Abstract
Brassinosteroids are a class of phytohormones that play crucial roles in improving stress tolerance in plants. Many biochemical and physiological changes in response to abiotic stress are related to regulation of gene expression and accumulation of associated proteins. MicroRNAs (miRNAs) are class of small non-coding RNAs that regulate gene expression post-transcriptionally. Roles of these regulatory RNAs in brassinosteroid (BR) signalling have however remained elusive. In this study using high-throughput small RNA sequencing method, we present a comprehensive compilation of BR-induced differentially expressed microRNAs in root and shoots of Arabidopsis thaliana seedlings. We identified 229 known miRNAs belonging to 102 families and 27 novel miRNAs that express in response to exogenous BR treatment. Out of 102 families, miRNAs belonging to known 48 families and out of 27 novel miRNAs, 23 were observed to be differentially expressed in response to BR treatment. Among the conserved miRNAs, all members of miR169 were observed to be downregulated in both shoot and root samples. While, auxin-responsive factors were predicted to be direct targets of some novel miRNAs that are upregulated in shoots and suppressed in roots. The BR-responsive tissue-specific miRNome characterized in this study can be used as a starting point by investigators for functional validation studies that will shed light on the underlying molecular mechanism of BR-mediated stress tolerance at the level of post-transcriptional gene regulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brassinosteroids (BRs) are a class of polyhyroxylated steroidal hormones that function as growth-promoting molecules and occur widely across the plant kingdom. A number of studies have shown that BRs promote cell division and expansion; regulate senescence, male sterility, pollen development and fruit ripening; and modulate plant responses under numerous biotic and abiotic stresses (Sreeramulu et al. 2013; Sharma et al. 2015; Alptekin et al. 2017; Anwar et al. 2018). Studies on interactive roles of BR-responsive elements with transcription factors and signalling components of other phytohormones are also documented (Choudhary et al. 2012; Gruszka 2013; Divi et al. 2015; Yuan et al. 2015). Recently, it has been shown that drought/dehydration stress induces miRNA expression in a tissue-specific manner, and comprehensive data on understanding miRNomes in members of Triticeae and other grasses has been compiled (Budak and Akpinar 2011; Akpinar et al. 2015; Alptekin et al. 2017). Exogenous application of BR is known to increase tolerance in plants towards various abiotic stresses (temperature, water, salt, radiation and heavy metals) by improving their photosynthetic capacity, increasing activities of antioxidant enzymes and affecting other physiological processes (Anuradha and Rao 2001; Yuan et al. 2012; Wei et al. 2015; Li et al. 2016). Majority of these biochemical and physiological changes are a reflection of underlying alterations in transcript abundance and their associated proteins. Hence, an understanding of post-transcriptional gene regulatory processes is crucial for gaining insight into the underlying molecular mechanisms of stress tolerance in plants. In this study, we have focused on establishing a direct link between BR and miRNA induction that will provide insight into microRNA-BR crosstalk in A. thaliana seedlings.
MicroRNA-mediated post-transcriptional regulation of gene expression is crucial for both stress recognition and responses (Mallory and Vaucheret 2006; Ergen and Budak 2009; Budak et al. 2015). A number of studies in Arabidopsis and other plants have demonstrated the importance of miRNA-based stress responses where defects in miRNA metabolism and their biogenesis cause plants to become hypersensitive to abiotic stress-associated hormones such as abscisic acid (Lu and Fedoroff 2000; Kusumanjali et al. 2012; Kumari et al. 2013; Gim et al. 2014; Sirohi et al. 2018). To date, a number of in silico and experimental studies on drought, UV and heat stresses have identified and characterized abiotic stress-associated miRNAs (Budak and Akpinar 2011). However, direct involvement of phytohormones such as BR in modulating miRNA expression at genome-wide levels has not been studied. Though, recently regulation of root development in Populus euphratica by ABA-responsive miRNA processes was reported. This study provided insight into crosstalk between ABA regulation of root growth through miRNA-mediated pathways (Lian et al. 2018). Since mechanism of BR regulation especially through miRNA-mediated pathways is poorly understood so, in this study, we report tissue-specific BR-responsive miRNA population using the next-generation sequencing method.
Small RNA sequencing is a powerful tool for genome-wide identification and quantification of small RNA species in plants. This methodology has opened up tremendous possibilities to explore involvement of small RNAs in response to various treatments in plant species as it helps in identifying large number of small RNAs in short time duration (Song et al. 2010; Ge et al. 2013). Using this powerful technique, large body of data has been generated in numerous plant species (Xu et al. 2010; Wang et al. 2011; Xie et al. 2017).
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana, ecotype Columbia (Col-0), was used as the plant material. For analysing BR dose-dependent effects on seed germination and seedling growth, four different concentrations of 24-epibrassinolide (Sigma) 0 nM, 10 nM, 100 nM and 1 μM were selected on the basis of literature study. Stock solution (10−2 M) of 24-epibrassinolide (Sigma) was prepared in 50% (v/v) ethanol. Seeds were surface sterilized with 1% bleach (sodium hypochlorite) and 0.5% Triton X-100 followed by five to six times washing with sterile water. These were then incubated at 4 °C for 48 h for vernalization. Imbibed seeds were inoculated (20 seeds/plate) on one-half-strength Murshige and Skoog (MS) plates prepared with 1% sucrose and 0.8% agar and supplemented with different concentrations of BR (0 nM, 10 nM, 100 nM and 1 μM). Plates were oriented vertically and placed in climate-controlled conditions (22 °C ± 2 °C temperature, 55% ± 5% relative humidity with 16-h light of intensity 60–90 μmol/m2 s and 8-h dark cycle). Throughout the experiment, petri plates were sealed with gas-permeable tapes to avoid ethylene accumulation. Seed germination was visually scored after 2 days, and percent germination was calculated. Images were captured using stereozoom microscope (Olympus, SZX16). For analysing effects on seedling growth, 5-day-old seedlings propagated on one-half-strength MS plates were transferred to fresh plates supplemented with 0 nM, 10 nM, 100 nM and 1 μM BR. To monitor the effects on primary root elongation, root tips were marked with a black dot immediately after transfer to BR-supplemented plates. Morphological analysis was performed after 7 days of growth on these plates. For biomass calculations, fresh weight of 30 seedlings per treatment was determined. Images of seedlings were captured using Canon D70 DSLR camera. Reproducibility and authenticity of dose-dependent experiments were confirmed by repeating the experiments thrice.
