Abstract
We have previously reported that reliable detection of 2-hydroxyglutarate (2HG) in isocitrate dehydrogenase (IDH)-mutant WHO grade 2 and 3 gliomas is possible utilizing 3.0-T single-voxel magnetic resonance spectroscopy (SVMRS). We set out to determine whether the same method could be applied to detect 2HG in IDH-mutant glioblastoma. Forty-four patients harboring glioblastoma underwent pre-operative MRS evaluation to detect 2HG and other metabolites. Presence of IDH-mutations was determined by IDH1 R132H immunohistochemical analysis and DNA sequencing of surgically obtained tissues. Six out of 44 (13.6%) glioblastomas were IDH-mutant. IDH-mutant glioblastoma exhibited significantly higher accumulation of 2HG (median 3.191 vs. 0.000 mM, p < 0.0001, Mann-Whitney test). A cutoff of 2HG = 0.897 mM achieved high sensitivity (100.0%) and specificity (92.59%) in determining IDH-mutation in glioblastoma. Glioblastoma with high 2HG accumulation did not have significantly longer overall survival than glioblastoma with low 2HG accumulation (p = 0.107, log-rank test). Non-invasive and reliable detection of 2HG in IDH-mutant glioblastoma was possible by 3.0-T SVMRS.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
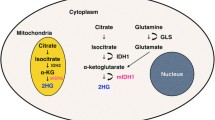
Glioblastoma is the most common and most malignant primary brain tumor. Despite recent advances, the median overall survival is 14.6 months. Recent whole-genome studies have shown that about 10% of glioblastoma harbor isocitrate dehydrogenase (IDH) mutations [1,2,3,4]. Glioblastomas with these mutations are known to have a better prognosis than their wildtype countertypes, and in the most recent, WHO classifications of tumors of the central nervous system [5] are distinguished from IDH-wildtype glioblastoma. In IDH-mutant gliomas, 2-hydroxyglutarate (2HG) is produced by conversion from α-ketoglutarate (α-KG). 2HG is known to competitively inhibit α-KG, causing DNA and histone hypermethylation, thus leading to glioma genesis [6]. We have previously shown that quantification of 2HG by single-voxel magnetic resonance spectroscopy (SVMRS) can be used to reliably distinguish between IDH-mutant and wildtype WHO grade 2 and 3 gliomas pre-operatively [7]. In that study, we excluded cases of WHO grade 4 glioblastomas from the analysis to minimize possible effects of necrosis on the spectra. In the current study, we analyzed whether glioblastoma could be pre-operatively divided into IDH-mutant and wildtype groups by SVMRS.
Materials and methods
Participants
Forty-seven consecutive patients harboring World Health Organization (WHO) grade IV gliomas, receiving magnetic resonance spectroscopy (MRS) evaluation at the Center for Integrated Brain Science, University of Niigata, before surgery and surgical treatment at the Department of Neurosurgery, University of Niigata, from July 2007 to September 2015 were included in the study. The patients underwent MRS evaluation a median of 6 days before surgery (range 1–22 days). A total of three patients were excluded from the study: two patients because of low signal-to-noise ratios of less than four and one patient because creatine (Cr) was not detected. Thus, a total of 44 patients were ultimately analyzed. Written informed consent was obtained from all of the participants in accordance with the human research guidelines of the Institutional Review Board of University of Niigata.
MRS analysis
MRI/1H-MRS analysis was performed using a 3.0-T system (Signa LX, General Electric, Waukesha, WI) as previously described [7]. Briefly, proton density images (fast spin echo; TR/TE = 5000/40; FOV 20 × 20 mm; matrix 256 × 256; slice thickness 5 mm; interslice gap 2.5 mm) were taken. The slice with the largest depiction of tumor on proton density images was selected for SVMRS. A point-resolved spectroscopic sequence (PRESS), with chemical-shift-selective water suppression, was used with the following parameters: (TR 1.5 s; TE 30 ms; data point 512; spectral width 1000 Hz; number of acquisitions 128–196; volume of interest (VOI) 12–20 × 12–20 × 12–20 mm). The volume of interest (VOI) was designed to minimize suspected areas of necrosis and hemorrhage. Suspected glioblastoma cases with both large cystic components and no solid component or cases with intratumoral hemorrhage on pre- and post-contrast MR images were not assessed.
