Abstract
Heat shock transcription factors (Hsfs) play an essential role as transcriptional regulatory proteins against heat stress by controlling the expression of heat-responsive genes. Common bean is a highly thermosensitive crop, and, therefore, its genome sequence information is segregated, characterized here in terms of heat shock transcription factors and its evolutionary significance. In this study, a complete comprehensive set of 29 non-redundant full-length Hsf genes were identified and characterized from Phaseolus vulgaris L. (PvHsf) genome sequence. Detailed gene information such as chromosomal localization, domain position, motif organization, and exon–intron identification were analyzed. All the 29 PvHsf genes were mapped on 8 out of 11 chromosomes, indicating the gene duplication occurred in the common bean genome. Motif analysis and exon–intron structure were conserved in each group, which showed that the cytoplasmic proteins highly influence the conserved structure of PvHsfs and heat-induced response. The HSF genes were grouped into three classes, i.e., A to C and 14 groups, based on structural features and phylogenetic relationships. Only one pair of paralog sequences suggests that it may be derived from the duplication event during evolution. A comparative genomics study indicated the influence of whole-genome duplication and purifying selection on the common bean genome during development. In silico expression analysis showed the active role of class A and B family during abiotic stress conditions and higher expression in floral organs. The qRT-PCR analysis revealed PvHSFA8 as the master regulator and PvHSFB1A and PvHSFB2A induction during heat exposure in French beans.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Plants are ancient living creatures that have been confronted with various biotic and abiotic stresses during evolution through a regulatory network of inherent adaptation measures. As sessile organisms, plants cannot avoid different biotic and abiotic stresses by changing their positions (Zhu 2016; Duan et al. 2019). Abiotic stress conditions such as constantly increasing high temperature, low temperature, drought, salinity, high and low light intensity, exposure to ozone (Oȝ), and exposure of the cell to toxins (ethanol, arsenic, trace metals, UV light) have considerable impact in plant yield and reduce more than 50% productivity in many species (Cramer et al. 2011). Heat stress is significant stress which causes the protein denaturation affecting the cytoskeleton arrangement of the cell and induces disruption in the metabolism, photosynthesis activity, and cell membrane structure (Martin et al. 1992). The stressful environmental condition causes morpho-anatomical, physicochemical, and biochemical changes at the cellular level, affecting plant biomass production and economic yield worldwide (Fahad et al. 2017). Therefore, the primary objective of agronomic research is to increase crop productivity under biotic and abiotic stresses.
All living organisms possess a common heat shock response mechanism that initiates the synthesis of “heat shock proteins” known as “stress-induced proteins” required for the development of short-term stress (Li et al. 2019). Among all 64 identified transcription factor groups in plants, the “Heat shock transcription factor family is ubiquitous and responsible for the expression of the heat shock proteins (Nover et al. 2001). Stress-induced gene expression leads to the rapid accumulation of heat shock proteins which belong to 11 conserved multi-protein families. As the major HSPs are highly homologous among eukaryotes, the evolutionary conservation of the heat shock response is evidence that justifies that the production of HSPs is a fundamental and imperative process (Vierling 1991).
A characteristic feature of transcription factors is that they contain one or more DNA binding domain (DBD) which binds to the promoter region for abiotic responsive gene expression (Scharf et al. 2012). In general, HSF proteins have a core structure consisting of an N-terminal DBD consisting of a four-stranded antiparallel β-sheet and three helical bundles. It is characterized by a central helix-turn-helix (HTH) motif, which recognizes and binds to the conserved cis-acting elements known as “heat shock element” (HSEs; 5-AGAAnnTTCT-3). An adjacent domain commonly known as the oligomerization domain with a heptad hydrophobic repeat (HR-A/B) required for HSF trimerization is connected to the DBD region by a linker group of amino acids. Arginine (R) & lysine (K)-rich nuclear localization signal (NLS) region and leucine (L)-rich nuclear export signal (NES) regions are responsible for the dynamic distribution of HSF between the nucleus and cytoplasm (Kotak et al. 2007; Dirk and Kutay 1999).
In most cases, it has been seen that class A and C HSFs have monopartite and bipartite NLSs adjacent to the oligomerization domain. The AHA (aromatic large hydrophobic amino acid residues) motif or the activation domain is the least defined part of HSF, consisting of activators and repressor elements. It is a distinct region of class A HSFs as class B and C lacks the AHA motif. It is required for the transcription of HSPs, by binding to some essential transcription protein complexes (Nover et al. 2001; Soares-Cavalcanti et al. 2012).