Superoxide dismutase assay
Total protein was extracted from 100 mg tissue using extraction buffer [sodium phosphate buffer (100 mM, pH 7.0), EDTA (0.4 mM), glycerol (10%), Mgcl2 (5 mM), phenazinemetho sulfate (1 mM), dithiothreitol (2 mM) and HEPES (50 mM, pH 7.5)]. The homogenate was transferred to fresh tubes and centrifuged at 10,000 rpm for 20 min at 4 °C. Protein concentration in supernatants obtained from each sample was estimated using Bradford method. Superoxide dismutase (SOD) activity was analysed following protocol by Kakkar et al. (1984). Briefly, reaction mixture containing 1.2 ml of 0.052 M tetrasodium pyrophosphate buffer (pH 8.3), 0.3 ml of 1 mM phenazinemetho sulfate (PMS) and 0.2 ml of 1 mM nitroblue tetrazolium (NBT), 0.2 ml of 1 mM NADH and 1 ml distilled water were mixed. To this, 0.1 ml of protein extract was added and the reaction was allowed to occur at 30 °C for 90 s. Reaction was stopped by adding 1 ml glacial acetic acid. Mixture was vigorously shaken after adding 4 ml of n-butanol and after 10-min incubation was centrifuged at 10,000 rpm for 5 min. The n-butanol layer was taken, and colour intensity of the chromogen in this fraction was measured at 560 nm against n-butanol taken as control (blank). SOD activity was measured and expressed as unit per milligram of protein.
Small RNA library construction, sequencing and data mining
For small RNA analysis, seedlings grown for 7 days on 100-nM BR-supplemented plates were harvested. Roots and shoots were separately sampled and collected as No BR-R and BR-R (for roots) and No BR-S and BR-S (for shoots) and processed for small RNA sequencing. Total RNA was isolated using TRiZol reagent (Invitrogen, USA). Small RNA library was prepared using the Small RNA Sample Preparation Kit (Illumina Technologies) following manufacturer’s instructions. Libraries were sequenced on Illumina HiSeq 4000 platform. Raw reads generated were first subjected to NGS QC Toolkit to remove low-quality reads keeping 70% length cut-off and a Phred score of 20 without adapter filtering. Adapter sequences were then removed from the filtered reads using Cutadapt tool (Illumina Small RNA 3′ Adapter). The adapter-trimmed reads were again subjected to NGS QC Toolkit to obtain a final set of adapter-trimmed high-quality (HQ) reads keeping 80% length cut-off and a Phred score of 30. The data so obtained contained > 99% high-quality reads with good phred score and was further analysed. Unique tags (non-redundant reads), abundance and read length of the HQ reads were then calculated using AWK Script. The sequenced dataset generated in this study has been deposited at Sequence Read Archive (SRA) of National Center for Biotechnology Information (NCBI) under the accession number SRP152755.
Data analysis
Pre-processed and cleaned sequence data was further analysed. Unique tags in the HQ reads were subjected to BLASTN against a curated RNA central database (RNA central database without miRNA, pre-miRNA and miRNA-like molecules). The unmapped tags were annotated using Rfam ID and Rfam class and categorized as different types of small non-coding RNAs like rRNA, tRNA, snoRNA, snRNA, etc. These non-coding RNAs and other classes of RNAs were removed from the sRNA sequences. Remaining unmapped reads were subjected to size filtering using 16–28-nt filter. These size-filtered reads were the putative miRNA candidates and were taken forward for miRNA identification. These reads were subjected to UEA Small RNA Workbench. miRProf analysis was undertaken for identification of known miRNAs. For this, alignments were performed against the miRBase database (Release 21; http://www.mirbase.org; Kozomara and Griffiths-Jones 2011). Conserved/known miRNAs were identified from perfectly matched sequences. For identification of novel miRNA, miRCat analysis was done using Arabidopsis thaliana genome as reference. Conditions for filtering miRCat result and identification of novel miRNAs included (a) number of consecutive mismatches between miRNA and miRNA* not more than 3, (b) loci must contain no more than four non-overlapping sRNAs and (c) hairpin must be at least 50 nt in length. Candidate pre-miRNAs were identified by folding the flanking genome sequences of distinct miRNAs using MIREAP followed by prediction of secondary structures using mFold v3.5 (Zuker 2003). Criteria used for stem-loop hairpin and genome mapping were similar to that mentioned in Meyers et al. (2008).
Intersample and intrasample normalization was performed, and absolute expression levels of miRNAs in each library were calculated. For differential expression analysis, absolute expression levels of miRNAs were compared between two libraries. Differences in expression were calculated using log2 fold change of absolute expression levels in different samples. MicroRNAs showing ≥ 1 fold change were considered as differentially expressed and were carried for further analysis.
Target prediction and functional classification based on GO analysis
Once a final set of miRNAs were obtained, potential targets of differentially expressed miRNAs were identified using psRNATarget server (http://plantgrn.noble.org/psRNATarget; Dai and Zhao 2011). Gene ontology (GO) analysis was performed to classify gene features with respect to molecular functions, cellular components, biological processes and biological pathways.