Spectral analysis was performed using LCModel version 6.3 (Stephen Provencher, Oakville, Ontario, Canada) [8]. Nineteen metabolites were included in this LCModel basis set, and glutathione (GSH), 2-hydroxyglutarate (2HG), myo-inositol (Ins), total NAA (tNAA: the sum of N-acetylaspartate (NAA) and N-acetylaspartylglutamate (NAAG)), total creatine (tCr: the sum of Cr and phosphocreatine (PCr)), and total choline (tCho: the sum of glycerophosphocholine (GPC) and phosphocholine (PC)) were noted. The following lipids and macromolecules were also noted: MM09, Lip09, and Lip 13.
Quantification estimates of metabolites were considered unreliable and excluded when Cramer-Rao lower bounds, returned as the percentage of standard deviation (%SD) by LCModel, were greater than 50% for 2HG [9, 10] and 35% for GSH and Ins, 30% for Glx, and 20% for tNAA, tCho, and tCr [7] as previously described. Because low estimates yielded large %SDs (i.e., when 2HG = 0, %SD = ∞), the above exclusion criteria were applied only when the estimated amount for metabolites was greater than 1.0 mM.
Pathological analysis
Surgical specimens were analyzed by two pathologists (H. T. and K. A.) according to the WHO classification 2016 [5]. IDH1 R132H (H09 clone, Dianova, Hamburg, Germany; 1:100) immunohistochemical analysis and DNA sequencing of IDH1 and IDH2 were performed as previously described [7].
Statistical analysis
Corrected metabolite concentrations of tumors in IDH-mutant glioma patients were compared those in IDH-wildtype glioma patients using the Mann-Whitney U test. Kaplan-Meier analysis was used to compare overall survival. Tests for associations between different parameters were carried out by the Fisher’s exact test for 2 × 2 contingency tables. p < 0.05 was considered significant. Statistical analyses were performed using GraphPad Prism 7 software (GraphPad Software, http://www.graphpad.com). Receiver-operating characteristic (ROC) curve was used to determine a cutoff for 2HG concentration to obtain maximal sensitivity and specificity to identify IDH mutations was calculated using MedCalc (version 17.8) software. In order to obtain a robust 95% confidence interval, bootstrap method was employed using 1000 replications.
Results
A summary of the patient characteristics of mutant and wildtype IDH glioblastoma groups is provided in Table 1. Six out of 44 or 13.6% of glioblastoma were IDH mutant, comparable to the reported approximately 5–15% [1,2,3,4]. IDH-mutant glioblastoma patients were significantly younger (median 36 years) than IDH-wildtype glioblastoma patients (median 65 years) (p < 0.0001; Mann-Whitney test). There was a higher proportion of newly diagnosed glioblastoma patients in the IDH-wildtype group (34/38, 89.4%) compared to IDH-mutant (3/6, 50.0%) (p = 0.0419, Fisher’s exact test). None of the three IDH-mutant glioblastoma patients underwent MRS before their initial surgery. 2HG values were consistent with IDH mutation status in all recurrent cases, whether IDH-mutant or wildtype. A higher proportion of patients were alive at follow-up in the IDH-mutant glioblastoma group (3/6, 50.0%) compared to IDH-wildtype glioblastoma group (5/38, 13.2%), although not statistically significant (p = 0.0632).
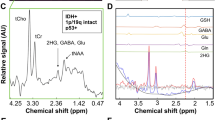
Representative SVMRS spectra of IDH-mutant and IDH-wildtype glioblastoma are provided in Fig. 1. Small peaks were detected at a chemical shift of about 2.25 ppm in IDH-mutant glioblastoma, reflecting the presence of 2HG. A higher lipid peak was noticed in both IDH-mutant and IDH-wildtype glioblastoma compared to those in WHO grade 2 and 3 gliomas [7].
IDH-mutant glioblastoma showed a significantly higher accumulation of 2HG (median 3.191 vs. 0.000 mM, p < 0.0001, Mann-Whitney test) (Fig. 2a). IDH mutant gliomas also expressed lower levels of Glx (median 7.090 vs. 9.393, p = 0.033), reflecting glutamine metabolism to produce 2HG in IDH-mutant gliomas. IDH-mutant glioblastoma exhibited less GSH accumulation, which is made from glutamine, although not statistically significant (median 1.653 vs. 1.924 mM, p = 0.2977). Levels of Ins, tNAA, tCho, and tCr were not significantly different between the two groups. 2HG was detected in all IDH-mutant glioblastoma, but not detected (2HG = 0) in 18 out of 38 (47.4%) (p = 0.0668, Fisher’s exact test) IDH-wildtype glioblastomas (Fig. 3a). This data was remarkably consistent with our previous data in WHO grade 2 and 3 gliomas [7], further validating our methodology.
