Abstract
Isochrysis galbana is valuable in aquaculture due to its production of eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA). However, achieving high yields of polyunsaturated fatty acids (PUFAs) presents challenges, leading to exploration of innovative approaches. This study investigated the influence of Bacillus jeotgali on the growth of I. galbana and its fatty acid composition. Co-culturing I. galbana with B. jeotgali significantly increased chlorophyll a content and cell abundance, particularly at higher bacterial population densities (algae-to-bacteria ratio of 1:10). Physiological and biochemical analyses found elevated soluble protein content in microalgae co-cultured with B. jeotgali, accompanied by decreased superoxide dismutase (SOD) activity. Fatty acid composition analysis demonstrated a distinctive profile in co-cultured I. galbana, characterized by increased PUFAs, especially EPA and DHA. Gene expression analysis indicated an upregulation of desaturase genes (d4FAD, d5FAD, d6FAD, and d8FAD) associated with PUFA synthesis pathway in I. galbana during co-culturing with B. jeotgali. This study advances our understanding of bacteria-microalgae interactions and presents a promising strategy for enhancing the production of DHA and EPA.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Microalgae are unicellular organisms and they represent the smallest living organisms, exhibiting the ability to convert solar energy into chemical energy through photosynthesis (Xie et al. 2022). Currently, microalgae have extensive applications across various sectors, including aquatic feed, food production, cosmetics, environmental remediation, and biofuel generation. Isochrysis galbana, a species of marine unicellular microalgae, is notable for synthesizing of docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA), long chain polyunsaturated fatty acids (PUFAs) with significant health and nutritional value (Liu et al. 2013). It is widely utilized in hatcheries to nourish fish larvae and clams. Nevertheless, the large-scale cultivation of I. galbana for DHA and EPA production has not been realized, primarily due to the unsatisfactory low yields of DHA and EPA in the cultured strains of I. galbana. Therefore, innovative approaches are crucial to enhance DHA and EPA production in I. galbana.
Researches have shown that abiotic factors such as light intensity, nitrogen levels, temperature, and salinity play a crucial role in influencing the growth and fatty acid composition of microalgae. Additionally, utilizing mixed carbon sources, including CO2, glucose, and amino acids, has been identified as a means to improve the growth and enhance the accumulation of DHA in I. galbana (Zheng et al. 2022). The synthesis of DHA in Dunaliella sp. was notably influenced by the interactive effects of salinity, temperature, and light. The peak DHA content was recorded in Dunaliella sp. under conditions of 0.5 M salinity, 25 °C temperature, and 11,200 μmol photon m−2 s−1 light intensity (Gharajeh et al. 2020). Cultivating Nannochloropsis oceanica at a low temperature significantly enhanced DHA production (Sirisuk et al. 2018). In addition to regulating abiotic conditions, genetic engineering represents a viable approach to augmenting DHA and EPA content in microalgae. For instance, the overexpression of the Δ6-desaturase gene in Phaeodactylum tricornutum led to an increased EPA content (Zhu et al. 2017). The utilization of CRISPR as a biotechnological tool for microalgae has further facilitated the enhancement of fatty acid content (Lin and Ng 2020). Nevertheless, its implementation in microalgae currently presents challenges. Mutagenesis represents an alternative approach for augmenting the fatty acid content of microalgae. Physical mutagens such as ultraviolet, X-rays, gamma radiation, or atmospheric sources can induce genetic changes. In one study, Aurantiochytrium sp. subjected to UV irradiation (50 W, 30 s) exhibited a DHA content up to 1.9 times higher than the wild Aurantiochytrium sp. (Liu et al. 2020a). However, these methods for enhancing DHA and EPA accumulation incur high costs and are time-consuming in practical microalgae culture applications. To enhance DHA and EPA production in microalgae, identifying a more environmentally friendly, cost-effective, and feasible method is imperative.