Validation of BR-responsive miRNAs and target gene expression
For validating expression of BR-responsive miRNAs identified as differentially expressing in NGS study, stem-loop RT-qPCR method was used, while for validating expression of target genes, RT-qPCR method was employed. Total RNA isolated from tissue samples sent for small RNA sequencing were used as templates for expression studies. Experiments were performed using three biological replicates with three technical replicates for each biological sample. Stem-loop RT-qPCR was performed using SYBR Green (Fermentas, USA) as per protocol mentioned in Gasic et al. (2007). Primers were designed according to previously described protocol (Chen et al. 2005). Quantitative variation between different samples was calculated using 2−ΔΔCt method (Livak and Schmittgen 2001). Amplification of 5SrRNA was used as internal control to normalize data. Primer sequences used for this study are listed in Supplementary Table 1.
For RT-qPCR of selected target genes, first-strand complementary DNA (cDNA) was synthesized using 1 μg of total RNA in a 20-μL reaction using the RevertAid First-Strand cDNA synthesis kit (Thermo Scientific) following manufacturer’s protocol. Primers were designed preferentially from the 3′ ends of genes using NetPrimer software (http://www.premierbiosoft.com/netprimer) with default parameters (Supplementary Table 1). Forward and reverse primers at 5 μM were mixed with SYBR Green PCR master mix as per manufacturer’s instructions. Reaction was carried out using the real-time PCR system (Agilent). To normalize variance among the samples, A. thaliana actin gene was used as endogenous control. Transcript levels for candidate genes in different samples were determined using the delta delta cycle threshold method (Livak and Schmittgen 2001). Relative expression values were calculated after normalizing against the maximum expression value.
Results
Dose-dependent effects of 24-epibrassinolide
To determine the optimal concentration of BR for analysing tissue-specific BR-responsive miRNA population, a dose-dependent study to analyse effects of BR on seed germination, seedling growth and activity of the antioxidant enzyme SOD was undertaken. It was observed that exogenous application of 0 nM, 10 nM, 100 nM and 1 μM BR did not affect germination efficiency of A. thaliana seeds (Fig. 1a). However, effects on hypocotyl elongation on 2-day-old seedlings were more prominent (Fig. 1a). As compared to seedlings on 0 nM BR, increased hypocotyl elongation was observed in light-grown seedlings at 10 nM, 100 nM and 1 μM concentration (Fig. 1a). Similar effects on hypocotyl growth have been previously reported in Arabidopsis seedlings grown on 10 nM, 100 nM and 1 μM BR-supplemented MS plates (Gupta et al. 2015). To see morphological effects at later stages of seedling development, 5-day-old seedlings grown on one-half-strength Murashige and Skoog media were transferred to fresh media plates supplemented with different BR concentrations (0 nM, 10 nM, 100 nM and 1 μM). It was observed that seedlings grown on 0 nM developed primary roots with fewer lateral roots. In contrast, seedlings propagated on BR-supplemented media developed longer primary roots that showed prominent curling (Fig. 1b). Interestingly, seedlings propagated on 100 nM BR showed more proliferative root and shoot growth as compared to seedlings grown on 0 nM, 10 nM or 1 μM BR. Determination of fresh weight showed that seedlings propagated on 100 nM BR responded maximally by accumulating highest biomass as compared to seedlings grown in other concentrations. Further, analysis of SOD activity in seedlings also showed maximal activation at 100 nM concentration (Fig. 1c). Hence, on the basis of positive effects of 100 nM 24-epibrassinolide on shoot and root growth and increased activity of SOD, 100 nM BR was selected for tissue-specific BR-responsive miRNA study.
Analysis of seed germination and seedling growth in A. thaliana in response to BR treatment. a Images of 2-day-old seeds germinated on MS media supplemented with different concentrations of BR. Effects on primary root growth and root hair formation were analysed. b Five-day-old seedlings grown on MS media and transferred to fresh media plates supplemented with different concentrations of BR. Black dots were marked at the position showing tips of elongated primary root at the end of 5 days immediately after transferring on BR-supplemented MS plates. Morphological and biochemical analysis was performed on these seedlings after 7 days of growth on BR-supplemented media. c Bar graphs showing changes in fresh weight of seedlings and activity of superoxide dismutase in seedlings. Statistical analysis using one-way ANOVA followed by Dunnett’s t test was performed and expressed as mean ± SD. ***P > 0.001, **P > 0.01, *P > 0.05; ns not significant. Asterisks indicate significant differences in fresh weight biomass between control (0 nM) and BR-treated seedlings
Annotation of sequenced small RNA reads
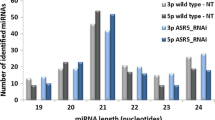
Root and shoot samples from seedlings grown for 7 days on 100 nM BR-supplemented plates were separately harvested, and total RNA was isolated (Fig. 2). To perform small RNA sequencing, four libraries were prepared and sequenced using Illumina HiSeq platform. These included No-BR-R (root samples grown on MS media without BR), No-BR-S (shoot samples without BR), BR-R (root tissues with 100 nM exogenously supplied BR) and BR-S (shoot samples with 100 nM exogenously supplied BR). A total of 24,816,172, 25,902,123, 25,210,454 and 25,691,643 raw reads were obtained from No-BR-R, No-BR-S, BR-R and BR-S libraries, respectively (Table 1). After removal of low-quality reads and adaptor sequences, reads shorter than 15 nt and those with poly-A sequences accounted for 0.12% in No-BR-R, 0.13% in No-BR-S, 0.11% in BR-R and 0.15% in BR-S samples, respectively. Thus, 24 to 25 million clean reads of high-quality (> 99%) were obtained (Table 1). These high-quality reads were then annotated to ascertain the abundance of different categories of small RNAs in the four samples (Fig. 3a). Out of tRNAs, rRNAs and snRNAs, percent distribution of miRNAs in each of the four libraries was observed to be 31% in BR-R, 21% in BR-S, 20% in No-BR-R and 33% in No-BR-S (Fig. 3a). The range of length distribution of small RNA sequences was observed to be between 15 and 51 nucleotides with 23 and 24 nt small RNA sequences dominating the small RNA population (Fig. 3b). Among these, 23 nucleotide small RNAs constituted 15% in all the four libraries, while abundance of 24 nt RNAs was observed to be > 35% in No-BR-S, ~ 25% in No-BR-R, ~ 32% in BR-S and ~ 31.5% in BR-R samples, respectively. Mean frequencies of sRNAs among the libraries were taken as the average number of common sequence frequency which was observed to be 302.68, while library-specific reads varied from 1.37 in BR-R to 19.50 in BR-S (Table 2). While, frequencies of sRNAs in No-BR-R and No-BR-S were observed to be 1.45 and 1.53, respectively.