Glioblastoma is known to have prominent lipid peaks, as demonstrated in Fig. 1. We next compared metabolites, including lipid and macromolecules, between six IDH-mutant glioblastomas and eight IDH-mutant WHO grade 3 gliomas (Fig. 2b). Higher levels of Lip 13 were detected in IDH-mutant glioblastoma (median 25.420 vs. 7.061 mM, p = 0.0173).
ROC curve analysis obtained an optimal cutoff of 2HG = 0.897 mM, with a sensitivity of 100.0%, specificity of 92.6%, likelihood ratio of 13.5, and area under curve of 0.981 (Fig. 3a, b). A 95% confidence interval of 0.553 to 1.993 mM was obtained with the bootstrap method. A cutoff of 1.673 mM yielded the highest likelihood ratio, 22.5, with a sensitivity of 83.3% and specificity of 96.3%. At a cutoff of 1.489 mM, which was determined to be the optimal cutoff for grade 2 and 3 gliomas [7], sensitivity was 83.3%, specificity was 92.6%, and likelihood ratio was 11.25. Median overall survival was longer in glioblastoma with high accumulation of 2HG (2HG > 1.103 mM) (738 days), compared to low 2HG glioblastoma group (566 days). However, overall survival was not significantly longer in high 2HG group (p = 0.17, log-rank test, Fig. 4). All three recurrent IDH-mutant glioblastomas had poor prognoses with overall survival of 100 to 302 days; however, all three newly diagnosed IDH-mutant glioblastoma patients were alive at last follow-up (Table 1, Fig. 4).
These tumors included cases in which necrosis was evident on post-contrast MR images and Lip13 accumulation was high on MRS. Also, there were non-necrotic cases the VOI was selected to encompass a solid component or the lesion was invasive with heterogeneous enhancement on post-contrast MR images but without necrosis. The percentage of solid/invasive and necrotic lesions and median 2HG and Lip13 values is summarized in Table 2. Somewhat surprisingly, median 2HG was less in solid/invasive IDH-mutant glioblastomas compared to IDH-wildtype glioblastomas (2.902 vs. 4.119 mM), probably reflecting the reduced cellularity in invasive lesions or secondary glioblastoma with solid low-grade components. As expected, Lip13 was much higher in the necrotic lesions both IDH-mutant and IDH-wildtype.
Discussion
Several studies, including ours, have shown that measurement of 2HG by MRS can be a reliable method to distinguish between IDH-mutant and IDH-wildtype gliomas [7, 11,12,13]. Almost all of these reports focus on grade 2 and grade 3 gliomas, partially because of the relative rarity of IDH-mutant glioblastoma. Over an 8-year period, we were able to assess six cases of IDH-mutant glioblastoma. In the current study, we have shown that by minimizing areas of necrosis by careful selection of VOI, 2HG can be reliably measured in WHO grade 4 glioblastomas as well.
We detected less Glx (the sum of glutamine and glutamate) in IDH-mutant glioblastoma. In vitro studies which placed isotope-labeled glutamine into media-growing IDH-mutant cells showed that the labeled carbon is ultimately used by 2HG [14], suggesting that 2HG is primarily derived from glutamine in mutant IDH gliomas. Glutamine is hydrolyzed by glutaminase to produce glutamate, which is subsequently converted to α-KG [14, 15]. Furthermore, a study injecting hyperpolarized 13C α-KG into rats injected with IDH-mutant glioblastoma cells found that glutamine production is reduced in IDH-mutant gliomas, mainly due to the decrease of branched-chain amino acid transaminase 1 (BCAT1) enzyme, which catalyzes the transamination of branched-chain amino acids while converting α-KG to glutamate [16]. This decrease in Glx was consistent with findings of our previous report on grade 2 and grade 3 gliomas, and also in line with a report from Nagashima et al. suggesting that elevated 2HG as well as decreased Glu is diagnostic for IDH-mutant gliomas [13]. Thus, the concentration of Glx was thought to be good surrogate marker to 2HG in determining possible false positive IDH-wildtype glioblastoma.