In recent years, the interactions between algae and bacteria have gained considerable attention. Studies have shown that microalgae can contribute oxygen to bacteria growth through photosynthesis, while bacteria reciprocate by providing microalgae with essential metabolites and inorganic carbon necessary for growth. The synergy between microalgae and bacteria extends to the creation of optimal environments for each other, involving the provision of vital cofactors such as iron, vitamins, indole-3-acetic acid, and tryptophan (Seymour et al. 2017). While the influence of bacteria on the growth of microalgae has been extensively studied, its role in the biosynthesis of algal DHA and EPA has not been sufficiently explored. Among the limited studies, the impact of growth-promoting bacteria on EPA production of Nannothropsis oceanica was investigated. Co-culturing with probiotic bacteria significantly enhanced the growth of N. oceanica, leading to a 2.25-fold increase in EPA content (Liu et al. 2020b). Furthermore, Marinobacter sp. was found to significantly promote DHA production in I. galbana (Wang et al. 2022). These early evidences indicate that co-cultivating bacteria with microalgae emerges as a viable approach to augment the production of DHA or EPA in microalgae.
In this study, we investigated the influence of the phycosphere bacterium B. jeotgali on the growth of I. galbana and its fatty acid composition. Additionally, we explored the impact of the optimal bacterial concentration on the growth of I. galbana. Furthermore, we analyzed the gene expression associated with fatty acid synthesis to elucidate the underlying mechanisms governing fatty acid metabolism regulation. This study contributes to our understanding of bacteria-microalgae interactions and demonstrates the potential of co-culturing as a promising strategy for enhancing large-scale production of DHA and EPA.
Materials and Methods
Cultivation Conditions for Microalgae
Axenic I. galbana 3011 was obtained from the Marine Biotechnology Laboratory at Ningbo University, China. In this study, algae were cultivated using NMB3 medium, which consisted of KNO3 (0.1 g/L), KH2PO4 (0.01 g/L), MnSO4-H2O (2.50 mg/L), FeSO4-7H2O (2.50 mg/L), EDTA-Na2 (10.00 g/L), vitamin B1 (6.00 µg/L), and vitamin B12 (0.05 µg/L). The incubation conditions were maintained at a photoperiod of 12/12 h day/night, a temperature of 25 °C, and a light intensity of 100 μmol photon m−2 s−1, achieved with a cool light fluorescent lamp.
Co-cultivation of Microalgae and Bacteria
The B. jeotgali strain used in this study was isolated from I. galbana cultures in our laboratory and is presently archived in the Marine Biotechnology Experiment at Ningbo University. Monoclonal bacteria were specifically chosen and subjected to overnight cultivation in fresh 2216E liquid medium, which consisted of peptone (5.00 g/L), yeast extract (1.00 g/L), ferric citrate (0.10 g/L), sodium chloride (19.45 g/L), magnesium chloride (5.98 g/L), sodium sulfate (3.24 g/L), calcium chloride (1.80 g/L), potassium chloride (0.55 g/L), sodium carbonate (0.16 g/L), potassium bromide (0.08 g/L), strontium chloride (34.00 mg/L), boric acid (22.00 mg/L), sodium silicate (4.00 mg/L), sodium fluoride (2.40 mg/L), ammonium nitrate (1.60 mg/L), disodium phosphate (8.00 mg/L). The bacterial solution was cultured at 28 °C and 180 rpm until the bacterial concentration reached OD600 = 0.4–0.6. Subsequently, the bacterial solution underwent centrifugation at 8000 rpm and was subjected to three washes with sterile seawater. The bacteria were resuspended using NMB3 medium. At the exponential growth phase of I. galbana with an initial cell density of approximately 1 × 106 cells/mL, B. jeotgali was introduced to I. galbana culture at varying ratios of algae to bacteria, specifically 1:1, 1:2, and 1:10 (cell count: cell count). The control group consisted of axenic I. galbana culture. The densities of microalgae and bacteria were determined using cell counting chambers and the plate count method, respectively. I. galbana and B. jeotgali were co-cultured for 14 days. For chlorophyll a, cell abundance, soluble protein and SOD activity determination, samples were collected on days 0 and every other day thereafter.
Chlorophyll a, Soluble Protein and SOD Activity Determination
I. galbana and B. jeotgali were co-cultured at a ratio of 1:10, and the chlorophyll a content of I. galbana was measured every two days. The methods employed in this study were based on the procedures described in previous research by Wang et al. (2022). Algal culture was centrifugated at 16,000 × g for 2 min at room temperature and the supernatant was discarded. Glass beads and 1 mL of 90% ice-cold acetone were added to a 2 mL cryotube, and the mixture was homogenized three times (65 Hz, 60 s/time) using a homogenizer. After homogenization, the mixture was centrifuged at 14,000 × g for 2 min, and the resulting supernatant contained the extracted chlorophyll. Subsequently, 200 µL of the extracted chlorophyll solution was aliquoted, and its absorbance at 630 nm, 664 nm, and 750 nm was measured using a spectrophotometer. The concentration of chlorophyll a was calculated using the following formula:
Soluble protein was extracted by homogenization and quantified using the bicinchoninic acid (BCA) protein assay. SOD activity was determined according to the instructions provided with the SOD kit from Nanjing Jiancheng Bioengineering Institute.