In plants, the ubiquity of heat shock proteins was first observed in Glycine max and tobacco, using the cell culture technique, which revealed the synthesis of specific proteins under heat conditions. To our knowledge, HSFs have been investigated in many plants, including Lycopersicon peruvianum (tomato), Arabidopsis thaliana, Zea mays (maize), Glycine max (soybean), Medicago truncatula (barrel clover), Lotus japonicus, Triticum aestivum (wheat), Oryza sativa (rice), Vigna radiata (mung bean), and Populus trichocarpa (black cottonwood). Here, well-described 21 Arabidopsis HSF genes were used to identify and characterize the pool of HSF genes present in the Phaseolus vulgaris genome.
Material and methods
Identification of Hsfs
Amino acid sequences corresponding to the 15 AtHsfs (A. thaliana) genes were retrieved from TAIR (The Arabidopsis Information Resource; http://www.Arabidopsis.org/). These protein sequences were used as queries against the database of Phytozome (http://www.phytozome.net) in a standalone blast search with e values 0.001. Self-BLAST was performed for sequences to remove redundancy. Based on the HMM profile, all the protein sequences thus obtained were confirmed with HMMER3.0 (http://hmmer.org/). Results from BLAST and HMMER hits were matched and parsed manually.
Characterization of Hsf genes, chromosomal localization, and protein domains
The ProtParam tool (www.expacy.org/tools/protparam.html) was used to predict physicochemical properties. This tool helps compute the molecular weight, theoretical pI, amino acid composition, atomic composition, extinction coefficient, estimated half-life, instability index, aliphatic index, and grand average of hydropathicity (GRAVY). The subcellular localization is predicted by CELLO for eukaryotic sequences. PROSITE was used for documentation and information about the protein domains, functions, families, patterns, and profiles (http://prosite.expacy.org/). MacDraw is a Raster-based and vector graphic drawing application used for drawing chromosomal maps. A Gene Structure Display Server (GSDS) illustrated the gene structure.
Conserved domains, motif prediction, and promoter region identification
PSort and NetNES results were used for predicting the NLS (nuclear localization signal) and NES (nuclear export signal) regions, respectively. Using the amino acid sequences as a query, the servers predicted possible regions in the sequence. DBD region and coiled-coil regions were identified from the COIL server and HMMER server results. Using the MEME suite server, the conserved motifs were predicted in 29 putative identified proteins. The advance options menu decided the minimum and maximum width for motifs at 10 and 50. The highly conserved motifs are identified in species using MAST algorithms (http://meme.nbcr.net/).
The PLANTCARE database analyzed class A HSF genes in GTF/GFF3 format for finding the cis-acting elements. PLACE database was used to understand the functionality of the promoter regions.
Multiple sequence alignment and phylogeny analysis
Phylogeny analysis has always been an essential factor for understanding the evolutionary relationship among species. Total 100 HSFs from A. thaliana (21), G. max (26), V. radiate (24), and P. Vulgaris (29) were aligned using the Clustal Omega program. The alignment file was used for constructing the phylogeny tree using the Neighbor-Joining algorithm with 1000 bootstrap replications and wrapped in MEGA 7.
Comparative analysis
Ortholog and paralog genes are two different homologous genes derived from the ancestral DNA by speciation and duplication events, respectively. Paralog sequences were identified by performing a similarity search among the PvHSF proteins. GALAXY server was used for predicting the ortholog genes present in other species. By completing the BLAST program among the HSF proteins retrieved from different species, ortholog sequences were found in Vigna radiata, Glycine max, and Arabidopsis thaliana. CoGe database was used for analyzing and visualizing the synteny between Phaseolus vulgaris and Arabidopsis thaliana.
Expression analysis
In silico analysis
For expression analysis of identified PvHsfs, the GEO dataset (Accession: GSE 123,381) was retrieved from NCBI. The EST sequence information was used for tissue-specific expression analysis retrieved from the Phytozome database. Heat map generation and further editing were carried out using the “gplot” package of R studio.
