Abstract
Hepatocellular carcinoma (HCC) is a leading cause of cancer-related mortality globally. Many herbal medicines and their bioactive compounds have shown anti-tumor properties. This study was conducted to examine the effect of psilostachyin C (PSC), a sesquiterpenoid lactone isolated from Artemisia vulgaris L., in the malignant properties of HCC cells. CCK-8, flow cytometry, wound healing, and Transwell assays revealed that 25 μM PSC treatment significantly suppressed proliferation, cell cycle progression, migration, and invasion of two HCC cell lines (Hep 3B and Huh7) while promoting cell apoptosis. Bioinformatics prediction suggests CREB binding protein (CREBBP) as a promising target of PSC. CREBBP activated transcription of GATA zinc finger domain containing 2B (GATAD2B) by binding to its promoter. CREBBP and GATAD2B were highly expressed in clinical HCC tissues and the acquired HCC cell lines, but their expression was reduced by PSC. Either upregulation of CREBBP or GATAD2B restored the malignant properties of HCC cells blocked by PSC. Collectively, this evidence demonstrates that PSC pocessess anti-tumor functions in HCC cells by blocking CREBBP-mediated transcription of GATAD2B.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In 2020, liver cancer ranked as the sixth most frequently diagnosed cancer globally and stood as the third primary contributor to cancer-related fatalities, recording around 906,000 newly diagnosed cases and 830,000 deaths (Sung et al. 2021). Hepatocellular carcinoma (HCC) stands out as the predominant subtype, encompassing over 80% of cases of primary liver cancer on a global scale (Chidambaranathan-Reghupaty et al. 2021). While there have been significant strides in treatment options like liver transplantation, surgical resection, percutaneous ablation, radiation, transarterial, and systemic therapies, as evidenced by studies (Forner et al. 2018; Vogel et al. 2022), the overall 5-year survival rate for HCC still lingers at a discouraging level (Grandhi et al. 2016; Villanueva 2019). The demand for effective treatments for HCC has persistently remained unmet.
Initiatives in HCC management increasingly explore traditional herbal medicines and their bioactive components as therapeutic options. This is due to their multi-level, multi-target, and coordinated intervention effects (Chen et al. 2016; Luk et al. 2007). Artemisia vulgaris L. (common mugwort) holds significant historical importance in medicine, earning it the title “mother of herbs” during the Middle Ages (Ekiert et al. 2020). Accumulating evidence indicates its diverse properties, including antioxidant, anti-inflammatory, antinociceptive, anti-hypertensive, hepato-protective, antifungal, and antimicrobial characteristics (Blagojevic et al. 2006; Jiang et al. 2019a; Obistioiu et al. 2014; Pires et al. 2009; Temraz and El-Tantawy 2008; Tigno et al. 2000). This species is notably characterized by the presence of sesquiterpenoid lactones (STLs), such as psilostachyin, psilostachyin C (PSC), and vulgarin, as indicated in the literature (Ekiert et al. 2020). STLs, a feature not exclusive to the Asteraceae family, are known for their diverse biological activities, with anti-tumor, anti-inflammatory, and anti-parasitic effects particularly prominent in reported studies (Laurella et al. 2022). Remarkably, among numerous STLs assessed for their impact on a murine lymphoma cell line BW5147, PSC stood out as the most bioactive and least toxic compound (Martino et al. 2015). Moreover, it demonstrated the induction of reactive oxygen species and led to cell cycle arrest at the S phase (Martino et al. 2015). Beyond this, PSC displayed cytotoxic effects on a human colonic adenocarcinoma cell line Colo 205 (Kovacs et al. 2022). These findings collectively highlight PSC’s potential as a promising anti-tumor agent.
In this investigation, bioinformatics predictions pinpoint CREB binding protein (CREBBP, also known as CBP) as a promising target for PSC, while GATA zinc finger domain containing 2B (GATAD2B) emerges as a potential transcription target of CREBBP. CREBBP and its closely related paralog p300 function as widely expressed transcriptional coactivators and major lysine acetyltransferases in metazoans (Attar and Kurdistani 2017). Transcription factors (TFs) such as CREB and AP-1 bind to specific DNA sites, recruiting coactivator and histone acetyltransferase proteins (CREBBP and p300). This catalyzes histone acetylation, establishing an open chromatin architecture that facilitates gene transcription (Bowlt Blacklock et al. 2018; Thiel and Rossler 2022). Additionally, CREBBP/p300 serves as a bridge to the basal transcriptional apparatus, supporting transcriptional activation (Thiel and Rossler 2022). As for GATAD2B, it has been implicated in critical roles related to tumorigenesis and metastasis in KRAS-driven lung cancer (Grzeskowiak et al. 2018), but its precise functions in HCC remain unclear. This study was conducted to investigate whether PSC demonstrates anti-tumor properties in HCC and if these processes involve the regulation of the CREBBP/GATAD2B axis.
Materials and methods
Bioinformatics analyses
To explore the downstream targets of PSC, the chemical structure of PSC (PubChem SID: 447274617) was retrieved from the NCBI’s PubChem Substance database (https://www.ncbi.nlm.nih.gov/pcsubstance/?term=). Next, the structure was input into SwissTargetPrediction (http://swisstargetprediction.ch/) for target prediction. Human transcription factors and transcription co-factors were sourced from the Human TFDB database (http://bioinfo.life.hust.edu.cn/HumanTFDB#!/), and their intersection with the downstream targets of PSC was determined using Jvenn system (https://jvenn.toulouse.inrae.fr/app/example.html). Following this, the expression of the intersecting genes in HCC was forecasted using the cancer analysis database UALCAN (https://ualcan.path.uab.edu/index.html) for screening.
The downstream targets of the TF CREBBP were acquired from the hTFtarget system (http://bioinfo.life.hust.edu.cn/hTFtarget/#!/). The Jvenn system was applied again to cross-reference the candidate downstream targets and genes positively correlated with CREBBP (Pearson correlation coefficient > 0.5) in UALCAN.
Clinical samples
Twelve pairs of tumor tissues and the adjacent non-involved normal tissues were collected from HCC patients diagnosed and underwent surgery at Honghui Hospital, Xi’an Jiaotong University from June 2018 to December 2022. All patients had not undergone any anti-cancer treatments before surgery. The collection and use of human samples were approved by the Ethics Committee of Honghui Hospital, Xi’an Jiaotong University. All procedures were adhered to the guidelines of the Declaration of Helsinki. All patients signed an informed consent form.