Fatty Acid Determination and Analysis
For fatty acid analysis, samples from mono-culture of I. galbana, mono-culture of B. jeotgali, and their co-cultures were collected on the 14th day post-co-culturing. The samples were centrifuged at 6000 rpm for 10 min, and the supernatant was discarded. The resulting pellets were frozen at –80 °C and subsequently dried for 48 h using a vacuum freeze dryer. For further analysis, the lyophilized samples (10 mg) were weighed according to the protocol described by Wang et al. (2022). Hexane, an internal standard nonadecanoic acid (1 mg/mL), and formyl chloride were added to the weighed samples. The samples were incubated in a water bath at 70 °C for 2 h. After incubation, the samples were allowed to cool before the addition of 6% K2CO3 and hexane. The lower liquid layer was collected in a centrifuge tube and centrifuged at 3000 rpm for 10 min. A sterile syringe was then used to aspirate the supernatant, which was then filtered through an organic phase filter membrane. The resulting purified fatty acids underwent methylation, and the filtrate was collected into a sample bottle. The method for determining fatty acids followed the procedure described by Wang et al. (2022).
Total RNA Extraction
Cell cultures were gathered by discarding the supernatant through centrifugation at 8000 rpm for 10 min at 4 °C. The collected cultures in the centrifuge tubes were subsequently sealed and preserved in liquid nitrogen. Total RNA extraction from the samples was performed using the E. Z. N. A.® Plant RNA Kit from OMEGA Bio-Tek. The purified RNA was then subjected to concentration measurement using a Thermo Scientific NanoDrop One spectrophotometer (Thermo Fisher Scientific, USA).
Real-Time Quantitative PCR
The cDNA was synthesized through reverse transcription using the PrimeScript™ RT kit from TaKaRa, Japan. Gene expression analysis was conducted by real-time quantitative PCR (RT-qPCR) utilizing the LongGene Q2000A qPCR system in Hangzhou, China, and TB Green® Premix Ex TaqTM II (Tli RNaseH Plus) from TaKaRa, Japan. Ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcL) served as the reference gene. Primers for the delta-4 desaturase genes (d4FAD), delta-5 desaturase genes (d5FAD), delta-6 desaturase genes (d6FAD), and delta-8 desaturase genes (d8FAD) were listed in Table S1. The relative expression of the target genes was calculated using the 2^−∆∆Ct method (Cao et al. 2016).
Statistical Analysis
The data were analyzed using SPSS for Windows (IBM Corp., Armonk, NY, USA), and Student’s t test was performed to determine the significant difference between two groups. All analyses were carried out in triplicate, and differences were considered statistically significant at a p value < 0.05. Data in the paper are presented as mean ± standard deviation (SD).
Results
Effects of B. jeotgali on the Growth of I. galbana
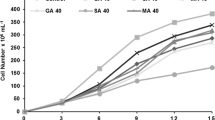
To investigate the impact of B. jeotgali on the growth of I. galbana, co-culture experiments were conducted with varying density ratios for the two organisms. Algae bacteria were co-cultured at different density ratios for 14 days, during which the growth status of the algal cells was monitored by assessing their chlorophyll a content (Fig. 1A). The findings revealed that co-culturing I. galbana with B. jeotgali at a cell density of 1:1 resulted in a significantly higher chlorophyll a content in I. galbana compared to the control group. Furthermore, at bacterial cell density ratios of 1:2 and 1:10, the chlorophyll a content of algal cells exhibited even more substantial increases compared to the control group. This result suggests that B. jeotgali significantly enhanced the growth of algal cells, and the degree of promotion depended on the population density of the bacteria. To further assess the impact of B. jeotgali on the growth of I. galbana with an algae-to-bacteria ratio of 1:10, the cell numbers of I. galbana were quantified. The cell abundance of I. galbana in co-cultures was significantly higher than in the control group (Fig. 1B). Furthermore, as the co-culture time extended, the cell abundance of I. galbana continued to increase, accompanied by a darker coloration of the algal fluid (Fig. 1B). This outcome provided additional confirmation of the promoting effect of B. jeotgali on the growth of I. galbana.