Quantitative real-time PCR (qRT-PCR) analysis
The 10-day-old seedlings of the French bean variety “Arka Arjun” (grown in portrays under homogenous conditions) were treated with heat stress of 48 °C for 0 min (control), 15 min, 30 min, and 1 h in three replicates. Total RNA was isolated from leaf tissues of all four treatments using an RNA extraction kit (RNeasy Mini Kit, QIAGEN) per the manufacturer’s instruction. DNA contamination in the total RNA was removed using DNase I enzyme, New England Biolabs. Furthermore, 1.5 µg of DNase I-treated RNA was used for first-strand cDNA synthesis using qScript cDNA SuperMix kit, QUANTABIO. Real-time quantitative PCR was performed using 1 µl of cDNA in a 10 µl reaction volume with QuantiTect SYBR® Green PCR Kits by QIAGEN in CFX96 Touch Deep Well Real-Time PCR Detection System. Pvhsf gene-specific primers were designed (Supplementary Table 1) using IDT Primer Quest Tool and outsourced for synthesis. Two housekeeping genes (Actin 11 and Skip 16) were used as an internal control to normalize the expression level of the target gene. Each treatment was replicated thrice independently. The protocol for the thermal cycler was an initial incubation at 95 °C for 15 min, followed by 40 cycles at 94 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. The relative quantification of PvHsfs transcription levels was calculated using the delta-delta ct method described previously by Livak and Schmittgen (2001).
Results
Identification and characterization of PvHsfs
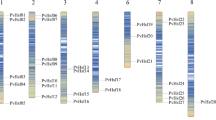
The Hidden Markov Model (HMM) search and Standalone BLAST+ (version 2.2.23) software packages were used to search for members of Hsfs in common bean. Sixty-five Hsf loci were reported from standalone BLAST against database Phytozome based on e value and identity percentage and score with significant alignments. Further, with threshold e value and removal of incomplete, unanchored, and redundant sequences, 29 PvHsf non-redundant candidate genes were identified, similar to the results of HMM search and characterized for biochemical properties and subcellular localization (Table 1). All the 29 PvHsf genes were nomenclature as unified orthologous concerning Arabidopsis (AtHsf) as adopted for abiotic and biotic stress tolerance mechanism accounted genes (CDPKs: Wankhede et al. 2017, MLOs: Kumari et al. 2017, Kumari Meenu and Verma Pooja 2019) and mapped on 11 Phaseolus vulgaris chromosomes (Fig. 1). PvHsf genes were widely distributed in every chromosome except chromosomes V, X, and XI. Chromosomes I and II contain the maximum number of genes, i.e., 6 (20.68%) and 7 (24.13%), respectively. Five Hsfs were detected on chromosome VII and four on chromosome III apart from single pair on chromosome IV, VI, and IX and chromosome VIII occupied only one Hsf gene.
The key domain is characterized by PROSITE results, which provide information about the HSF domain. The DBD domain is located towards the N-terminal of the sequence, followed by the oligomerization domain. PSort resulted in the absence of the NLS region in class B, including PvHSFA8 and PvHSFA3. However, the NES region is present in all the classes except for a few members. Through comparison analysis, conserved AHA motif was observed towards the C-terminal only in class A HSFs. Using COILS and HMMER results identified the coiled-coil region, a characteristic of the HR-A/B region (Table 2). The MEME suite server predicted conserved motifs of PvHsfs proteins. Thirty corresponding consensus motifs were predicted (Fig. 2). The members of class A contained the most conserved motifs, with the largest number of 11 detected in PvHsfA1A and PvHsfA1D. Class B members possessed 3–7 motifs, while four motifs aligned in members of class C family, i.e., PvHsfC1. Motifs 1, 2, and 4 represent the Hsf DBD domains found in all 29 PvHsfs. Motif 3 represents the HR-A/B domain from the class A and C HsFs. All class B Hsf proteins exhibited the coiled-coil region of the HR-A/B domain by motif 5. Motif 6 represents the HR-A/B domain from class A Hsfs. Motif 7 conserved for NLS domains in class A, while motif 8 conserved for NLS of B class families. Furthermore, motifs 10, 11, 14, 16, 17, 18, 23, 26, 28, and 30 represented NES domains with (L) leucine-rich. AHA motifs identified by 9, 15, 16, 20, 21, 22, 24, and 29 with W (tryptophan) rich domain.
Gene structure and phylogeny analysis
The structure of the Hsf gene with one intron was found to be mostly conserved throughout subfamilies among 72.41% of PvHSF members (Fig. 3). However, the length and location of the intron vary among the Hsfs. Noticeably, two introns were observed among 24.13% of Hsfs genes, all belonging to class A subfamilies (PvHSFA1B, PvHSFA2A, PvHSFA4C, PvHSFA7A, PvHSFA8, PvHSFA9A) except PvHSFB2C of class B. PvHSFA2B is comprised of 3 introns with a comparatively smaller length.