Cell culture
Human HCC cell lines Hep 3B (CL-0102) and Huh7 (CL-0120) were acquired from Procell Life Science & Technology Co., Ltd. (Wuhan, Hubei, China). A normal liver epithelial cell line THLE-2 (CRL-270) was acquired from American Type Culture Collection (Manassas, VA, USA). Hep 3B cells were cultured in minimum essential medium (containing NEAA) supplemented with 10% fetal bovine saline (FBS) and 1% penicillin/streptomycin (P/S) (PM150410, Procell). Huh7 cells were cultured in RPMI-1640 (iCell-0002, Cellverse Bioscience Technology Co., Ltd., Shanghai, China) containing 10% FBS and 1% P/S. THLE-2 cells were cultured in BEGM (CC-3170, Lonza/Clonetics Corporation, Bend, OR, USA). All cells were cultured in a 37℃ incubator containing 5% CO2 with 70-80% humidity.
3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay
Following the manufacturer’s instructions, the toxicity of PSC on HCC cells and THLE-2 was analyzed using the MTT cytotoxicity assay kit (C0009M, Beyotime Biotechnology Co., Ltd, Shanghai, China). In brief, cells were added to 96-well plates at approximately 5000 cells per well. Subsequently, PSC was added in increasing concentrations of 0, 10, 25, 50, 100, and 200 μM. After 24 h, 10 μL 5 mg/mL MTT solution was added to each well, and the plate was further incubated in a cell culture incubator for 4 h. Afterward, 100 μL formazan solubilization solution was added to each well, and the plate was again incubated at 37℃ for 4 h in the cell culture incubator. Finally, the absorbance at 570 nm wavelength was measured using a microplate reader.
Cell transfection
Lentiviral vectors containing short hairpin RNA targeting CREBBP (CREBBP-KD), overexpression plasmids of CREBBP and GATAD2B (CREBBP-OE and GATAD2B-OE), or the negative control (NC) plasmids (KD-NC, OE-NC) were acquired from OriGene (https://www.origene.com/) and used to affect the HCC cells. Stably infected (transfected) cells were screened using puromycin. PSC ( > = 98%, 6466-67-7, Hölzel Diagnostika Handels GmbH, Hilden, Germany) was used to treat cells at a dose of 25 μM for 24 h (Kovacs et al. 2022). An equal volume of dimethyl sulphoxide (DMSO) solution was used as controls.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
A TRIzol kit (B511321, Sangon Biotech Co., Ltd., Shanghai, China) was used to extract total RNA from cells, and the RNA was reverse-transcribed into cDNA using the Reliance Select cDNA Synthesis Kit (12012802, Bio-Rad Laboratories, Hercules, CA, USA). The cDNA was used for qPCR analysis using the PTC Tempo 96 Thermal Cycler (12015382, Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control for mRNA quantification using the 2−ΔΔCt method. The primers used were as follows: CREBBP: forward: 5ʹ-AGTAACGGCACAGCCTCTCAGT-3ʹ, reverse: 5ʹ-CCTGTCGATACAGTGCTTCTAGG-3ʹ; GATAD2B: forward: 5ʹ-GGAGACAACAGGACTGCTGGAA-3ʹ, reverse: 5ʹ-GATGTCTGGTGAGGGAGTTAGC-3ʹ; GAPDH: forward: 5ʹ-GTCTCCTCTGACTTCAACAGCG-3ʹ, reverse: 5ʹ-ACCACCCTGTTGCTGTAGCCAA-3ʹ.
Western blot (WB) analysis
Total protein from cells was extracted using the RIPA lysis buffer (89901, Thermo Fisher Scientific, Rockford, IL, USA), and the protein concentration was determined using the Quick Start™ Bradford Protein Assay Kit (5000201, Bio-Rad). The protein was denaturized by heating, separated by 10% gel electrophoresis, and transferred onto polyvinylidene fluoride membranes. Thereafter, the membranes were incubated with the antibodies against CREBBP (1:1,000, ab253202, Abcam Inc., Cambridge, MA, USA) and GATAD2B (1:1000, ab308631, Abcam) at 4℃ overnight. After that, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:20,000, ab205718, Abcam). The protein signals were determined using enhanced chemiluminescence kit (abs920, Absin Biotechnologies Co., Ltd., Shanghai, China). Protein levels relative to GAPDH (1:10,000, ab181602, Abcam) were analyzed using Image J.
Immunohistochemistry (IHC)
The clinical tissue samples were fixed in 10% formalin, embedded in paraffin, and then made into 4-mm sections. The sections were then dewaxed, rehydrated, blocked with normal goat serum and 3% H2O2 solution, and reacted with the antibodies against CREBBP (1:2000, ab253202, Abcam) and GATAD2B (1:100, ab308631, Abcam) at 4℃ overnight, followed by incubation with HRP-conjugated goat anti-rabbit IgG (1:20,000, ab205718, Abcam). Subsequently, the staining was developed using 3,3’-diaminobenzidine, and nuclei were counter-stained using hematoxylin. The staining was observed under the fluorescence microscope.
Cell proliferation detection
Proliferation of cells was determined following the instructions of the cell counting kit-8 (CCK-8) kit (CK04-100T, Shanghai Yanjin Biological Co., Ltd., Shanghai, China). In short, 100 μL of cell suspension was added into 96-well plates for 24 h of pre-incubation at 37℃ with 5% CO2. Subsequently, each well was added with 25 μM PSC or DMSO, followed by further incubation for 0, 24, 48, 72, and 96 h, respectively. Following this, each well was added with 10 μL of CCK-8 solution for 2 h. The optical density (OD) at 450 nm was determined. The OD value at 0 h was used for normalization.
Cell cycle analysis
Cell cycle distribution in cells was determined following the instructions of the Cell Cycle Assay Kit Plus (C6078S, Ueland Biotechnologies Co., Ltd., Suzhou, Shanghai, China). In brief, 24 h after 25 μM PSC treatment, approximately 1 × 105 HCC cells were centrifuged at 1,000 g for 3 min. After the removal of supernatant, the cells were resuspended in 1 mL pre-cooled staining buffer. Each tube of cell samples was added with 4 μL RedNucleus I staining solution and incubated at room temperature (20 ∼ 25℃) in the dark for 20 min. Subsequently, the samples were subjected to flow cytometric analysis at an excitation wavelength of 638 nm to analyze cellular DNA concentrations.
Cell apoptosis detection
Apoptosis of cells was examined using a fluorescein isothiocyanate (FITC)/propidium iodide (PI) kit (A211, Vazyme Biotech Co., Ltd., Nanjing, Jiangsu, China). Briefly, stably transfected HCC cells, 24 h after 25 μM PSC treatment, were seeded into six-well plates at 2 × 106 cells per well. The cells were lysed in trypsin, resuspended to 1 × 106 cells/mL, and stained with 5 μL Annexin V-FITC and 5 μL PI at room temperature in the dark for 10 min. Apoptosis of cells was analyzed by a flow cytometer.