Effects of Bacillus jeotgali on the growth of Isochrysis galbana. A Effects of B. jeotgali with three different initial algae/bacteria density ratios (1:1, 1:2, 1:10) on chorophyll a content of I. galbana. B Effects of B. jeotgali (1:10) on the cell abundance of I. galbana. Error line represented standard deviation (SD), * represented significant difference, p < 0.05
Physiological and Biochemical Effects of B. jeotgali on I. galbana
The physiological and biochemical effects of B. jeotgali on I. galbana were investigated by comparing soluble protein contents and SOD activity of I. galbana in co-cultures and mono-cultures (Fig. 2). The total concentration of soluble proteins in the microalgal cells showed a steady increase over time, with the soluble protein content in the co-culture consistently higher than that in the mono-culture from day 2 to day 14 (Fig. 2A). Additionally, the SOD activity in the co-cultures exhibited a significant decrease compared to that in mono-cultures except on the second day (Fig. 2B).
Effect of B. jeotgali on the Fatty Acid Composition of I. galbana
The influence of B. jeotgal on the fatty acid composition of I. galbana was examined in detail (Fig. 3 and Table 1). The results revealed the presence of tridecanoic acid (C13:0), which is not typically found in I. galbana but is present in B. jeotgal, among the saturated fatty acids. Myristic acid (C14:0) and palmitic acid (C16:0) were identified as the predominant saturated fatty acids in I. galbana, while pentadecanoic acid (C15:0) was the main saturated fatty acid in B. jeotgal. Under co-culturing conditions, I. galbana exhibited lower levels of saturated fatty acids compared to when cultured alone, with the exception of C14:0 and C15:0. Notably, heptadecanoic acid (C17:0) was not detected in the co-culture (Fig. 3A, Table 1).
In B. jeotgal, only two monounsaturated fatty acids, palmitoleic acid (C16:1 n-7) and heptadecenoic acid (C17:1 n-7), were detected. Palmitoleic acid (C16:1 n-7) and elaidic acid (C18:1 n-9t) were identified as the major monounsaturated fatty acids in I. galbana cultured alone and co-cultured, with elaidic acid (C18:1 n-9t) at a significantly lower level under co-culturing conditions compared with I. galbana cultured alone. Notably, oleic acid (C18:1 n-9c) was significantly higher in co-cultured I. galbana than when cultured alone (Fig. 3B, Table 1).
Polyunsaturated fatty acids were not detected in B. jeotgal. However, co-cultured I. galbana exhibited significantly higher levels of hexadecadienoic acid (C16:2 n-6), α-linolenic acid (C18:3 n-3), octadecatetraenoic acid (C18:4 n-3), and docosapentaenoic acid (C22:5 n-6) compared to I. galbana cultured alone. Particularly, the significant elevations of EPA (C20:5 n-3) and DHA (C22:6 n-3) in the microalgae under co-culturing were observed, increased from 0.97 ± 0.02% to 1.19 ± 0.03% and 9.09 ± 0.12% to 15.53 ± 0.07%, respectively. Under co-culturing conditions, the total polyunsaturated fatty acid content of I. galbana (48.45 ± 0.36%) was significantly higher than that observed under mono-culturing conditions (36.08 ± 0.31%) (Fig. 3C, Table 1).
Effect of B. jeotgali on the Expression of Genes Involved in the Polyunsaturated Fatty Acid Synthesis Pathway of I. galbana
The impact of B. jeotgali on the expression of four desaturase genes (d4FAD, d5FAD, d6FAD, and d8FAD) were examined by RT-qPCR, with rbcL gene selected as the internal reference. The results unveiled a noticeable up-regulation in the expression of the four genes associated with the polyunsaturated fatty acid synthesis pathway in I. galbana after 2 days of co-culturing. This upregulation was observed in comparison to the expression levels of the same genes in I. galbana under mono-culturing conditions (Fig. 4). Specifically, d4FAD, d5FAD, d6FAD, and d8FAD exhibited increases in expression by 3.5, 3.1, 5.3, 2.5 times, respectively, highlighting the dynamic response of these genes to the presence of B. jeotgali in the co-culture environment.