Identification and differential expression analysis of BR-induced miRNAs
Known miRNAs in the sequenced data were identified using BLASTN function in miRBase database using A. thaliana genome as reference. A total of 229 known miRNAs were identified in all the four libraries, and these belong to 102 known miRNA families. As shown in the Venn diagram, a total of 171 miRNAs identified were observed to be common in all the four libraries, and this accounted for 74.7% of the total miRNAs identified in all the four samples (Fig. 4a). Whereas five miRNAs (2.2%) were identified as BR-R specific, two miRNAs each (0.9%) were identified in BR-S and No-BR-R-specific libraries while four miRNAs (1.7%) were identified in No-BR-S sample. In order to identify which of the known miRNA species were expressed in response to BR treatment, differential expression of miRNAs was analysed using normalized reads in all the four samples. Scatter plots by transforming the data onto M (log ratio) and A (mean average) scales and then plotting the values to generate MA plots showed differential and baseline-expression patterns of miRNAs in root and shoot tissues where each dot represents individual miRNA (Fig. 4b, c). MicroRNAs showing changes in expression levels ≥ 1.0-fold were considered as differentially expressed while those with expression levels below 1-fold were considered as being in baseline-expression zone. Cluster heatmap of differentially expressed known miRNAs was generated using absolute expression values of treated and control samples. The miRNA families miR169 (miR169d, miR169e, miR169f, miR169g), miR156 (miR156a, miR156g), miR172 and those belonging to unknown families, such as miR5648, miR5653, miR5654 and miR6563, were observed to be deferentially expressed in response to BR treatment (Table 3; Fig. 4d). For example, miR172d-3p, miR172c, miR829p-3p, miR846-5p, miR5666 and miR5643a were observed to be most differentially expressed between root and shoot samples (Fig. 4d marked as ##), while miR169g-5p, miR169f-5p, miR169e and miR169d (marked as *# in Fig. 4d) showed decreased expression upon BR treatment in roots.
Distribution and expression pattern of known miRNAs in different tissue libraries. a Venn diagram showing distribution of known miRNAs in BR-S, No-BR-S, BR-R and No-BR-R libraries. b MA plot of differentially expressed known miRNAs in roots. c MA plot of differentially expressed known miRNAs in shoot samples. d Heat map of differentially expressed known miRNAs in all four libraries. Asterisk indicates downregulated miRNAs in BR-R; double asterisk indicates upregulated miRNAs in BR-R; number sign indicates downregulated miRNAs in BR-S; double number sign indicated upregulated miRNAs in BR-S; asterisk and number sign indicates downregulated miRNAs in both roots and shoots in response to exogenous BR (BR-R and BR-S)
Out of 229 known miRNAs belonging to 102 families identified in this study, 48 miRNA families showed differential expression in response to BR treatment in root and shoot tissues (Table 3). The miR5663-3p belonging to an unknown family with 7.924813 fold change showed highest expression in root, followed by miR8121 (7.857981 fold change), miR830-3p belonging to miR830 family (7.667606 fold change), miR850 (7.636625 fold change), miR5637 (7.554589 fold change) of unknown family and miR859 (7.276124), respectively. On the other hand, miR169d, miR169e, miR169f-5p, miR169g-5p belonging to miR169 family with expression values of − 8.810572 showed strong downregulation in root, followed by miR156a-3p belonging to miR156 family (− 7.584963 fold change), miR173-3p (− 7.118941 fold change), miR447c-5p, belonging to miR447 family (− 7.118941 fold change), miR865-5p (− 6.882643 fold change) and miR833a-5p belonging to miR833 family (− 6.741467 fold change), respectively. The highest upregulated miRNA in response to BR treatment in the shoot tissue was miR865-5p belonging to miR865 family with a fold change of 8.276124 with respect to control (No-BR-S). Whereas, miR835-5p belonging to miR835 family was observed to be significantly downregulated in shoots with a fold change − 8.076816 when compared with No-BR-S (Table 3).
Identification and expression analysis of BR-responsive novel miRNAs
After removal of known/conserved miRNAs, sequences were subjected to miRCat analysis to identify novel miRNAs expressed in response to BR treatment. Using the Arabidopsis genome database, a total of 27 novel miRNAs were identified (Table 4). Mature sequences of these novel miRNAs were observed to be 21–22 nucleotide in length, and 5′ nucleotide was observed to be ‘U’ in most of the miRNAs. Stem-loop sequences for each novel miRNAs were marked as per analysis with reference genome, and fold-back structures were predicted through Mfold software (Fig. S1). Majority of the identified novel miRNA candidates were observed to be located on chromosomes 1, 2 and 4, while only two miRNAs (miR-20 and miR-05) are present on chromosome 3, while four miRNAs (miR-09, miR-10, miR-14 and miR-15) were observed to be localized on chromosome 5 (Fig. 5a).
