8) modulates TNFalpha- and IL-1beta-triggered NF-kappaB activation by targeting TAK1 for K63-linked polyubiquitination. Proc Natl Acad Sci USA 108:19341–19346
Nisole S, Stoye JP, Saib A (2005) TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol 3:799–808
Bieniasz PD (2004) Intrinsic immunity: a front-line defense against viral attack. Nat Immunol 5:1109–1115
Yang K, Shi HX, Liu XY et al (2009) TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J Immunol 182:3782–3792
Boutou E, Matsas R, Mamalaki A (2001) Isolation of a mouse brain cDNA expressed in developing neuroblasts and mature neurons. Brain Res Mol Brain Res 86:153–167
Yang B, Wang J, Wang Y (2013) Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J Immunol 190:3613–3619
Peters CJ, Rees JR, Hardwick RH et al (2010) A 4-gene signature predicts survival of patients with resected adenocarcinoma of the esophagus, junction, and gastric cardia. Gastroenterology 139(e1915):1995–2004
Jarvinen AK, Autio R, Kilpinen S et al (2008) High-resolution copy number and gene expression microarray analyses of head and neck squamous cell carcinoma cell lines of tongue and larynx. Genes Chromosom Cancer 47:500–509
Kashimoto K, Komatsu S, Ichikawa D et al (2012) Overexpression of TRIM44 contributes to malignant outcome in gastric carcinoma. Cancer Sci 103:2021–2026
Hu Y, Wang J, Yang B (2011) Guanylate binding protein 4 negatively regulates virus-induced type I IFN and antiviral response by targeting IFN regulatory factor 7. J Immunol 187:6456–6462
Beck G, Yard BA, Schulte J et al (2003) Secreted phospholipases A2 induce the expression of chemokines in microvascular endothelium. Biochem Biophys Res Commun 300:731–737
Chen L, Chen DT, Kurtyka C et al (2012) Tripartite motif containing 28 (Trim28) can regulate cell proliferation by bridging HDAC1/E2F interactions. J Biol Chem 287:40106–40118
Zhou ZY, Yang GY, Zhou J et al (2012) Significance of TRIM29 and beta-catenin expression in non-small-cell lung cancer. J Chin Med Assoc 75:269–274
Liotta LA, Steeg PS, Stetler-Stevenson WG (1991) Cancer metastasis and angiogenesis: an imbalance of positive and negative regulation. Cell 64:327–336
Giancotti FG, Ruoslahti E (1999) Integrin signaling. Science (New York) 285:1028–1032
Stetler-Stevenson WG (1990) Type IV collagenases in tumor invasion and metastasis. Cancer Metastasis Rev 9:289–303
Zeng ZS, Cohen AM, Guillem JG (1999) Loss of basement membrane type IV collagen is associated with increased expression of metalloproteinases 2 and 9 (MMP-2 and MMP-9) during human colorectal tumorigenesis. Carcinogenesis 20:749–755
Papathoma AS, Zoumpourlis V, Balmain A et al (2001) Role of matrix metalloproteinase-9 in progression of mouse skin carcinogenesis. Mol Carcinog 31:74–82
Cockett MI, Murphy G, Birch ML et al (1998) Matrix metalloproteinases and metastatic cancer. Biochem Soc Symp 63:295–313
Bianco FJ Jr, Gervasi DC, Tiguert R et al (1998) Matrix metalloproteinase-9 expression in bladder washes from bladder cancer patients predicts pathological stage and grade. Clin Cancer Res 4:3011–3016
Lee JM, Dedhar S, Kalluri R et al (2006) The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol 172:973–981
Tsuruta D (2009) NF-kappaB links keratinocytes and lymphocytes in the pathogenesis of psoriasis. Recent Pat Inflamm Allergy Drug Discov 3:40–48
Wang S, Uchi H, Hayashida S et al (2009) Differential expression of phosphorylated extracellular signal-regulated kinase 1/2, phosphorylated p38 mitogen-activated protein kinase and nuclear factor-kappaB p105/p50 in chronic inflammatory skin diseases. J Dermatol 36:534–540
Acknowledgments
This work was supported by grants from the Shanghai Chest Hospital Science and Technology Fund Planning Project (YZ-11-28).
Conflict of interest
The authors have no financial conflicts of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Q. Luo and H. Lin contributed equally to the work.
About this article
Cite this article
Luo, Q., Lin, H., Ye, X. et al. Trim44 facilitates the migration and invasion of human lung cancer cells via the NF-κB signaling pathway. Int J Clin Oncol 20, 508–517 (2015). https://doi.org/10.1007/s10147-014-0752-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10147-014-0752-9