Discussion
I. galbana is a vital food source for mollusk larvae, early fish, crustaceans, and various aquatic organisms. Furthermore, I. galbana is renowned for its production of DHA and EPA, which are crucial for health and thus hold a niche in the nutraceutical industry. Traditional methods of enhancing EPA and DHA accumulation in microalgae have focused on modifying abiotic conditions such as light intensity, temperature, and nutrient levels. However, these methods are time-consuming and often lead to inhibition of cell growth, ultimately reducing the total yield of EPA and DHA (Jakhwal et al. 2022). Interestingly, our study has discovered that B. jeotgali enhances both cell growth and the production of DHA and EPA in I. galbana, presenting an innovative approach to boosting DHA and EPA production.
Our findings indicate a significant enhancement in the growth of I. galbana facilitated by B. jeotgali, with the promotion effect increasing as the proportion of bacteria increased. Previous studies have shown that the ratio of cell density between bacteria and microalgae significantly impacts the bacteria’s growth-promoting effects on microalgae. For instance, the presence of Marinobacter sp. at algae/bacteria ratios of 1:50 and 1:100 notably increased the cell growth of I. galbana compared to the control group. However, Marinobacter sp. did not affect the growth of I. galbana at a ratio of 1:1 (Wang et al. 2022). Additionally, Alteromonas macleodii was found to promote the growth of Prochlorococcus at low densities but inhibit its growth at high densities (Aharonovich and Sher 2016). Some studies have highlighted the growth-promoting effects of bacteria belonging to the Bacillus genus on microalgae (Amavizca et al. 2017; Sonowal et al. 2022; Sauvage et al. 2022; Yee et al. 2021). For instance, the endosymbiont B. tequilensis has been shown to improve the cell density, dry weight, chlorophyll content, and astaxanthin content of Haematococcus lacustris (Jeon et al. 2023).
Numerous mechanisms have been reported to promote the growth of microalgae by bacteria, one of which involves bacterial regulation of the intracellular redox balance by scavenging reactive oxygen species (ROS). For example, Peng et al. found that the plant growth-promoting bacteria, Azospirillum brasilense, can reduce the accumulation of ROS in Chlorella sorokiniana cells, thus completely mitigate oxidative stress in C. sorokiniana resulting from nitrogen limitation (Peng et al. 2021). SOD is an important antioxidant enzyme that is involved in the first line of defense against ROS within cells (Bui and Ki 2023). It helps maintain the balance of ROS and protects cells from damage and potential cell death caused by ROS accumulation. In this study, we observed a significant decrease in SOD activity in I. galbana when co-cultured with B. jeotgali compared to the control group. Similarly, co-culturing I. galbana with A. macleodii or Marinobacter sp. also resulted in a substantial inhibition of SOD activity. This effect could potentially be attributed to the bacteria’s capacity to produce extracellular antioxidants (Cao et al. 2021; Wang et al. 2022). Therefore, we speculated that B. jeotgali might promote the growth of I. galbana by removing ROS. However, additional research is required to test this hypothesis and confirm the underlying mechanisms involved. Bacillus sp. has also been reported to notably enhance the growth of Scenedesmus sp. through the secretion of indole-3-acetic acid (IAA) (Dao et al. 2018). Similarly, our research also indicates the capability of B. jeotgali to produce IAA, potentially exerting a beneficial influence on algal growth (data not shown). Further investigation is warranted to ascertain whether B. jeotgali promotes the growth of I. galbana by supplying it with IAA.
Notably, these studies on the effects of Bacillus sp. on microalgae predominantly focus on growth-related mechanisms, while their role in algal DHA and EPA biosynthesis remains unclear. Recent studies have found that certain bacteria have the capacity to promote the accumulation of DHA or EPA in microalgae (Liu et al. 2020b; Wang et al. 2022). However, to the best of my knowledge extends, no research has investigated the promotion of both DHA and EPA production by bacteria in microalgae. In this study, what is new is that a notable enhancement in the content of both DHA and EPA was observed in I. galbana when co-cultivated with B. jeotgali. It was reported that higher protein synthesis in microalgae results in elevated expressions of omega-3/6 fatty acid biosynthetic genes, namely desaturase and elongase, which subsequently synthesize long-chain PUFAs such as EPA (Jeyakumar et al. 2020). As higher protein synthesis was found in co-culture, we conjectured that the high protein content might be one reason for the increased DHA content. Similarly, the proportion of unsaturated fatty acids in C. vulgaris-Mesorhizobium sangaii co-culture was significantly increased compared to algae monoculture (Wei et al. 2019). This enhancement in co-cultures might be due to the metabolic regulation and nutrient exchange between microalgae and bacteria. We are also concerned about the increase of PUFA content and the decrease of MUFA content in I. galbana when co-cultivated with B. jeotgali. It has been reported that increasing IAA concentration could significantly increase PUFA proportion in N. oceanica cultures, while MUFA decreased (Udayan and Arumugam 2017). Given that B. jeotgali also has the ability to produce IAA, it is worth investigating whether the effect of B. jeotgali on the accumulation of fatty acids in I. galbana is related to its ability to produce IAA.