Distribution and expression pattern of novel miRNAs in the root and shoot BR treated and untreated tissue libraries. a Distribution of novel miRNA precursor genes on different chromosomes of A. thaliana. b Distribution of novel miRNAs among the four libraries. c Heat map of differentially expressed novel miRNAs. High and low expression values are shown in colour code with red indicating strong expression while green indicating weak expression. d MA plot of differentially expressed novel miRNAs in roots. e MA plot of differentially expressed novel miRNAs in shoot samples. Single asterisk indicates upregulated miRNAs in BR-R; double asterisk indicates upregulated miRNAs in BR-S; triple asterisk indicates downregulated miRNAs in BR-R; quadruple asterisk indicates downregulated miRNAs in BR-S
Among the 27 novel miRNAs identified, 2 of them, i.e. miR-08 and miR-21, were observed to be expressed in all the four samples (Table S2; Fig. 5b). While, four miRNAs, i.e. miR-01, miR-03, miR-04 and miR-05, were observed to be BR-R specific and miR-12, miR-13, miR-14 and miR16 were identified as BR-S specific. On the other hand, miR-23, miR-24, miR-25, miR-26 and miR-27 were exclusively expressed in No-BR-S, while miR-17, miR-18, miR-19, miR-20 and miR-22 were observed to be No-BR-R specific (Table S2; Fig. 5b). Differential expression analysis shows seven miRNAs miR-01, miR-02, miR-03, miR-04, miR-05, miR-07 and miR-10 in roots and six miRNAs miR-02, miR-09, miR-12, miR-13, miR-14 and miR-16 in shoots to be significantly upregulated (Fig. 5c; Table 4). Similarly, six putative novel miRNAs, namely miR-06, miR-17, miR-18, miR-19, miR-20 and miR-22, in roots and six miRNAs miR-07, miR-23, miR-24, miR-25, miR-26 and miR-27 in shoots were observed to be significantly downregulated (Fig. 5c; Table 4). The most significantly upregulated miRNAs in roots are miR-10 (10.943 fold change) followed by miR-01 (10.586 fold change), miR-07 (9.8602 fold change), miR-04 (7.5554 fold change) and miR-03 (6.7898 fold change), while in shoots, these are miR-02 (14.628 fold change), miR-16 (9.4429 fold change), miR-09 (8.7345 fold change) and miR-13 (7.5970 fold change) (Table 4). On the other hand, significantly downregulated miRNAs in roots are miR-20 (− 14.618 fold change), miR-17 (− 14.053 fold change) and miR-18 (− 13.105 fold change), while in shoots, these are miR-26 (− 11.800 fold change), miR-24 (− 8.8600 fold change) and miR-07 (− 7.826 fold change), respectively (Table 4). MA plot for root and shoot samples was generated separately by using the log2 fold change values, and those showing ≥ 1 fold change were categorized as differentially expressed while those with < 1 fold change as baseline-expressed (Fig. 5d, e).
Target prediction of differentially expressed miRNAs and their functional categorization
Target genes of differentially expressed miRNAs were predicted on the basis of psRNATarget analysis using A. thaliana as genome reference. Target genes of most of the known and conserved miRNAs were similar to known targets identified in different studies (Table S3). While, among the novel miRNAs identified in this study, target genes for only six could be predicted (Table 5). Among these, auxin-responsive factors (ARFs) were predicted to be direct targets of miR-17 that shows positive BR-responsive expression in shoots and is downregulated in roots (Fig. 5; Table 5).
GO analysis of all the differentially expressed miRNAs was performed. This analysis showed that predicted target genes are mainly involved in transcriptional processes and regulation. With respect to cellular component, the targets were mainly functional in the nucleus and these were largely involved in metabolic pathways (Fig. 6). This analysis therefore showed that target genes of BR-induced miRNAs were mainly involved in distinct molecular, cellular and metabolic processes in plants.
Expression validation of BR-responsive miRNAs and their target genes
To validate differential expression patterns of BR-responsive miRNAs, six known and novel miRNAs were randomly selected and their expression patterns was analysed using stem-loop RT-qPCR. The result of stem-loop RT-qPCR was compared with abundance of each selected miRNAs in the small RNA libraries (Fig. 7a). It was observed that expression patterns for miR169a, miR167a, miR156f, miR393a, miR-01 and miR-16 correlated with results obtained in the small RNA sequencing study.
Expression validation using stem-loop RT-qPCR and RT-qPCR. Expression validation of selected BR-responsive miRNAs and their target genes. a Relative expression of known (miR169a, miR167a, miR156f and miR393a) and novel miRNAs (miR-01 and miR-16) as determined by stem-loop RT-qPCR. b Relative transcript abundance of selected target genes of conserved miRNAs, miR169a, miR167a and miR156f identified as BR-responsive in this study by RT-qPCR. Statistical analysis using one-way ANOVA followed by Dunnett’s t test is expressed as mean ± SD where ***P > 0.001, **P > 0.01 and *P > 0.05. Asterisks indicate significant differences in relative expression levels of miRNAs between control and BR-treated tissue samples
We also validated expression of three genes AthNFYA3, AthARF10 and AthSPL2 that are known targets of miR169a, miR167a and miR156f identified as BR-responsive in this study (Fig. 7b). Comparison of expression patterns of miRNAs with their respective targets showed that steady-state levels of miRNA transcripts were inversely related to expression patterns of their respective target genes (Fig. 7a, b). For instance, expression of AthNFYA3 and AthSPL2 was observed to be upregulated in contrast to expression of their targeting miRNAs miR169a and miR156f that were downregulated in both root and shoot samples. Similarly, AthARF10 that is known to be targeted by miR167 members was observed to be downregulated in contrast to miR167a expression that showed increased expression in response to BR.