Wound healing assay
To analyze the migration of cells, the treated HCC cells were seeded into six-well plates. When a 90% cell confluence was reached, a sterile pipette tip was used to generate scratches (wounds) on the cell monolayer. After recording the width of the wounds, the wells were added with 25 μM PSC or DMSO. After 36 h, the wound width was recorded again. The wound healing rate within 36 h was then calculated using Image J software.
Transwell assay
Transwell chambers pre-coated with 40 μL Matrigel (356230, Solarbio Science & Technology Co., Ltd., Beijing, China) were used to examine the invasion ability of cells. Briefly, the chambers were inserted into 24-well basolateral chambers. HCC cells were resuspended in serum-free medium containing 25 μM PSC or DMSO and seeded into the apical chambers, while the basolateral chambers were added with complete medium containing 10% FBS. After 36 h, cells invaded the basolateral chambers were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and observed under the microscope.
Chromatin immunoprecipitation (ChIP)
Crosslinking reactions were performed by adding 37% formaldehyde to the HCC cells, and the reaction was terminated with glycine. After washing with PBS, the cells were lysed in Pierce IP Lysis Buffer (87787, Thermo Fisher Scientific) containing a protease inhibitor mixture (P1006, Beyotime), followed by the addition of Protein A agarose beads (70804-100, BEAVER Biotechnologies Co., Ltd., Suzhou, Jiangsu, China) overnight at 4 °C. The beads were then incubated with anti-CREBBP (1:100, ab253202, Abcam) and an isotype control anti-rabbit IgG (1:30, ab313801, Abcam) at 4 °C for 12 h. The beads were collected, washed with elution buffer (H5413, Merck KGaA, Darmstadt, Germany), and DNA was extracted using a genomic DNA purification kit (K0512, Thermo Fisher Scientific). Subsequently, PCR amplification was performed using specific primers for the GATAD2B promoter.
Dual luciferase reporter assay
The GATAD2B promoter sequence was obtained from UCSC (Genome Browserhttps://genome.ucsc.edu/). According to the instructions of the Dual Luciferase Reporter Assay Kit (DL101-01, Vazyme), the sequence was inserted into the firefly luciferase plasmid, which was then transfected into HCC cells. After 48 h of incubation, the cells were lysed using cell lysis buffer, and the cell lysate was centrifuged at 12,000× g for 2 min. Luciferase substrate (100 μL at room temperature) was added to the assay plate, and 20 μL of cell lysate was added to each well for the detection of luciferase reporter activity.
Statistical analysis
Statistical analyses were conducted using Prism 8.0.2 software (GraphPad, La Jolla, CA, USA). Quantitative data are presented as mean ± standard deviation. Student’s t-tests were used for comparisons between two groups, while one-way or two-way analysis of variance (ANOVA) followed by Tukey’s post hoc test was used for comparisons among multiple groups. Half maximal inhibitory concentration (IC50) of PSC in cells was calculated using the nonlinear fitting analysis. Correlation between CREBBP expression and GATAD2B was analyzed using nonlinear regression and correlation analyses. *p < 0.05 indicates statistical significance.
Results
PSC treatment represses malignant phenotype of HCC cells
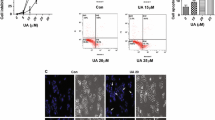
First, MTT assay was performed to examine the toxicity of PSC on HCC cells and THLE-2 cells. Notably, the PSC treatment exhibited a dose-dependent cytotoxicity to HCC cell lines. Its IC50 in Hep 3B cells was 51.93 μM and that in Huh7 cells was 48.20 μM. However, THLE2 cells showed a stronger tolerance, with an estimated PSC IC50 of 201.6 μM (Fig. 1A). To minimize potential toxic effects while still observing drug efficacy, doses below the IC50 value have been deemed reasonable (Hailan et al. 2022; Sun et al. 2020). Therefore, we applied a dose of approximately half of the IC50 in HCC cells (25 μM) for subsequent experiments. This dose would significantly affect HCC cells while having little psytotoxicity to normal cells.
PSC exerts anti-tumor functions on HCC cells in vitro. HCC cells (Hep 3B and Huh7) and normal THLE-2 cells were exposed to PSC at different concentrations (0, 10, 25, 50, 100, and 200 μM) for 24 h. A, dose-dependent toxicity and IC50 of PSC in these cells determined by MTT assay. In the subsequent experiments, Hep 3B and Huh7 cells were treated with 25 μM PSC for 24 h or treated with an equal volume of DMSO solution. B, proliferation of cells determined by CCK-8 assay; C, cell cycle distribution of cells determined by flow cytometry; D, apoptosis of cells determined by flow cytometry; E, migration ability of cells analyzed by wound healing assay; F, invasion ability of cells determined by Transwell assay. Three biological replicates were conducted. Differences were compared by the two-way ANOVA (B-F). IC50 of PSC in cells was calculated using the nonlinear fitting analysis (A) *p < 0.05 vs. DMSO
Compared to DMSO, the PSC treatment significantly suppressed proliferation of Hep 3B and Huh7 cells according to the CCK-8 assay (Fig. 1B). Flow cytometric analysis showed that the PSC treatment decreased the proportion of cells at G0/G1 phases while increasing cell populations at S and G2/M phases (Fig. 1C). Meanwhile, apoptosis of Hep 3B and Huh7 cells was substantially increased by the PSC treatment (Fig. 1D). Wound healing and Transwell assays revealed a decrease in cell migration and invasion abilities caused by PSC (Fig. 1E-F). This evidence supports that PSC has anti-tumor properties on HCC in vitro. Exact p-values and the mean and SD values are presented in Supplemental Materials.
PSC reduces CREBBP protein level
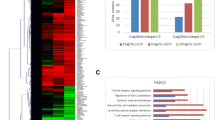
To analyze the downstream molecular mechanisms of PSC in HCC, we obtained the chemical structure of PSC (PubChem SID: 447274617) from PubChem Substance system (https://www.ncbi.nlm.nih.gov/pcsubstance/?term=) (Fig. 2A) and predicted its possible downstream targets from SwissTargetPrediction. Subsequently, we downloaded human TFs and co-factors from Human TFDB system. The predicted TFs were cross screened with the PSC’s targets using Jvenn system, resulting in 12 intersecting TFs (Fig. 2B). Among the differentially expressed TFs, HMOX1, LRRK2, SRC, CREBBP, PIN1, NR1H3, NR1H2, and JAK2 were suggested to show significant aberrant expression in liver HCC (LIHC) according to data in the UALCAN system. LRRK2 and CREBBP were the only two TFs whose exact roles in HCC remain unclear (previous research concerning the roles of other TFs in HCC is discussed in the later text). Both LRRK2 (Jiang et al. 2019b; Wang et al. 2023) and CREBBP (Hu et al. 2021) have been demonstrated to play oncogenic functions in several other cancer types. However, LRRK2 presented a low expression pattern whereas CREBBP presented a high expression pattern in LIHC according to the UALCAN data (Fig. 2C). Finally, we chose CREBBP as the subject for further research.