Bacillus sp. is widely employed as an environmental probiotic in aquaculture due to its multifunctionality, including enzyme production, antimicrobial compound synthesis, and disease protection within aquaculture systems (Soltani et al. 2019). Notably, it exhibits bacteriostatic activity against various shrimp pathogens (Ringø 2020). Bacillus sp. as a valuable probiotic for shrimp, revealing a dose-dependent enhancement in shrimp survivals and immune parameters over a 28-day feeding experiment (Rahiman et al. 2010). The study demonstrates that the inclusion of 0.2% isomaltooligosaccharides with 108 CFU/g feed PB significantly boosts shrimp immune responses, leading to improved disease resistance (Li et al. 2009). Bacillus sp. isolated from green crabs displays antagonistic activity against Vibrio parahaemolyticus (Wu et al. 2014). The potential antagonistic effects of B. jeotgali in this study on harmful pathogens require further investigation. If our strain demonstrates antagonistic effects on aquatic pathogens, combined with its capacity to enhance the accumulation of EPA and DHA in microalgae, it holds significant application value.
Data Availability
No datasets were generated or analysed during the current study.
Abbreviations
- d4FAD :
-
Delta-4 desaturase gene
- DHA:
-
Docosahexaenoic acid
- EPA:
-
Eicosapentaenoic acid
- FAMEs:
-
Fatty acid methyl esters
- MUFAs:
-
Monounsaturated fatty acids
- PUFA:
-
Polyunsaturated fatty acid
- RT-qPCR:
-
Real-time quantitative PCR
- rbcL:
-
Ribulose-1,5-bisphosphate carboxylase/oxygenase
- SOD:
-
Superoxide dismutase
- SFAs:
-
Saturated fatty acids
References
Aharonovich D, Sher D (2016) Transcriptional response of Prochlorococcus to co-culture with a marine Alteromonas: differences between strains and the involvement of putative infochemicals. ISME J 10:2892–2906
Amavizca E, Bashan Y, Ryu CM, Farag MA, Bebout BM, De-Bashan LE (2017) Enhanced performance of the microalga Chlorella sorokiniana remotely induced by the plant growth-promoting bacteria Azospirillum brasilense and Bacillus pumilus. Sci Rep 7:1–11
Bui QTN, Ki JS (2023) Two novel superoxide dismutase genes (CuZnSOD and MnSOD) in the toxic marine dinoflagellate Alexandrium pacificum and their differential responses to metal stressors. Chemosphere 313:137532
Cao JY, Xu YP, Zhao L, Li SS, Cai XZ (2016) Tight regulation of the interaction between Brassica napus and Sclerotinia sclerotiorum at the microRNA level. Plant Mol Biol 92:39–55
Cao JY, Wang YY, Wu MN, Kong ZY, Lin JH, Ling T, Xu SM, Ma SN, Zhang L, Zhou CX, Yan XJ, Xu JL (2021) RNA-seq insights into the impact of Alteromonas macleodii on Isochrysis galbana. Front Microbiol 12:711998
Dao GH, Wu GX, Wang XX, Zhang TY, Zhan XM, Hu HY (2018) Enhanced microalgae growth through stimulated secretion of indole acetic acid by symbiotic bacteria. Algal Res 33:345–351
Gharajeh NH, Valizadeh M, Dorani E, Hejazi MA (2020) Dunaliella sp. ABRIINW-I1 as a cell factory of nutraceutical fatty acid pattern: an optimization approach to improved production of docosahexaenoic acid (DHA). Chem Eng Process 155:108073
Jakhwal P, Biswas JK, Tiwari A, Kwon EE, Bhatnagar A (2022) Genetic and non-genetic tailoring of microalgae for the enhanced production of eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA)-a review. Bioresour Technol 344:126250
Jeon MS, Han SI, Ahn JW, Jung JH, Choi JS, Choi YE (2023) Endophyte Bacillus tequilensis improves the growth of microalgae Haematococcus lacustris by regulating host cell metabolism. Bioresour Technol 387:129546