Discussion
MicroRNAs play an important role in regulating gene expression at post-transcriptional levels. In the last few years, deep sequencing technology has been used to identify miRNAs in plant genomes. By employing this technique, researchers have been able to identify and characterize miRNAs involved in specific signalling pathways. To date, many miRNAs have been functionally characterized and demonstrated to be involved in various mechanisms like phytohormone signalling, biotic and abiotic stresses, plant development and nutrient signalling pathways (Pant et al. 2009; Akpinar et al. 2015; Jian et al. 2016). Among these, phytohormones such as brassinosteroids have been the focus of many studies because of their diverse roles in plant development and stress-adaptive responses. However, there are still many gaps in our understanding related to brassinosteroid research that needs attention.
The aim of this study was to identify brassinosteroid-responsive miRNAs expressed in tissue-specific manner in the model plant A. thaliana. To achieve this, small RNA sequencing was carried out using control (No-BR) and BR-supplemented root and shoot samples. Saliently, brassinosteroid treatment was observed to significantly affect miRNA population as evident from dominance of 24-nt small RNA molecules in filtered libraries. A total of 229 known and 27 novel miRNAs have been identified that were observed to be differentially expressed in response to BR. The 229 known miRNAs belonging to 102 families included the most conserved miRNAs reported in A. thaliana (Sunkar and Zhu 2004). Moreover, conserved miRNAs were observed to be more abundant in comparison to non-conserved miRNAs. The most abundant miRNA family in both root and shoot libraries was observed to be miR169. Interestingly, all members of miR169 were observed to be downregulated in root and shoot tissues in response to BR application. Previous studies have shown that miR169 is mainly involved in abiotic stress and AUX/ABA signalling (Ni et al. 2013). These results therefore suggest miRNA-based crosstalk between brassinosteroid and auxin signalling pathways. Auxins are known to be key players in plant development process (Ubeda-Tomás et al. 2012). Brassinosteroids are also known to regulate many aspects of plant development and stress responses (Sahni et al. 2016). However, clear genetic evidence of their integrated effects on plant growth and stress tolerance is still lacking.
Characterization and comparative profiling of the miRNAs between BR and No-BR samples have provided insight into the possible involvement of miRNAs in BR signalling. Differential expression analysis showed 59 miRNAs in root library while only 50 in shoots to be differentially expressed. This indicates that root microRNA biome is more sensitive to BR treatment than shoot microRNA population. Plants exposed to BR showed increase in miRNA population by 8% in comparison to No-BR root samples. On the other hand, a 12% decrease of miRNAs in BR-exposed shoot tissues was observed in comparison to No-BR shoot samples. Previous studies have shown that proliferation and differentiation root cells are highly influenced by interactive role of auxins and brassinosteroids (Chaiwanon and Wang 2015). Many biochemical assays have also suggested a synergistic interaction between auxins and BRs (Goda et al. 2004).
Relationship between miRNAs and its target genes has been the focus of miRNA-associated genetic regulation. Prediction of target genes of differentially expressed miRNAs provides valuable information about miRNA-mediated biological processes interlinked with phytohormone signalling pathways in general. Target gene annotations of potential miRNAs have provided insight into gene regulation in response to BR exposure. The precursor sequence of novel miRNAs identified in this study showed fold-back hairpin structures and as expected had low free energy (− 30 to − 57 kcal/mol). A majority of miRNAs are known to have lower free energies as compared to tRNAs and rRNAs (Bonnet et al. 2004). In silico analysis showed that some miRNAs may be involved in targeting genes affecting hormone signalling and metabolism. GO analysis showed that BR-responsive miRNAs could also be involved in regulating genes affecting plant development, hormone signalling and stress responses (Fig. 8). Validation of expression of three of the genes AthARF10, AthNFYA3 and AthSPL2 that are targets of miR167, miR169 and miR156 and identified as BR-responsive in this study showed that expression of these genes was inversely related to levels of targeting miRNAs. ARFs mediate auxin signalling and regulate plant development; AthNFYA3 is known to be involved in abiotic stress responses, while AthSPL2 affects different aspects of plant development as well (Shikata et al. 2009; Leyva-Gonza’lez et al. 2012; Nicholas and Naden 2017). This observation suggests a direct role of BR in affecting miRNA-regulated transcript levels of genes regulating plant development and responses to various stress conditions.
Many recent studies support the role of BR and miRNAs in modulating plant development and metabolic processes in responses to environmental cues (Jay et al. 2010; Sharma et al. 2012). Differential expression of miRNAs in response to drought/dehydration stress has been documented in plants such as barley, Brachypodium distachyon and wild emmer wheat (Budak and Akpinar 2011; Akpinar et al. 2015; Alptekin et al. 2017). These studies together with the results presented in this study support integrated roles of BR and miRNA-based post-transcriptional gene regulatory mechanisms that can be further investigated for better understanding of molecular basis of BR-mediated stress tolerance in plants.
Conclusions
-
This study provides a comprehensive compilation of tissue-specific BR-responsive miRNomes in A. thaliana seedlings.
-
A number of conserved miRNAs were observed to be differentially expressed in roots and shoots in response to exogenously supplied 24-epibrassinolide.
-
Twenty-seven novel miRNAs were identified that show tissue-specific differential expression in a BR-responsive manner. These novel miRNAs did not show any sequence conservation with known miRNAs in databases. Auxin response factors were predicted as target genes for one of the novel miRNA, miR-17.