PSC reduces CREBBP protein level in HCC cells. A, chemical structure of PSC (PubChem SID: 447,274,617) obtained from the PubChem Substance system (https://www.ncbi.nlm.nih.gov/pcsubstance/?term=); B, intersections of human TFs and co-factors with the targets of PSC predicted from the SwissTargetPrediction system; C, expression patterns of LRRK2 and CREBBP predicted using UALCAN system; D, mRNA expression of CREBBP in clinical HCC samples and the adjacent non-involved tissue samples examined by RT-qPCR; E, positive staining of CREBBP in clinical HCC samples and the adjacent non-involved tissue samples examined by IHC; F-G, mRNA (F) and protein (G) levels of CREBBP in HCC cell lines (Hep 3B and Huh7) and normal THLE-2 cells determined by RT-qPCR and WB analysis, respectively; H-I, mRNA (H) and protein (I) levels of CREBBP in HCC cell lines (Hep 3B and Huh7) and normal THLE-2 cells after PSC or DMSO treatment determined by RT-qPCR and WB analysis, respectively. Three biological replicates were conducted. Differences were compared by the paired t test (D-E) or by the one-way (F-G) or two-way (H-I) ANOVA. *p < 0.05 vs. Normal liver/THLE-2/DMSO.
Indeed, RT-qPCR and IHC assays revealed an increase in the mRNA and protein expression of CREBBP in the clinical HCC samples compared to the adjacent tissues (Fig. 2D-E). Similarly, RT-qPCR and WB assays revealed that the Hep 3B and Huh7 cells exhibited increased mRNA and protein levels of CREBBP compared to the non-cancer THLE-2 cells (Fig. 2F-G). Notably, upon PSC (25 μM) treatment, the mRNA expression of CREBBP, both in HCC cells and non-cancer THLE-2 cells, was not significantly changed (Fig. 2H). Nevertheless, the protein level of CREBBP was substantially decreased in these cell lines (Fig. 2I). Exact p-values and the mean and SD values are presented in Supplemental Materials.
Overexpression of CREBBP reverses the biological functions of PSC on HCC cells
To investigate whether PSC suppresses malignant behaviors of HCC cells by affecting CREBBP levels, we established three groups of HCC cells with different treatment procedures: PSC + OE-NC, PSC + CREBBP-OE, and DMSO + CREBBP-OE. In the presence of PSC treatment, the lentiviral vector-encapsulated CREBBP-OE significantly elevated CREBBP mRNA expression in Hep 3B and Huh7 cells, while no major difference was found in CREBBP mRNA expression between the PSC + CREBBP-OE and DMSO-CREBBP-OE groups (Fig. 3A). WB analysis revealed that the CREBBP protein level was elevated in the PSC + CREBBP-OE group compared to the PSC + OE-NC group. Notably, further higher CREBBP protein level was observed in the DMSO + CREBBP-OE group (Fig. 3B). These observations indicate the successful administration of CREBBP-OE and substantiate a suppressive effect of PSC on CREBBP protein level. In the presence of PSC, the restoration of CREBBP level significantly rescued proliferation of Hep 3B and Huh7 cells (Fig. 3C), reduced the cell cycle arrest at S and G2/M phases (Fig. 3D), and reduced PSC-induced cell apoptosis (Fig. 3E). In addition, wound healing and Transwell assays revealed an increase in migration and invasion abilities of Hep 3B and Huh7 cells upon CREBBP restoration (Fig. 3F-G). Furthermore, compared to the PSC + CREBBP-OE group, malignant behaviors of Hep 3B and Huh7 cells, including proliferation (Fig. 3C), cell cycle progression (Fig. 3D), apoptosis resistance (Fig. 3E), migration (Fig. 3F), and invasion (Fig. 3G), were augmented in the DMSO + CREBBP group. These results further validate that ectopic CREBBP overexpression counteracts the suppressive effects of PSC on HCC cells. However, PSC still presents a tumor inhibiting role upon CREBBP overexpression, as compared to DMSO treatment. Exact p-values and the mean and SD values are presented in Supplemental Materials.
Overexpression of CREBBP negates the suppressive effects of PSC on HCC cells. Hep 3B and Huh7 cells were transfected with lentiviral vectors-encapsulated CREBBP-OE or NC-OE, followed by treatment with 25 μM PSC or DMSO. A-B, mRNA (A) and protein (B) levels of CREBBP in cells determined by RT-qPCR and WB analysis, respectively; C, proliferation of cells determined by CCK-8 assay; D, cell cycle distribution of cells determined by flow cytometry; E, apoptosis of cells determined by flow cytometry; F, migration ability of cells analyzed by wound healing assay; G, invasion ability of cells determined by Transwell assay. Three biological replicates were conducted. Differences were compared by the two-way ANOVA (A-G). *p < 0.05 vs. PSC + OE-NC; #p < 0.05 vs. PSC + CREBBP-OE
CREBBP activates GATAD2B transcription
To further delve into the molecular targets of CREBBP, we predicted the top 50 downstream targets of CREBBP using the hTFtarget system (http://bioinfo.life.hust.edu.cn/hTFtarget/#!/). These predicted targets were intersected with genes positively correlated with CREBBP (Pearson’s correlation coefficient > 0.5) predicted using the UALCAN system. This resulted in nine intersected genes (Fig. 4A). Among them, only four genes have not been studied in the field of HCC, and GATAD2B stood out as the one with the highest Pearson’s correlation coefficient (0.73). Data in the UALCAN system suggests a high expression profile of GATAD2B in LIHC (Fig. 4B). Indeed, we observed that GATAD2B exhibited increased mRNA expression in the clinical HCC tissues compared to the normal tissues (Fig. 4C), which presented a significant positive correlation with the CREBBP mRNA expression level in the clinical HCC tissues (Fig. 4D). Similarly, IHC revealed an increase in the positive staining of GATAD2B in the HCC tissues compared to the normal tissues (Fig. 4E). Subsequently, we treated Hep 3B and Huh7 cells with lentiviral vectors-carried CREBBP-KD. This successfully reduced the CREBBP protein levels in both cell lines (Fig. 4F), which resulted in a decrease in the GATAD2B mRNA level (Fig. 4G). Importantly, the CREBBP knockdown significantly reduced the enrichment of GATAD2B promoter fragments in the immunoprecipitants pulled down by the CREBBP antibody (Fig. 4H), and it reduced the luciferase activity of the reporter vector containing the GATAD2B promoter in the dual luciferase reporter gene assay (Fig. 4I). Exact p-values and the mean and SD values are presented in Supplemental Materials.