It is well known that glioblastomas have high lipid peaks, likely reflecting necrosis, and a previous report shows that glioblastomas have a higher lipid and macromolecule peaks at chemical shifts of 1.3 ppm (LM13) and 0.9 ppm (LM09) compared to normal brain [17]. We found the same tendency in IDH-mutant glioblastoma, as higher levels of Lip 13 (median 25.420 vs. 7.061 mM, p = 0.0173) were noted compared to IDH-mutant WHO grade 3 gliomas.
With careful selection of VOI and parameters, we were able to achieve a 100% specificity and over 92% specificity of 2HG detection by short-echo MRS with modulation of 2HG resonances by spectral fitting. During the study period, newer methods for detection of 2HG in IDH-mutant gliomas have emerged. Long-echo MRS with TE at 97 ms with the use of 3D volume-localized basis (VLB) spectra has been shown to be optimal for detection of 2HG [18, 19]. A comparative study of PRESS sequences at short- (35 ms) and long-TE (97 ms) found long-TE to be superior by minimizing the effect of macromolecule signals [19]. Unambiguous detection of 2HG in mutant IDH gliomas was achieved by 2D correlation spectroscopy (COSY) [12, 20, 21] and J-difference spectroscopy [20]. However, these methods are less available clinically and involve longer acquisition time; 2D correlation MRS involves complex quantification and has less sensitivity [22]. Pre-clinical trials involve intravenous injection of hyperpolarized 13C-labeled α-KG to detect decreased glutamate production in IDH-mutant tumors [16] and hyperpolarized pyruvate to monitor the conversion of pyruvate to lactate, which is decreased in IDH-mutant tumors [23]. These newer techniques are outlined in two important review articles [22, 24].
In the new WHO classification [5], IDH-mutant glioblastomas are distinguished from IDH-wildtype glioblastoma. IDH-mutant glioblastomas occur in younger patients and carry a significant better prognosis than their IDH-wildtype counterparts [2, 25]. However, overall survival was not significantly longer in the high 2HG group in this study (p = 0.17, log-rank test, Fig. 4), partly due to the small number of IDH-mutant glioblastoma and because three out of six IDH-mutant cases were recurrent. Evidence suggests that IDH-mutant secondary glioblastomas are more susceptible to temozolomide [26], and IDH-mutant malignant astrocytomas benefit from radical surgical resection [27]. In vivo measurement of 2HG has many potential therapeutic implications in IDH-mutant gliomas, such as indication for radical surgery, indication for pre-surgical chemotherapy in select cases, determination of therapeutic response after treatments [28], and detection of rare IDH1 and IDH2 mutations. Pre-operative diagnosis of IDH-mutation by SVMRS also opens the door for intraoperative-targeted imaging and treatments of IDH-mutant gliomas.
This study was not designed to include a validation cohort. Thus, the cutoff may not be optimal when it comes to reproduction in an independent cohort. The same methods employed for WHO grade 2 and 3 gliomas were employed in the current study for glioblastoma, yielding a similar cutoff of 2HG = 1.489 mM. Ninety-five percent confidence interval determined by the bootstrap method was 0.553 to 1.993 mM. Careful selection of cases by discarding cystic lesions and lesions with marked intratumoral hemorrhage and by minimizing selection of necrosis when determining the VOI were essential to obtain reproducible results.
We have previously reported that quantification of 2HG by SVMRS can be used to reliably distinguish between IDH-mutant and IDH-wildtype WHO grade 2 and 3 gliomas pre-operatively. In the current study, we found that the same methods can be utilized to distinguish between IDH-mutant and IDH-wildtype glioblastoma. It has become increasingly clear that IDH-mutant gliomas have completely different biological backgrounds than IDH-wildtype gliomas. Thus, non-invasive diagnosis of IDH-mutation will increasingly become vital for the treatment of these tumors.