-
This work lays the foundation for further investigation into understanding the underlying molecular mechanisms of BR-mediated stress tolerance by post-transcriptional gene regulation.
References
Akpinar BA, Kantar M, Budak H (2015) Root precursors of microRNAs in wild emmer and modern wheats show major differences in response to drought stress. Funct Integr Genomics 15:587–598
Alptekin B, Langridge P, Budak H (2017) Abiotic stress miRNomes in the Triticeae. Funct Integr Genomics 17:145–170
Anuradha S, Rao SSR (2001) Effect of brassinosteroids on salinity stress induced inhibition of seed germination and seedling growth of rice (Oryza sativa L.). Plant Growth Regul 33:151–153
Anwar A, Liu Y, Dong R, Bai L, Yu X, Li Y (2018) The physiological and molecular mechanism of brassinosteroid in response to stress. Biol Res 51:46
Bonnet E, Wuyts J, Rouze P, Van de Peer Y (2004) Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20:2911–2917
Budak H, Akpinar BA (2011) Dehydration stress-responsive miRNA in Brachypodium distachyon: evident by genome-wide screening of microRNAs expression. OMICS 15:791–799
Budak H, Kantar M, Bulut R, Akpinar BA (2015) Stress responsive miRNAs and isomiRs in cereals. Plant Sci 235:1–13
Chaiwanon J, Wang ZY (2015) Spatiotemporal brassinosteroid signalling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr Biol 25:1031–1042
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33:179
Choudhary SP, Yu JQ, Yamaguchi-Shinozaki K, Shinozak K, Tran LS (2012) Benefits of brassinosteroid crosstalk. Trends Plant Sci 17:594–605
Curaba J, Singh MB, Bhalla PL (2014) miRNAs in the crosstalk between phytohormone signalling pathways. J Exp Bot 65:1425–1438
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39:155–159
Divi UK, Rahman T, Krishna P (2015) Gene expression and functional analyses in brassinosteroid-mediated stress tolerance. Plant Biotechnol J 14:419–432
Ergen NZ, Budak H (2009) Sequencing over 13 000 expressed sequence tags from six subtractive cDNA libraries of wild and modern wheats following slow drought stress. Plant Cell Environ 32:220–236
Gasic EV, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:12
Ge A, Shangguan L, Zhang X, Dong Q, Han J, Liu H, Wang X, Fang J (2013) Deep sequencing discovery of novel and conserved microRNAs in strawberry (Fragaria×ananassa). Physiol Plantarum 148:387–396
Gim JA, Ha HS, Ahn K, Kim DS, Kim HS (2014) Genome-wide identification and classification of microRNAs derived from repetitive elements. Genomics Inform 12:261–267
Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S (2004) Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134:1555–1573
Gruszka D (2013) The brassinosteroid signalling pathway-new key players and interconnections with other signalling networks crucial for plant development and stress tolerance. Int J Mol Sci 14:8740–8774
Gupta A, Singh M, Laxmi A (2015) Multiple interactions between glucose and Brassinosteroid signal transduction pathways in Arabidopsis are uncovered by whole-genome transcriptional profiling. Plant Physiol 168:1091–1105
Hewezi T, Maier TR, Nettleton D, Baum TJ (2012) The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol 159:321–335
Huang J, Ghosh R, Bankaitis VA (2016) Sec14-like phosphatidylinositol transfer proteins and the biological landscape of phosphoinositide signaling in plants. Biochim Biophys Acta 186:1352–1364
Jay F, Renou JP, Voinnet O, Navarro L (2010) Biotic stress-associated microRNAs: identification, detection, regulation, and functional analysis. Methods Mol Biol 592:183–202
Jian H, Wang J, Wang T, Wei L, Li J, Liu L (2016) Identification of rape seed microRNAs involved in early stage seed germination under salt and drought stresses. Front Plant Sci 7:658
Kakkar P, Das B, Viswanathan PN (1984) A modified spectrophotometric assay of superoxide dismutase. Indian J Biochem Biophys 21:130–132
Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39:152–157
Kumari G, Kusumanjali K, Srivastava PS, Das S (2013) Isolation and expression analysis of miR165a and REVOLUTA from Brassica species. Acta Physiol Plant 35:399–410
Kusumanjali K, Kumari G, Srivastava PS, Das S (2012) Sequence conservation and divergence in miR164C1 and its target, CUC1, in Brassica species. Plant Biotechnol Rep 6:149–116
Leyva-Gonza’lez MA, Ibarra-Laclette E, Cruz-Ramı’rez A, Herrera-Estrella L (2012) Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS ONE 7:e48138
Li X, Tan DX, Jiang D, Liu F (2016) Melatonin enhances cold tolerance in drought-primed wild-type and abscisic acid-deficient mutant barley. J Pineal Res 61:328–339
Lian C, Yao K, Duan H, Li Q, Liu C, Yin W, Xia X (2018) Exploration of ABA responsive miRNAs reveals a new hormone signaling crosstalk pathway regulating root growth of Populus euphratica. Int J Mol Sci 19:1481
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Lu C, Fedoroff N (2000) Amutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12:2351–2366
Ma Z, Hu X, Cai W, Huang W, Zhou X, Luo Q, Yang H, Wang J, Huang J (2014) Arabidopsis miR171-targeted scarecrow-like proteins bind to GT cis-elements and mediate gibberellin-regulated chlorophyll biosynthesis under light conditions. PLoS Genet 10:1004519
Mallory AC, Vaucheret H (2006) Functions of microRNAs and related small RNAs in plants. Nat Genet 38:31–37
Mallory AC, Bartel DP, Bartel B (2005) MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17:1360–1375
Marsit CJ, Eddy K, Kelsey KT (2006) MicroRNA responses to cellular stress. Cancer Res 66:10843–10848