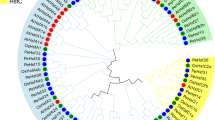
The phylogenetic tree was constructed using MEGA 7 software with the neighbor-Joining method using multiple sequence alignment of 2 Leguminosae members (Glycine max; GmHsfs and Vigna radiata; VrHsfs), 21 AtHSF, and 29 PvHSF genes which categorized the 29 PvHSFs into three classes (A, B, and C) and 14 groups (Fig. 4). Class A was the largest and contained 17 PvHSF, again divided into nine groups. Class B had 11 members, and class C was the smallest, consisting of only one gene.
Phylogeny analysis of Hsf members from Phaseolus vulgaris L. (PvHsfs), Arabidopsis thaliana (AtHsfs), Vigna radiata (VrHsfs), and Glycine max L. (GmHsfs). A different class of families denoted as follows: class A, yellow; class B, red; and class c, blue. Amino acid sequences were aligned using ClustalW with default alignment parameters. The neighbor-joining tree was prepared with 1000 bootstraps
Regulatory cis-acting elements
Sixty-four different promoter regions are responsible for various expressions identified among members (Supplementary Table 2). The PvHSFA5.2 contented maximum number of promoter regions, while PvHSFA4C identified for least. CAAT-box and MYC promoter region is present in all the class A Hsfs gene, while the MYB promoter region is present in all the members except the PvHSFA4 sub-group. All members carried the ABRE (ABA-responsive element) promoter region except PvHSFA8 and A6 group. DRE region was only seen in PvHSFA4A, and LTRE was observed in PvHSFA5.2 and PvHSFA9A. P-box, W-box, box S, G-box, and I-box were observed in members of class A. Light-responsive element, 3-af1 binding site was only monitored in PvHSFA1B (Fig. 5).
Comparative genomics
For synteny analysis, 11 HSF genes from 4 groups (A1, A2, A6, B2) were compared with their orthologs sequences present in Arabidopsis Thaliana. The synteny analysis (Fig. 6) revealed that PvHSFA2B, PvHSFA6A, and PvHSFB2D are in synteny with their orthologs through microsynteny that can be observed in maximum sequences. However, perfect synteny was observed in PvHSFB2D. For PvHSFA2A and A6B, the synteny was observed between 100–110 kb and 85–185 kb, respectively. The least synteny was observed in PvHSFA1B/E, in which the regions from AtHSFA1E (70–130 kb) were distributed over a 110 kb region (20–190 kb). In PvHSFB2C, the widely distributed genes present in AtHSFB2B were conserved from 85 to 100 kb. The comparative analysis found 14 orthologs present in Arabidopsis Thaliana, 22 in mung bean, and 23 in soybean (Table 3). Only one pair of paralog sequences were found in the common bean HSF family.
In silico expression analysis for abiotic stress
The expression data showed that 19 Hsfs out of 29 PvHsf genes were upregulated under drought stress and the highest expression of PvHSFB2A, however, was downregulated indicating its master role under abiotic stress (Fig. 7a). The ubiquitous expression of PvHSFB2A throughout plant organs from the ESTs dataset again supports its crucial importance in the gene regulatory network of common bean (Fig. 7b). Additionally, the relative expression of PvHSFA6B and PvHSFB2D were identified as having no or almost negligible expression in plant tissues. Based on this analysis, three genes (PvHSFA8, PvHSFB1A, and PvHSFB2A were selected for qRT-PCR expression analysis.
qRT-PCR analysis of PvHSF genes in response to heat stress
This study selected three PvHSF genes from the HSF family main classes based on in silico expression analysis for abiotic stress (EST sequences from Phytozome database). These selected genes were examined for their function under temporal progressive heat stress conditions (48 °C). The genes included in the study were from classes A, B1A, and B2A. The qRT-PCR was carried out using 10-day-old seedlings of French bean exposed to heat treatment at 0 min, 15 min, 30 min, and 1 h.
The qRT-PCR analysis provided expression patterns for selected PvHSFs under progressive heat stress conditions. The experiment data showed that transcript levels of PvHSFA8 were sequentially upregulated from control to 1 h. A contrasting pattern was found in PvHSFB1A, where the initial expression level was increased on exposure of 15 min, while induction in the transcript level was weakened as heat stress progressed. However, the strong induction of PvHSFB2A was observed only after exposure for 1 h and remained idled in other treatments (Fig. 8).