References
Parsons DW, Jones C, Zhang X, Cheng-Ho Lin J, Leary RJ, Angenendt P, Mankoo P, Carter H, Sui I, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA Jr, Hartigan J, Smith DR, Strausberg RL, Nagahashi M, Shinjo SMO, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Volgelstein B, Velculescu VE, Kinzler KW (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321:1807–1812
Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, VE V, Vogelstein B, Bigner DD (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773. https://doi.org/10.1056/NEJMoa0808710
Polivka J, Polivka J Jr, Rohan V, Pesta M, Repik T, Pitule P, Topolcan O (2014) Isocitrate dehydrogenase-1 mutations as prognostic biomarker in glioblastoma multiforme patients in West Bohemia. Biomed Res Int 2014:735659. https://doi.org/10.1155/2014/735659
Ogura R, Tsukamoto Y, Natsumeda M, Isogawa M, Aoki H, Kobayashi T, Yoshida S, Okamoto K, Takahashi H, Fujii Y, Kakita A (2015) Immunohistochemical profiles of IDH1, MGMT and P53: practical significance for prognostication of patients with diffuse gliomas. Neuropathology 35:324–335. https://doi.org/10.1111/neup.12196
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Ellison DW, Figarella-Branger D, Perry A, Reifenberger G, von Deimling A (2016) WHO classification of tumours of the central nervous system. IARC, Lyon
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19:17–30. https://doi.org/10.1016/j.ccr.2010.12.014
Natsumeda M, Igarashi H, Nomura T, Ogura R, Tsukamoto Y, Kobayashi T, Aoki H, Okamoto K, Kakita A, Takahashi H, Nakada T, Fujii Y (2014) Accumulation of 2-hydroxyglutarate in gliomas correlates with survival: a study by 3.0-tesla magnetic resonance spectroscopy. Acta Neuropathol Commun 7:158. https://doi.org/10.1186/s40478-014-0158-y
Provencher SW (1993) Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med 30:672–679. https://doi.org/10.1002/mrm.1910300604
Tkac I, Oz G, Adriany G, Ugurbil K, Gruetter R (2009) In vivo 1H NMR spectroscopy of the human brain at high magnetic fields: metabolite quantification at 4T vs. 7T. Magn Reson Med 62:868–879. https://doi.org/10.1002/mrm.22086
Takado Y, Igarashi H, Terajima K, Shimohata T, Ozawa T, Okamoto K, Nishizawa M, Nakada T (2011) Brainstem metabolites in multiple system atrophy of cerebellar type: 3.0-T magnetic resonance spectroscopy study. Mov Disord 26:1297–1302. https://doi.org/10.1002/mds.23550
Pope WB, Prins RM, Albert Thomas M, Nagarajan R, Yen KE, Bittinger MA, Salamon N, Chou AP, Yong WH, Soto H, Wilson N, Driggers E, Jang HG, Su SM, Schenkein DP, Lai A, Cloughesy TF, Kornblum HI, Wu H, Fantin VR, Liau LM (2012) Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neuro-Oncol 107:197–205. https://doi.org/10.1007/s11060-011-0737-8
Elkhaled A, Jalbert LE, Phillips JJ, Yoshihara HA, Parvataneni R, Srinivasan R, Bourne G, Berger MS, Chang SM, Cha S, Nelson SJ (2012) Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med 4:116ra5 1–116ra510. https://doi.org/10.1126/scitranslmed.3002796
Nagashima H, Tanaka K, Sasayama T, Irino Y, Sato N, Takeuchi Y, Kyotani K, Mukasa A, Mizukawa K, Sakata J, Yamamoto Y, Hosoda K, Itoh T, Sasaki R, Kohmura E (2016) Diagnostic value of glutamate with 2-hydroxyglutarate in magnetic resonance spectroscopy for IDH1 mutant glioma. Neuro-Oncology 18:1559–1568
Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462:739–744. https://doi.org/10.1038/nature08617
Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, Tsukamoto T, Rojas CJ, Slusher BS, Rabinowitz JD, Dang CV, Riggins GJ (2010) Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res 70:8981–8987. https://doi.org/10.1158/0008-5472.CAN-10-1666
Chaumeil MM, Larson PE, Woods SM, Cai L, Eriksson P, Robinson AE, Lupo JM, Vigneron DB, Nelson SJ, Pieper RO, Phillips JJ, Ronen SM (2014) Hyperpolarized [1-13C] glutamate: a metabolic imaging biomarker of IDH1 mutational status in glioma. Cancer Res 74:4247–4257. https://doi.org/10.1158/0008-5472.CAN-14-0680