CREBBP activates GATAD2B transcription. A, intersections of top 50 downstream targets of CREBBP predicted from hTFtarget and the genes positively correlated with CREBBP (Pearson’s correlation coefficient > 0.5) predicted from UALCAN; B, expression profile of GATAD2B in LIHC predicted using the UALCAN system; C, mRNA expression of GATAD2B in clinical HCC samples and the adjacent non-involved tissue samples examined by RT-qPCR; D, a positive correlation between GATAD2B mRNA and CREBBP mRNA expression in clinical HCC tissue samples; E, positive staining of GATAD2B in clinical HCC samples and the adjacent non-involved tissue samples examined by IHC; Hep 3B and Huh7 cells were administered lentiviral vectors-carried CREBBP-KD/KD-NC; F, protein level of CREBBP in cells determined by WB analysis; G, mRNA expression of GATAD2B in cells determined by RT-qPCR; H, binding between CREBBP and GATAD2B promoter fragments determined by ChIP-qPCR assay; I, transcriptional regulation of CREBBP on GATAD2B promoter determined by dual luciferase reporter gene assay. Three biological replicates were conducted. Differences were compared by the paired t test (C, E) or two-way ANOVA (F-I). In panel D, linear regression and correlation analyses were performed. *p < 0.05 vs. Normal liver/KD-NC
Concerning the malignant properties of HCC cells, functional assays demonstrated that the CREBBP knockdown significantly suppressed proliferation (Supplemental Fig S1A), migration (Supplemental Fig S1B), and invasion (Supplemental Fig S1C) of Hep 3B and Huh7 cells while promoting cell apoptosis (Supplemental Fig S1D). Notably, the CREBBP knockdown also promoted cell cycle arrest at S and G2/M phases in both cell lines (Supplemental Fig S1E). These results substantiated a tumor-promoting role of CREBBP in HCC. Exact p-values and the mean and SD values are presented in Supplemental Materials.
Overexpression of GATAD2B negates the suppressive effects of PSC on HCC cells
Interestingly, RT-qPCR unveiled a significant downregulation of GATAD2B mRNA and protein levels in Hep 3B and Huh7 cells following PSC treatment (Fig. 5A-B). This downregulation was also evident in THLE-2 cells (Fig. 5A-B). The important implications might be that PSC blocks CREBBP-mediated transcriptional activation of GATAD2B, thus blocking malignant properties of the HCC cells. Therefore, we treated the HCC cells with lentiviral vectors-encapsulated GATAD2B-OE, followed by treatment with 25 μM PSC or DMSO. Not surprisingly, in the presence of PSC treatment GATAD2B-OE successfully increased the mRNA expression and protein levels of GATAD2B while having no effect on the CREBBP expression (Fig. 5C-D). Concurrently, the GATAD2B levels were higher in the DMSO + GATAD2B-OE group compared to the PSC + GATAD2B-OE group (Fig. 5C-D). Regarding the malignant phenotype of HCC cells, it was found that the GATAD2B overexpression significantly restored proliferation of Hep 3B and Huh7 cells (Fig. 5E), alleviated the cell cycle arrest at S and G2/M phases (Fig. 5F), reduced apoptosis rate of the PSC-treated Hep 3B and Huh7 cells (Fig. 5G), and reinstated migration and invasion capabilities in Hep 3B and Huh7 cells (Fig. 5H-I). These observations reveal that GATAD2B overexpression also negates the inhibitory effects of PSC on HCC cells in vitro. Notably, cells in the PSC + GATAD2B-OE group still exhibited a less malignant phenotype compared to those in the DMSO + GATAD2B-OE group (Fig. 5E-I), confirming the tumor-suppressive role of PSC. Exact p-values and the mean and SD values are presented in Supplemental Materials.
Overexpression of GATAD2B restores malignant properties of HCC cells suppressed by PSC. A-B, mRNA (A) and protein (B) levels of GATAD2B in Hep 3B and Huh7 cells after PSC treatment determined by RT-qPCR and WB analysis, respectively; Hep 3B and Huh7 cells were transfected with lentiviral vectors-encapsulated GATAD2B-OE or NC-OE, followed by treatment with 25 μM PSC or DMSO; C-D, mRNA (C) and protein (D) levels of GATAD2B in cells determined by RT-qPCR and WB analysis, respectively; E, proliferation of cells determined by CCK-8 assay; F, cell cycle distribution of cells determined by flow cytometry; G, apoptosis of cells determined by flow cytometry; H, migration ability of cells analyzed by wound healing assay; I, invasion ability of cells determined by Transwell assay. Three biological replicates were conducted. Differences were compared by the two-way ANOVA (A-I). *p < 0.05 vs. DMSO/PSC + OE-NC; #p < 0.05 vs. PSC + GATAD2B-OE; ns, non-significant
Discussion
In an effort to investigate the roles of PSC in HCC, this study discerns that PSC impedes various malignant properties exhibited by two HCC cell lines in vitro. A deeper exploration into the functional mechanisms discloses that the inhibition of HCC progression by PSC is attributed to its suppression of CREBBP-mediated GATAD2B transcription.
STLs have established roles in cytotoxicity and antitumor functions due to the presence of the α-methylene-γ-lactone group (Laurella et al. 2022; Sulsen et al. 2019). For instance, Alantolactone, an allergic STL extracted from Inula helenium L roots, induces reactive oxygen species, activates the apoptotic pathway, and promotes apoptosis in various cancer cell types, including breast cancer (Jiang et al. 2022), lung cancer (Liu et al. 2019), gastric cancer (He et al. 2019), colorectal cancer (Ding et al. 2016), and liver cancer (Lei et al. 2012). Similarly, other types of STLs, such as vernolide-A and vernodaline (Nguyen et al. 2020), parthenolide (Sztiller-Sikorska and Czyz 2020), artemisinin (Chen et al. 2020; Efferth 2017), have been demonstrated to possess potent anti-tumor functions by blocking various oncogenic signaling pathways. In this investigation, PSC took center stage due to its distinction as the most potent and least toxic among various STLs displaying anti-tumor effects on the murine lymphoma cell line BW5147. (Martino et al. 2015). Specifically, it was observed to induce cell cycle arrest in BW5147 cells at the S phase (Martino et al. 2015). Additionally, prevailing evidence characterizes PSC as a G2 cell cycle checkpoint inhibitor, impeding cells in mitosis and inducing the formation of aberrant microtubule spindles (Sturgeon et al. 2005). Our study aligns with these findings, revealing that PSC treatment induced cell cycle arrest at S and G2/M phases in two HCC cell lines (Hep 3B and Huh7). Notably, it significantly curtailed cell proliferation, migration, and invasion, concurrently inducing cell apoptosis. This comprehensive evidence underscores the robust tumor-suppressive effects of PSC on HCC cell progression in vitro.