Discussion
Identification and characterization of Hsf genes in common bean
The Hsf genes are ubiquitous, and their numbers vary in organisms. However, the numbers of Hsfs in plants outnumber other microorganisms. Although from different studies on plants, it often counts from 16 to 82 (16, strawberry; 82, wheat). Cytoplasmic occurrence of PvHsfs is similar to other Hsfs like GmHsfs and LpHsfs (Chung et al. 2013). The 473 Mb of 587 Mb common bean genome have been sequenced and assembled (Schmutz et al. 2014). This crop is diploid (2n = 2x = 11), much smaller than the soybean genome (1115 Mb), and significantly higher than Arabidopsis (125 Mb). Subsequently, the number of protein-coding genes greatly varies among Arabidopsis (25 kb), soybean (46.43 kb), and common bean (31.64 kb). As mentioned above, the number of HSFs is lying very close from 21 to 26 (21, Arabidopsis; 26, soybean) likely to cereal crop rice 25 (genome size: 389 Mb). Howsoever, 29 PvHsfs reported in this study (Supplementary Table 1) are adjacent to other previously reported Leguminosae members, i.e., 24 Hsfs in mung bean (Li et al. 2019), 20 Hsfs in Populous trichofera, and 16 Hsfs in Medicago truncatula (Wang et al. 2012). These results indicate that difference in copies of Hsfs among these plant species is independent of genome size or protein-coding genes and conserved in plant species of the family with moderate evolutionary events like whole-genome duplication (WGD) followed by gene loss and purifying selection (Lin et al. 2014, Chalhoub et al. 2014). Considering the total chromosome number, it could be referred that the common bean has a moderate number of HSFs, being a member of the legume family.
Gene features such as exon–intron distribution, gene length, and GC content have acute impacts during evolutionary events like WGD. Various studies also indicated that introns are responsible for gene expression and evolution (Rose 2008). Similar to this, only one intron gene feature was also reported from mung beans (Li et al. 2019) and most of the Hsf members of the Rosaceae family (Qiao et al. 2015). However, introns ranged from zero to two in Brassica oleracea (Lohani et al. 2019), and zero to five in Brassica napus (Zhu et al. 2017), though at least one member of class C in Brassica napus and Brassica oleracea lacks intron. These results indicate that intron losses escaped during expansion and divergence of Leguminosae Hsfs. Consequently, at least one intron and the absence of an intron lacking are characteristics of the Leguminosae Hsf gene family.
Invariably, all the plant HSFs have three classes in the HSF family. The common bean has a comparatively higher number of members in each family class except class C as17 PvHsfs concerning 13 VrHsfs, while GmHsfs consists of 19 members in this class. Like Populus and Medicago, other legumes consisted of 15 and 10 members in class A, respectively (Nover et al. 2001, Chung et al. 2013). However, 11 members of class B PvHSFs are relatively conserved among the family, ranging from 6 to 12 in the above-discussed legume species. Complete conservation of class C PvHsfs with only one gene is observed likely to Arabidopsis and legumes. This report presents considerable evidence of tightly conserved class C and moderate conservation of class B members; however, evolutionary events are most frequently observed among class A members. The phylogeny tree and distance matrix indicated that the PvHsfs gene family is more closely related to the VrHsfs to GmHsfs.