Mewis K, Lenfant N, Lombard V, Henrissat B (2016) Dividing the large glycoside hydrolase family 43 into subfamilies: a motivation for detailed enzyme characterization. Appl Environ Microbiol 82:1686–1692
Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, Griffiths-Jones S, Jacobsen SE, Mallory AC, Martienssen RA, Poethig RS, Qi Y, Vaucheret H, Voinnet O, Watanabe Y, Weigel D, Zhu JK (2008) Criteria for annotation of plant microRNAs. Plant Cell 20:3186–3190
Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K (2006) Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 25:2537–2545
Ni Z, Hu Z, Jiang Q, Zhang H (2013) GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol Biol 82:113–129
Nicholas SC, Naden TK (2017) The role of auxin response factors in the development and differential growth of inflorescence stems. Plant Signal Behav 12:4-e1307492
Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR (2009) Identification of nutrient-responsive Arabidopsis and rape seed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150:1541–1555
Sahni S, Prasad BD, Liu Q, Grbic V, Sharpe A, Singh SP, Krishna P (2016) Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance. Sci Rep 6:28298
Sharma I, Bhardwaj R, Pati PK (2012) Mitigation of adverse effects of chlorpyrifos by 24-epibrassinolide and analysis of stress markers in a rice variety Pusa Basmati-1. Ecotoxicol Environ Saf 85:72–81
Sharma I, Bhardwaj R, Pati PK (2015) Exogenous application of 28-Homobrassinolide modulates the dynamics of salt and pesticides induced stress responses in an elite rice variety Pusa Basmati-1. J Plant Growth Regul 34:509–518
Shikata M, Koyama T, Mitsuda N, Ohme-Takagi M (2009) Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol 50:2133–2145
Sirohi G, Kusumanjali K, Kumar R, Jain A, Srivastava PS, Das S (2018) Syntenic analysis and functional characterization of miR165a from Brassica species. Acta Physiol Plant 40:16
Sobkowiak L, Karlowski W, Jarmolowski A, Szweykowska-Kulinska Z (2012) Non-canonical processing of Arabidopsis pri-miR319a/b/c generates additional microRNAs to target one RAP2.12 mRNA isoform. Front Plant Sci 3:46
Song C, Chen W, Zhang C, Korir NK, Yu H, Ma Z, Fang J (2010) Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genomics 11:431
Sreeramulu S, Mostizky Y, Sunitha S, Shani E, Nahum H, Salomon D, Hayun LB, Gruetter C, Rauh D, Ori N, Sessa G (2013) BSKs are partially redundant positive regulators of brassinosteroid signalling in Arabidopsis. Plant J 74:905–919
Sunkar R, Zhu JK (2004) Novel and stress-regulated micro-RNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Ubeda-Tomás S, Beemster GTS, Bennett MJ (2012) Hormonal regulation of root growth: integrating local activities into global behaviour. Trends Plant Sci 17:326–331
Wang T, Chen L, Zhao M, Tian Q, Zhang WH (2011) Identification of drought responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics 12:367
Wei LJ, Deng XG, Zhu T, Zheng T, Li PX, Wu JQ, Zhang DW, Lin HH (2015) Ethylene is involved in brassinosteroids induced alternative respiratory pathway in cucumber (Cucumis sativus L.) seedlings response to abiotic stress. Front Plant Sci 6:982
Williams L, Carles CC, Osmont KS, Fletcher JC (2005) A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci U S A 102:9703–9708
Wu MF, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133:4211–4218
Xie W, Adolf J, Melzig MF (2017) Identification of Viscum album L. miRNAs and prediction of their medicinal values. PLoS One 12:0187776
Xu Q, Liu Y, Zhu A, Wu X, Ye J, Yu K, Guo W, Deng X (2010) Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genomics 11:246
Yu N, Cai WJ, Wang S, Shan CM, Wang LJ, Chen XY (2010) Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22:2322–2335
Yu S, Galvao VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW (2012) Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24:3320–3332
Yuan L, Yuan Y, Du J, Sun J, Guo S (2012) Effects of 24-epibrassinolide on nitrogen metabolism in cucumber seedlings under Ca(NO3)2 stress. Plant Physiol Biochem 61:29–35
Yuan LB, Peng ZH, Zhi TT, Zho Z, Liu Y, Zhu Q, Xiong XY, Ren CM (2015) Brassinosteroid enhances cytokinin–induced anthocyanin biosynthesis in Arabidopsis seedlings. Biol Plant 59:99–105
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Acknowledgements
We acknowledge Bionivid Pvt. Ltd., Bangalore, India, for small RNA sequencing and preliminary data analysis.
Funding
This study was funded by Science and Engineering Research Board (SERB), Government of India, through grant #YSS/2014/000350 to GS.
Author information
Authors and Affiliations
Contributions
GS conceived, designed and performed the experiments along with AK. MK provided resources and mentored GS. Together, they analysed the data and wrote the manuscript. All authors read and approved the final version of paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Fig. 1
Fold-back structures of novel miRNAs identified in the study as predicted using MFOLD. The mature miRNAs are highlighted in blue while the miRNA* are shown in red colour. (PDF 25879 kb)
ESM 2
(DOC 108 kb)
Rights and permissions
About this article
Cite this article
Sirohi, G., Khandelwal, A. & Kapoor, M. High-throughput sequencing and differential expression analysis of miRNAs in response to Brassinosteroid treatment in Arabidopsis thaliana. Funct Integr Genomics 19, 597–615 (2019). https://doi.org/10.1007/s10142-019-00668-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-019-00668-1