Regarding the downstream molecular targets of PSC, a comprehensive bioinformatics analysis identified eight TFs or co-factors, including HMOX1, LRRK2, SRC, CREBBP, PIN1, NR1H3, NR1H2, and JAK2, that displayed altered expression patterns in HCC and were targeted by PSC. Among them, HMOX1, linked to ferroptosis promotion in HCC cells (Zheng et al. 2023), while SRC (Wang et al. 2020), PIN1 (Cheng et al. 2016), and JAK2 (Xiao et al. 2022) have been associated with metastasis of HCC cells. NR1H3 and NR1H2, liver X receptors, are correlated with hepatitis B virus-associated hepatic steatosis (Zhou et al. 2022), but they also exhibit tumor-suppressive effects on HCC (He et al. 2020; Hu et al. 2014). LRRK2 and CREBBP were the remaining molecules with unclear roles in HCC. CREBBP, selected for further investigation due to its high expression in HCC based on bioinformatics data, demonstrated elevated levels in HCC cell lines, which were suppressed by PSC treatment. As a transcriptional co-factor, CREBBP has been shown to suppress proliferation and chemo-resistance in ovarian cancer cells through the mediation of the PERK/ATF4/STC2 signaling pathway (Hu et al. 2021). Conversely, it has been implicated in promoting proliferation, angiogenesis, migration, and invasion in osteosarcoma cells (Feng et al. 2022). Silencing CREBBP has also been linked to G2/M phase cell cycle arrest in pancreatic cancer cells (Ono et al. 2021). Crucially, our study found that the upregulation of CREBBP restored the malignant phenotypes of HCC cells that were blocked by PSC treatment. This suggests that the inhibition of CREBBP is implicated in the tumor-suppressive effects mediated by PSC.
Bioinformatics predictions were additionally conducted to identify downstream targets of CREBBP. Among the candidates, GATAD2B emerged with the highest correlation coefficient with CREBBP, excluding genes with reported functions in HCC, such as SPAG9 (Lou et al. 2016), YY1 (Gao et al. 2023), NFAT5 (Qin et al. 2017), MDC1 (Hong et al. 2021), and NFIC (Zhang et al. 2019). Subsequently, our study confirmed that CREBBP bound to the GATAD2B promoter, initiating its transcription. GATAD2B has been recognized as an oncogenic factor in KRAS-driven lung cancer, implicated in tumor metastasis (Grzeskowiak et al. 2018). Additionally, the LRRC42-GATAD2B interaction has been suggested to play a pivotal role in lung carcinogenesis (Fujitomo et al. 2014). However, the role of GATAD2B in other cancer types is less explored. Our findings revealed increased GATAD2B expression in HCC samples, which was subsequently decreased by PSC treatment. Notably, the restoration of proliferation, migration, invasion, and cell cycle progression, coupled with a reduction in cell apoptosis upon GATAD2B overexpression, suggested its potential role as a downstream effector in CREBBP-mediated HCC progression.
In summary, our study establishes that PSC exerts robust tumor-suppressive effects on HCC through the inhibition of CREBBP-mediated GATAD2B transcription. These findings hold promise for the potential clinical application of PSC in managing HCC. However, it is essential to acknowledge certain limitations. Due to time and financial constraints, as well as ethical considerations, we did not conduct animal experiments to validate the biological functions of PSC, CREBBP, and GATAD2B in HCC tumorigenesis or metastasis in vivo. Additionally, we found that PSC treatment reduced CREBBP protein level without significantly altering its mRNA expression. This indicates that PSC possibly modulate CREBBP abundance through a post-transcriptional (Peng et al. 2023) or ubiquitin-proteasome or lysosomal pathways (Yang et al. 2023). To comprehensively address this issue, further experiments such as protein degradation assays or molecular docking approaches are necessary to gain a deeper understanding. We acknowledge the importance of addressing these gaps in future research endeavors.
Data availability
No datasets were generated or analysed during the current study.