It considered the importance of class A Hsf subfamilies with conserved features, which were observed to play an essential role during heat stress. Therefore, PvHSF members from class A only analyzed through the PLANTCARE database to understand the cis-acting elements (Fig. 5). CAAT-box and MYC were dominant elements present in all members of class A and an unknown promoter region. MYB, STRE, LTRE, ABRE, DRE, and MYC were some abiotic stress cis-acting elements found in class A PvHSFs. The regions mentioned above were reported to be necessary for heat shock response in plants. For instance, STRE was the first discovered cis-acting element to be responsive to stress and provided the binding site for HSFA1A in Arabidopsis Thaliana, and deletion of STRE (stress responsive elements) highly affects the promoter activity under stress conditions (Guo et al. 2015, Haralampidis et al. 2002). In mung beans, all the genes were reported to have multiple DREs (drought-responsive elements) and ABREs. PvHSFA4A contained DRE1 responsible for dehydration response. DRE was combined with DRE-binding proteins (DREB 2A) involved in transcriptional stress regulation in plants such as drought, salt, and low-temperature reactions (Ahuja et al. 2010, Ohama et al. 2017). LTR was observed in PvHSFA5.2 and PvHSFA9A, illustrating the low-temperature response provided under stress conditions (Maestrini et al. 2009). The absence of LTRE in maximum members suggests a lower response to low temperature than other plants. HSFa1a was reported to be the master regulator for thermotolerance in tomatoes (Mishra et al. 2002). In Arabidopsis, HSFA1a/b are responsible for the induction of class B genes under heat stress conditions (Lohmann et al. 2004, Von et al. 2007). ABA-responsive elements are also responsive to ABA and GA responses combined with ABRE-binding proteins. In PvHSFA1A, many ABA-responsive features like MYC and MYB were observed. Besides this, several members have observed W box and box S responsible for wound responsive elements. Therefore, cis-element analysis of identified PvHsf members presents evidence of these genes for differential gene regulation during abiotic stress, mostly thermotolerance and involvement in other regulatory functions like light responsive and wound responsive.
The HsfA1a was identified as a master regulator of induced thermotolerance in tomatoes and other members of class A (A2 and A6) and B2 acted as a synergistic co-activator. Therefore, considering their importance during heat stress response (Von et al. 2007), a detailed analysis was conducted for members of these four groups only. This study suggested that, during evolution, PvHSFA2B, PvHSFA6A, and PvHSFB2D had conserved their gene structure, indicating that the diversification took place due to speciation or duplication event. However, in PvHSFB2C, the syntenic genes were highly conserved in the 15 kb region. Maximum members showed microsynteny which can be correlated to whole-genome duplication, gene loss, or mutation. PvHSFB2D was observed as the most conserved sequence among the queries. The number of paralog sequences was reported as nine pairs in rice and maize, 12 pairs in apple, and two pairs in pepper (Guo et al. 2015, Verde et al. 2013). Therefore, only one pair (PvHSFA9A/PvHSFA9B) of paralog sequences indicated less ancestral genome duplication during evolution. A high number of orthologs sequences were observed in soybean (23) and mung bean (22), indicating the speciation event among these plants. The low number of ortholog sequences, i.e., 14 of Arabidopsis, indicates the phylogenetic distance occurred during the evolution. It can be inferred that the number of ortholog sequences decreases with increasing phylogenetic distance.
Expressions of PvHsf genes under abiotic stress (drought) conditions and tissue-specific expression were illustrated with the objective of validation of putatively identified genes. The digital expression data (RPKM) from GEO datasets were retrieved and expressed in the heatmap. This analysis shows that both class A and B families are directly involved in abiotic stress signaling pathways. Also, it may be inferred from this dataset that the B4 group (a, b, c, and d) of class B and A1 group (b and d), A3, A5.1, and A8 of class A along with PvHsfC1were upregulated during drought stress with wide variation (Fig. 7a and b). Therefore, to validate the expression of these putatively identified genes, three PvHSFs from A and B classes (PvHSFA8, PvHASFB1A, and PvHSFB2A) were selected for qRT-PCR analysis. All these three genes were induced under heat stress treatment. However, only PvHSFA8 showed a continuous increase in transcription level, which reached its highest on 1 h of heat stress exposure. The PvHSFB2A transcripts remain static and induced to peak level only upon maximum exposure (1 h).
Meanwhile, transcript expression downregulated as heat stress progressed in the case of PvHSFB1A (Fig. 8). Therefore, this study shows that PvHSFA8 is the crucial regulator for thermal stress in the case of French beans. Consequently, the transcript pattern of class B genes (PvHSFB2A and PvHSFB1A) indicated a partial role which includes various pathways for the heat stress reaction. Likely, class A and B family members played a key role during heat stress-driven experiments. For instance, LpHSFA1 was reported to be the master regulator for thermotolerance in tomatoes (Mishra et al. 2002). Also, two soybean Hsfs from class A2 (GmHsf12, GmHsf 28) and three from class B (GmHsf 34, GmHsf 35, and GmHsf 47) were upregulated during stress conditions (Chung et al. 2013). Therefore, class A and B family members’ activation in signaling pathways during abiotic stress is established for PvHsf genes also. Ubiquitous expression of PvHsfB2A in all plant organs and highest even downregulated expression during abiotic stress stipulated its prime importance among all identified PvHsfs. However, PvHsfB2D, B4C and D, PvHSFA6A and B had no expression in 1 to 4 organelles, likely to PbHSFA6C of Chinese white pear nill expression observed during its four developmental stages (Qiao et al. 2015). In common bean, PvHSFC1 has shown considerable indication in many samples and slight upregulation during drought stress showed its functional importance, but the information is still incomplete.