Crisi G, Orsingher L, Filice S (2013) Lipid and macromolecules quantitation in differentiating glioblastoma from solitary metastasis: a short-echo time single-voxel magnetic resonance spectroscopy study at 3 T. J Comput Assist Tomogr 37:265–271. https://doi.org/10.1097/RCT.0b013e318282d2ba
Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, Yang XL, Mashimo T, Raisanen JM, Marin-Valencia I, Pascual JM, Madden CJ, Mickey BE, Malloy CR, Bachoo RM, Maher EA (2012) 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med 18:624–629. https://doi.org/10.1038/nm.2682
Choi C, Ganji S, Hulsey K, Madan A, Kovacs Z, Dimitrov I, Zhang S, Pichumani K, Mendelsohn D, Mickey B, Malloy C, Bachoo R, Deberardinis R, Maher E (2013) A comparative study of short- and long-TE (1)H MRS at 3 T for in vivo detection of 2-hydroxyglutarate in brain tumors. NMR Biomed 26:1242–1250. https://doi.org/10.1002/nbm.2943
Andronesi OC, Kim GS, Gerstner E, Batchelor T, Tzika AA, Fantin VR, Vander Heiden MG, Sorensen AG (2012) Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med 4:116 ra4 1–116 ra410. https://doi.org/10.1126/scitranslmed.3002693
Esmaeili MVR, Bathen TF (2013) 2-hydroxyglutarate as a magnetic resonance biomarker for glioma subtyping. Transl Oncol 6:92–98. https://doi.org/10.1593/tlo.12424
Andronesi OC, Rapalino O, Gerstner E, Chi A, Batchelor TT, Cahill DP, Sorensen AG, Rosen BR (2013) Detection of oncogenic IDH1 mutations using magnetic resonance spectroscopy of 2-hydroxyglutarate. J Clin Invest 123:3659–3663. https://doi.org/10.1172/JCI67229
Chaumeil MM, Radoul M, Najac C, Eriksson P, Viswanath P, Blough MD, Chesnelong C, Luchman HA, Cairncross JG, Ronen SM (2016) Hyperpolarized (13)C MR imaging detects no lactate production in mutant IDH1 gliomas: implications for diagnosis and response monitoring. Neuroimage Clin 12:180–189. https://doi.org/10.1016/j.nicl.2016.06.018
Hu J, Salzillo TC, Sailasuta N, Lang FF, Bhattacharya P (2017) Interrogating IDH mutation in brain tumor: magnetic resonance and hyperpolarization. Top Magn Reson Imaging 26:27–32. https://doi.org/10.1097/RMR.0000000000000113
Nobusawa S, Watanabe T, Kleihues P, Ohgaki H (2009) IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res 15:6002–6007. https://doi.org/10.1158/1078-0432.CCR-09-0715
SongTao Q, Lei Y, Si G, YanQing D, HuiXia H, XueLin Z, LanXiao W, Fei Y (2012) IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci 103:269–273. https://doi.org/10.1111/j.1349-7006.2011.02134.x
Beiko J, Suki D, Hess KR, Fox BD, Cheung V, Cabral M, Shonka N, Gilbert MR, Sawaya R, Prabhu SS, Weinberg J, Lang FF, Aldape KD, Sulman EP, Rao G, McCutcheon IE, Cahill DP (2014) IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro-Oncology 16:81–91. https://doi.org/10.1093/neuonc/not159
Andronesi OC, Loebel F, Bogner W, Marjanska M, Vander Heiden MG, Iafrate AJ, Dietrich J, Batchelor TT, Gerstner ER, KJW G, Chi AS, Rosen BR, Cahill DP (2016) Treatment response assessment in IDH-mutant glioma patients by non-invasive 3D functional spectroscopic mapping of 2-hydroxyglutarate. Clin Can Res 22:1632–1641
Acknowledgements
We acknowledge Drs. Kimihiko Nakamura, Taro Nishikawa, Shinya Jinguji, Toshiharu Nomura, and others for the help with imaging.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Ethical approval was obtained in accordance with the human research guidelines of the Institutional Review Board of University of Niigata.
Informed consent
Written informed consent was obtained from all of the participants.
Rights and permissions
About this article
Cite this article
Natsumeda, M., Motohashi, K., Igarashi, H. et al. Reliable diagnosis of IDH-mutant glioblastoma by 2-hydroxyglutarate detection: a study by 3-T magnetic resonance spectroscopy. Neurosurg Rev 41, 641–647 (2018). https://doi.org/10.1007/s10143-017-0908-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10143-017-0908-y