References
Attar N, Kurdistani SK (2017) Exploitation of EP300 and CREBBP lysine acetyltransferases by cancer cold spring. Harb Perspect Med 7. https://doi.org/10.1101/cshperspect.a026534
Blagojevic P, Radulovic N, Palic R, Stojanovic G (2006) Chemical composition of the essential oils of Serbian wild-growing Artemisia absinthium and Artemisia vulgaris. J Agric Food Chem 54:4780–4789. https://doi.org/10.1021/jf060123o
Bowlt Blacklock K et al (2018) Identification of molecular genetic contributants to canine cutaneous mast cell tumour metastasis by global gene expression analysis. PLoS ONE 13:e0208026. https://doi.org/10.1371/journal.pone.0208026
Chen SR, Qiu HC, Hu Y, Wang Y, Wang YT (2016) Herbal medicine offered as an initiative therapeutic option for the management of hepatocellular. Carcinoma Phytother Res 30:863–877. https://doi.org/10.1002/ptr.5594
Chen GQ, Benthani FA, Wu J, Liang D, Bian ZX, Jiang X (2020) Artemisinin compounds sensitize cancer cells to ferroptosis by regulating iron homeostasis. Cell Death Differ 27:242–254. https://doi.org/10.1038/s41418-019-0352-3
Cheng CW, Leong KW, Tse E (2016) Understanding the role of PIN1 in hepatocellular carcinoma. World J Gastroenterol 22:9921–9932. https://doi.org/10.3748/wjg.v22.i45.9921
Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D (2021) Hepatocellular carcinoma (HCC): epidemiology, etiology and molecular classification. Adv Cancer Res 149:1–61. https://doi.org/10.1016/bs.acr.2020.10.001
Ding Y et al (2016) Induction of ROS overload by alantolactone prompts oxidative DNA damage and apoptosis in colorectal cancer cells. Int J Mol Sci 17:558. https://doi.org/10.3390/ijms17040558
Efferth T (2017) From ancient herb to modern drug: Artemisia annua and artemisinin for cancer therapy. Semin Cancer Biol 46:65–83. https://doi.org/10.1016/j.semcancer.2017.02.009
Ekiert H, Pajor J, Klin P, Rzepiela A, Slesak H, Szopa A (2020) Significance of Artemisia Vulgaris L. (Common Mugwort) in the history of medicine and its possible contemporary applications substantiated by phytochemical and pharmacological studies molecules. 25. https://doi.org/10.3390/molecules25194415
Feng D, Li Z, Yang L, Liang H, He H, Liu L, Zhang W (2022) BMSC-EV-derived lncRNA NORAD facilitates migration, invasion, and angiogenesis in osteosarcoma cells by regulating CREBBP via delivery of miR-877-3. Oxid Med Cell Longev 2022:8825784. https://doi.org/10.1155/2022/8825784
Forner A, Reig M, Bruix J (2018) Hepatocellular carcinoma. Lancet 391:1301–1314. https://doi.org/10.1016/S0140-6736(18)30010-2
Fujitomo T, Daigo Y, Matsuda K, Ueda K, Nakamura Y (2014) Identification of a nuclear protein, LRRC42, involved in lung carcinogenesis. Int J Oncol 45:147–156. https://doi.org/10.3892/ijo.2014.2418
Gao H, Fan H, Xie H (2023) The regulatory effect of the YY1/miR–HCC2/BAMBI axis on the stemness of liver cancer cells. Int J Oncol 62. https://doi.org/10.3892/ijo.2023.5507
Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, Kamel IR, Ghasebeh MA, Pawlik TM (2016) Hepatocellular carcinoma: from diagnosis to treatment. Surg Oncol 25:74–85. https://doi.org/10.1016/j.suronc.2016.03.002
Grzeskowiak CL et al (2018) In vivo screening identifies GATAD2B as a metastasis driver in KRAS-driven lung cancer. Nat Commun 9:2732. https://doi.org/10.1038/s41467-018-04572-3
Hailan WA, Al-Anazi KM, Farah MA, Ali MA, Al-Kawmani AA, Abou-Tarboush FM (2022) Reactive oxygen species-mediated cytotoxicity in liver carcinoma cells induced by silver nanoparticles biosynthesized using schinus molle extract. Nanomaterials (Basel) 12. https://doi.org/10.3390/nano12010161
He Y, Cao X, Kong Y, Wang S, Xia Y, Bi R, Liu J (2019) Apoptosis-promoting and migration-suppressing effect of alantolactone on gastric cancer cell lines BGC-823 and SGC-7901 via regulating p38MAPK and NF-kappaB pathways. Hum Exp Toxicol 38:1132–1144. https://doi.org/10.1177/0960327119855128
He J et al (2020) Liver X receptor inhibits the growth of hepatocellular carcinoma cells via regulating HULC/miR-134-5p/FOXM1 axis. Cell Signal 74:109720. https://doi.org/10.1016/j.cellsig.2020.109720
Hong H et al (2021) Falcarindiol enhances cisplatin chemosensitivity of hepatocellular carcinoma via down-regulating the STAT3-modulated PTTG1. Pathw Front Pharmacol 12:656697. https://doi.org/10.3389/fphar.2021.656697
Hu C et al (2014) LXRalpha-mediated downregulation of FOXM1 suppresses the proliferation of hepatocellular. Carcinoma Cells Oncogene 33:2888–2897. https://doi.org/10.1038/onc.2013.250
Hu H et al (2021) CREBBP knockdown suppressed proliferation and promoted chemo-sensitivity via PERK-mediated unfolded protein response in ovarian cancer. J Cancer 12:4595–4603. https://doi.org/10.7150/jca.56135
Jiang Z et al (2019a) The essential oils and Eucalyptol from Artemisia vulgaris L. prevent acetaminophen-induced liver injury by activating Nrf2-Keap1 and enhancing APAP clearance through non-toxic metabolic. Pathw Front Pharmacol 10:782. https://doi.org/10.3389/fphar.2019.00782
Jiang ZC, Chen XJ, Zhou Q, Gong XH, Chen X, Wu WJ (2019b) Downregulated LRRK2 gene expression inhibits proliferation and migration while promoting the apoptosis of thyroid cancer cells by inhibiting activation of the JNK signaling pathway. Int J Oncol 55:21–34. https://doi.org/10.3892/ijo.2019.4816
Jiang Y, Guo K, Wang P, Zhu Y, Huang J, Ruan S (2022) The antitumor properties of atractylenolides: molecular mechanisms and signaling pathways. Biomed Pharmacother 155:113699. https://doi.org/10.1016/j.biopha.2022.113699
Kovacs B et al (2022) Antiproliferative and cytotoxic effects of sesquiterpene lactones isolated from Ambrosia artemisiifolia on human adenocarcinoma and normal cell lines. Pharm Biol 60:1511–1519. https://doi.org/10.1080/13880209.2022.2103574
Laurella LC, Mirakian NT, Garcia MN, Grasso DH, Sulsen VP, Papademetrio DL (2022) Sesquiterpene lactones as promising candidates for cancer therapy: focus on pancreatic cancer. Molecules 27. https://doi.org/10.3390/molecules27113492
Lei JC, Yu JQ, Yin Y, Liu YW, Zou GL (2012) Alantolactone induces activation of apoptosis in human hepatoma cells. Food Chem Toxicol 50:3313–3319. https://doi.org/10.1016/j.fct.2012.06.014
Liu J, Yang Z, Kong Y, He Y, Xu Y, Cao X (2019) Antitumor activity of alantolactone in lung cancer cell lines NCI-H1299 and Anip973. J Food Biochem 43:e12972. https://doi.org/10.1111/jfbc.12972
Lou G et al (2016) Direct targeting sperm-associated antigen 9 by miR-141 influences hepatocellular carcinoma cell growth and metastasis via JNK pathway. J Exp Clin Cancer Res 35:14. https://doi.org/10.1186/s13046-016-0289-z