Conclusion
The comprehensive set of 29 full-length heat shock factor genes (PvHsfs) were identified and characterized in Phaseolus vulgaris with additional gene information such as chromosomal localization, exon–intron identification, and putative genes identified for heat stress response. All the genes were mapped onto 8 out of 11 chromosomes, indicating that the gene duplication occurred in the common bean genome. Motif analysis and exon–intron structure were sustained in each group which concludes that the structure of the HSF family is conserved in common bean. It can be inferred that the cytoplasmic proteins highly influence heat shock response in common beans. According to the structural characteristics of the proteins and phylogeny analysis, the HSF genes were grouped into three classes (A, B, and C) and 14 groups. Only one pair of paralog sequences suggests that it may be derived from the duplication event during evolution. From the synteny analysis, it could be referred to that whole-genome duplication and purifying selection have highly influenced the common bean genome. Expression analysis validated class A and B Hsfs involvement during abiotic stress and the critical importance of PvHsfBA8 for comparative higher occurrence under heat stress, while PvHSFB1A and PvHSFB2A induction verified their participation. This comprehensive analysis better understands PvHsf genes, facilitating further functional characterization in common beans.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Ahuja I, de Vos RCH, Bones AM, Hall RD (2010) Plant molecular stress responses face climate change. Trends Plant Sci 15:664–674. https://doi.org/10.1016/j.tplants.2010.08.002
Al-Whaibi MH. Plant heat-shock proteins: a mini review (2011) J King Saud Univ Sci. 23 (2):139–50 https://doi.org/10.1016/j.jksus.2010.06.022
Chalhoub B, Denoeud F, Liu S, Parkin IAP, Tang H, Wang X et al (2014) Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345:950–953
Chung E, Kim KM, Lee JH (2013) Genome-wide analysis and molecular characterization of heat shock transcription factor family in glycine max. J Genet Genomics 40:127–135. https://doi.org/10.1016/j.jgg.2012.12.002
Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol 11:163
Dirk G, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15(1):607–666
Duan S, Liu B, Zhang Y, Li G, Guo X (2019) Genome-wide identification and abiotic stress-responsive pattern of heat shock transcription factor family in Triticum. BMC Genomics 20:257. https://doi.org/10.1186/s12864-019-5617-1
Fahad S, Bajwa AA, Nazir U, Anjum SA, Farooq A, Zohaib A, Sadia S, Nasim W, Adkins S, Saud S, Ihsan MZ, Alharby H, Wu C, Wang D, Huang J (2017) Crop production under drought and heat stress: plant responses and management options. Front Plant Sci 8:1147. https://doi.org/10.3389/fpls.2017.01147
Guo M, Lu JP, Zhai YF, Chai WG, Gong ZH, Lu MH (2015) Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol 15:1–20. https://doi.org/10.1186/s12870-015-0512-7
Haralampidis K, Milioni D, Rigas S, Hatzopoulos P (2002) Combinatorial interaction of cis elements specifies the expression of the Arabidopsis AtHsp90-1 gene. Plant Physiol 129:1138–1149
Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10:310–316
Kumari M, Wankhede DP, Verma M, Verma P (2017) Genome wide identification of calcium dependent protein kinase and related kinase gene families in Solanum melongena L. Indian Journal of Horticulture 74(4):526–532
Meenu Kumari, Pooja Verma (2019) Genome-wide identification and characterization of the mildew resistance locus O (MLO) gene in Solanum melongena and Capsicum annuum. Research Journal of Biotechnology 14(7):94–110
Li S, Wang R, Jin H, Ding Y, Cai C (2019) Molecular characterization and expression profile analysis of heat shock transcription factors in mungbean. Front Genet 9:736. https://doi.org/10.3389/fgene.2018.00736
Lin Y, Cheng Y, Jin J, Jin X, Jiang H, Yan H et al (2014) Genome duplication and gene loss affect the evolution of heat shock transcription factor genes in legumes. PLoS ONE 9(7):e102825. https://doi.org/10.1371/journal.pone.0102825
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25(4):402–408
Lohani N, Golicz AA, Singh MB, Bhalla PL (2019) Genome-wide analysis of the Hsf gene family in Brassica oleracea and a comparative analysis of the Hsf gene family in B. oleracea, B. rapa and B. napus. Funct Integr Genomics 19:515–531
Lohmann C, Eggers-Schumacher G, Wunderlich M, Schöffl F (2004) Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol Genet Genomics 271:11–21
Maestrini P, Cavallini A, Rizzo M, Giordani T, Bernardi R, Durante M et al (2009) Isolation and expression analysis of low temperature-induced genes in white poplar (Populus alba). J Plant Physiol 166:1544–1556. https://doi.org/10.1016/j.jplph.2009.03.014
Martin J, Horwich AL, Hartl FU (1992) Prevention of protein denaturation under heat stress by the chaperonin Hsp60. Science 5084:995–998. https://doi.org/10.1126/science.1359644 (PMID: 1359644)
Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L et al (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16:1555–1567
Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf K-D (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6(3):177–189. https://doi.org/10.1379/1466-1268
Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K (2017) Transcriptional regulatory network of plant heat stress response. Trends Plant Sci 22:53–65. https://doi.org/10.1016/j.tplants.2016.08.015
Qiao X, Li M, Li L, Yin H, Wu J, Zhang S (2015) Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other Rosaceae species. BMC Plant Biol 15:12. https://doi.org/10.1186/s12870-014-0401-5
Rose AB (2008) Intron-mediated regulation of gene expression. Curr Top Microbiol Immunol 326:277–290
Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB, Grimwood J et al (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46:707–713. https://doi.org/10.1038/ng.3008
Scharf KD, Berberich T, Ebersberger I, Nover L (2012) The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim Biophys Acta - Gene Regul Mech 1819:104–119. https://doi.org/10.1016/j.bbagrm.2011.10.002
Soares-Cavalcanti NM, Belarmino LC, Kido EA, Pandolfi V, Marcelino-Guimaraes FC, Rodrigues FA et al (2012) Overall picture of expressed heat shock factors in Glycine max, lotus japonicus and medicago truncatula. Genet Mol Biol 35(SUPPL. 1):247–259
Verde I, Abbott AG, Scalabrin S, Jung S, Shu S, Marroni F et al (2013) The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet 45:487–494
Vierling E (1991) The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 42(1):579–620
Von Koskull-Doring P, Scharf KD, Nover L (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12(10):452–457. https://doi.org/10.1016/j.tplants.2007.08.014
Wang F, Dong Q, Jiang H, Rt-pcr HA (2012) Genome-wide analysis of the heat shock transcription factors in Populus trichocarpa and Medicago truncatula. Mol Biol Rep 39(2):1877–1886. https://doi.org/10.1007/s11033-011-0933-9
Wankhede DP, Meenu K, Tambi R, Aravind J, Rajkumar S (2017) Genome wide identification and characterization of Calcium Dependent Protein Kinase gene family in Cajanus cajan. J Environ Biol 38(1):167–177
Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167:313–324. https://doi.org/10.1016/j.cell.2016.08.029
Zhu X, Huang C, Zhang L, Liu H, Yu J, Hu Z et al (2017) Systematic analysis of Hsf family genes in the Brassica napus genome reveals novel responses to heat, drought and high CO2 stresses. Front Plant Sci 8:1174. https://doi.org/10.3389/fpls.2017.01174
Acknowledgements
Authors acknowledge the Director, ICAR-Indian Institute of Horticultural Research and ICAR-National Rice Research Institute for providing basic facilities.
Author information
Authors and Affiliations
Contributions
The research topic was conceived, and manuscript preparation including analysis was carried out by BM and MK, SKP and PC contributed result interpretation, PN and GCA supported editing and software scripts, and validation analysis was carried out by BD and PS. All authors approved this manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
Not applicable.
Consent for publication
All co-authors gave verbal consent for publication of this manuscript, and all images and data produced are original in this study.
Competing interests
The authors declare no competing interests.
Human and animal ethics
This article does not contain any studies with human participants or animals performed by any authors.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mallick, B., Kumari, M., Pradhan, S.K. et al. Genome-wide analysis and characterization of heat shock transcription factors (Hsfs) in common bean (Phaseolus vulgaris L.). Funct Integr Genomics 22, 743–756 (2022). https://doi.org/10.1007/s10142-022-00875-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-022-00875-3