Luk JM et al (2007) Traditional Chinese herbal medicines for treatment of liver fibrosis and cancer: from laboratory discovery to clinical evaluation. Liver Int 27:879–890. https://doi.org/10.1111/j.1478-3231.2007.01527.x
Martino R, Beer MF, Elso O, Donadel O, Sulsen V, Anesini C (2015) Sesquiterpene lactones from Ambrosia spp. are active against a murine lymphoma cell line by inducing apoptosis and cell cycle arrest. Toxicol Vitro 29:1529–1536. https://doi.org/10.1016/j.tiv.2015.06.011
Nguyen NH, Nguyen MT, Little PJ, Do AT, Tran PT, Vo XN, Do BH (2020) Vernolide-A and Vernodaline: Sesquiterpene Lactones with cytotoxicity against. Cancer J Environ Pathol Toxicol Oncol 39:299–308. https://doi.org/10.1615/JEnvironPatholToxicolOncol.2020034066
Obistioiu D, Cristina RT, Schmerold I, Chizzola R, Stolze K, Nichita I, Chiurciu V (2014) Chemical characterization by GC-MS and in vitro activity against Candida albicans of volatile fractions prepared from Artemisia dracunculus, Artemisia abrotanum, Artemisia absinthium and Artemisia vulgaris. Chem Cent J 8:6. https://doi.org/10.1186/1752-153X-8-6
Ono H et al (2021) C646 inhibits G2/M cell cycle-related proteins and potentiates anti-tumor effects in pancreatic cancer. Sci Rep 11:10078. https://doi.org/10.1038/s41598-021-89530-8
Peng Y et al (2023) Diosgenin inhibits prostate cancer progression by inducing UHRF1 protein degradation. Eur J Pharmacol 942:175522. https://doi.org/10.1016/j.ejphar.2023.175522
Pires JM, Mendes FR, Negri G, Duarte-Almeida JM, Carlini EA (2009) Antinociceptive peripheral effect of Achillea millefolium L. and Artemisia vulgaris L.: both plants known popularly by brand names of analgesic drugs. Phytother Res 23:212–219. https://doi.org/10.1002/ptr.2589
Qin X et al (2017) Upregulation of DARS2 by HBV promotes hepatocarcinogenesis through the miR-30e-5p/MAPK/NFAT5 pathway. J Exp Clin Cancer Res 36:148. https://doi.org/10.1186/s13046-017-0618-x
Sturgeon CM, Craig K, Brown C, Rundle NT, Andersen RJ, Roberge M (2005) Modulation of the G2 cell cycle checkpoint by sesquiterpene lactones psilostachyins a and C isolated from the common ragweed. Ambrosia artemisiifolia Planta Med 71:938–943. https://doi.org/10.1055/s-2005-873109
Sulsen VP et al (2019) Activity of estafietin and analogues on trypanosoma cruzi and leishmania braziliensis molecules. 24. https://doi.org/10.3390/molecules24071209
Sun X, Li J, Li Y, Wang S, Li Q (2020) Apatinib, a novel tyrosine kinase inhibitor, promotes ROS-dependent apoptosis and autophagy via the Nrf2/HO-1 pathway in ovarian cancer cells oxid. Med Cell Longev 2020:3145182. https://doi.org/10.1155/2020/3145182
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries CA. Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Sztiller-Sikorska M, Czyz M (2020) Parthenolide as cooperating agent for anti-cancer treatment of various malignancies. Pharmaceuticals (Basel). 13. https://doi.org/10.3390/ph13080194
Temraz A, El-Tantawy WH (2008) Characterization of antioxidant activity of extract from Artemisia vulgaris. Pak. J Pharm Sci 21:321–326
Thiel G, Rossler OG (2022) TRPM3-induced gene transcription is under epigenetic control. Pharmaceuticals (Basel) 15. https://doi.org/10.3390/ph15070846
Tigno XT, de Guzman F, Flora AM (2000) Phytochemical analysis and hemodynamic actions of Artemisia vulgaris. L Clin Hemorheol Microcirc 23:167–175
Villanueva A (2019) Hepatocellular Carcinoma N. Engl J Med 380:1450–1462. https://doi.org/10.1056/NEJMra1713263
Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A (2022) Hepatocellular carcinoma. Lancet 400:1345–1362. https://doi.org/10.1016/S0140-6736(22)01200-4
Wang T et al (2020) COL4A1 promotes the growth and metastasis of hepatocellular carcinoma cells by activating FAK-Src signaling. J Exp Clin Cancer Res 39:148. https://doi.org/10.1186/s13046-020-01650-7
Wang Y et al (2023) LRRK2 G2019S promotes the development of colon cancer via modulating intestinal inflammation. bioRxiv. https://doi.org/10.1101/2023.06.28.546897
Xiao Y et al (2022) MEX3C-mediated decay of SOCS3 mRNA promotes JAK2/STAT3 signaling to facilitate metastasis in hepatocellular. Carcinoma Cancer Res 82:4191–4205. https://doi.org/10.1158/0008-5472.CAN-22-1203
Yang C et al (2023) Tiliroside targets TBK1 to induce ferroptosis and sensitize hepatocellular carcinoma to. Sorafenib Phytomedicine 111:154668. https://doi.org/10.1016/j.phymed.2023.154668
Zhang L, Wang K, Deng Q, Li W, Zhang X, Liu X (2019) Identification of key hydroxymethylated genes and transcription factors associated with alpha-fetoprotein-negative hepatocellular carcinoma. DNA Cell Biol 38:1346–1356. https://doi.org/10.1089/dna.2019.4689
Zheng C et al (2023) Donafenib and GSK-J4 synergistically induce ferroptosis in liver cancer by upregulating HMOX1 expression. Adv Sci (Weinh) 10:e2206798. https://doi.org/10.1002/advs.202206798
Zhou L et al (2022) Integrated analysis highlights the immunosuppressive role of TREM2(+. Macrophages Hepatocellular Carcinoma Front Immunol 13:848367. https://doi.org/10.3389/fimmu.2022.848367
Acknowledgements
We thanks to the Key research and development project of Shaanxi Province (Approval no. 2021SF-181) for the funding support.
Funding
This study was supported by the Key research and development project of Shaanxi Province (Approval no. 2021SF-181).
Author information
Authors and Affiliations
Contributions
KJ conceived the study and conducted the experiments; NN is responsible for data collection, analyzed and interpreted the data; JH wrote the manuscript and revised the manuscript and important intellectual content. YC performed experiments and collected data; RW analyzed the results and reviewed the manuscript; JM performed experiments and statistical analysis. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The collection and use of human samples were approved by the Ethics Committee of Honghui Hospital, Xi’an Jiaotong University. All procedures were adhered to the guidelines of the Declaration of Helsinki. All patients signed an informed consent form.
Consent to participate
Written informed consent for publication was obtained from all participants.
Consent for publication
All authors read the journals guideline and agreed with consent for publication.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, K., Ning, N., Huang, J. et al. Psilostachyin C reduces malignant properties of hepatocellular carcinoma cells by blocking CREBBP-mediated transcription of GATAD2B. Funct Integr Genomics 24, 75 (2024). https://doi.org/10.1007/s10142-024-01353-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-024-01353-8