Jeyakumar B, Asha D, Varalakshmi P, Kathiresan S (2020) Nitrogen repletion favors cellular metabolism and improves eicosapentaenoic acid production in the marine microalga Isochrysis sp. CASA CC 101. Algal Res 47:101877
Li J, Tan B, Mai K (2009) Dietary probiotic Bacillus OJ and isomaltooligosaccharides influence the intestine microbial populations, immune responses and resistance to white spot syndrome virus in shrimp (Litopenaeus vannamei). Aquaculture 291:35–40
Lin WR, Ng IS (2020) Development of CRISPR/Cas9 system in Chlorella vulgaris FSP-E to enhance lipid accumulation. Enzyme Microb Technol 133:109458
Liu B, Eltanahy EE, Liu H, Chua ET, Thomas-Hall SR, Wass T, Pan K, Schenk PM (2020b) Growth-promoting bacteria double eicosapentaenoic acid yield in microalgae. Bioresour Technol 316:123916
Liu J, Sommerfeld M, Hu Q (2013) Screening and characterization of Isochrysis strains and optimization of culture conditions for docosahexaenoic acid production. Appl Microbiol Biotechnol 97:4785–4798
Liu L, Hu Z, Li S, Yang H, Li S, Lv C, Zaynab M, Cheng CHK, Chen H, Yang X (2020a) Comparative transcriptomic analysis uncovers genes responsible for the DHA enhancement in the mutant Aurantiochytrium sp. Microorganisms 8:529
Peng HX, De-Bashan LE, Higgins BT (2021) Azospirillum brasilense reduces oxidative stress in the green microalgae Chlorella sorokiniana under different stressors. J Biotechnol 325:179–185
Rahiman KMM, Jesmi Y, Thomas AP, Hatha AAM (2010) Probiotic effect of Bacillus NL110 and Vibrio NE17 on the survival, growth performance and immune response of Macrobrachium rosenbergii (de Man). Aquac Res 41:e120–e134
Ringø E (2020) Probiotics in shellfish aquaculture. Aquac Fish 5:1–27
Sauvage J, Wikfors GH, Dixon MS, Kapareiko D, Sabbe K, Li X, Joyce A (2022) Bacterial exudates as growth-promoting agents for the cultivation of commercially relevant marine microalgal strains. J World Aquac Soc 53:1101–1119
Seymour JR, Amin SA, Raina JB, Stocker R (2017) Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat Microbiol 2:17065
Sirisuk P, Sunwoo I, Kim SH, Awah CC, Hun Ra C, Kim JM, Jeong GT (2018) Enhan-cement of biomass, lipids, and polyunsaturated fatty acid (PUFA) production in Nannochloropsis oceanica with a combination of single wavelength light emitting diodes (LEDs) and low temperature in a three-phase culture system. Bioresour Technol 270:504–511
Soltani M, Ghosh K, Hoseinifar SH, Kumar V, Lymbery AJ, Roy S, Ringø E (2019) Genus Bacillus, promising probiotics in aquaculture: aquatic animal origin, bio-active components, bioremediation and efficacy in fish and shellfish. Rev Fish Sci Aquac 27:331–379
Sonowal S, Ahmed R, Chikkaputtaiah C, Basar E, Velmurugan N (2022) A comprehensive characterization of culturable endophytic bacteria of Paris polyphylla and their potential role in microalgal growth in co-culture. Appl Soil Ecol 174:104410
Udayan A, Arumugam M (2017) Selective enrichment of eicosapentaenoic acid (20:5n–3) in N. oceanica CASA CC201 by natural auxin supplementation. Bioresour Technol 242:329–333
Wang YY, Xu SM, Cao JY, Wu MN, Lin JH, Zhou CX, Zhang L, Zhou HB, Li YR, Xu JL, Yan XJ (2022) Co-cultivation of Isochrysis galbana and Marinobacter sp. can enhance algal growth and docosahexaenoic acid production. Aquaculture 556:738248
Wei ZJ, Wang HN, Li X, Zhao QQ, Yin YH, Xi LJ, Qin S (2019) Enhanced biomass and lipid production by co-cultivation of Chlorella vulgaris with Mesorhizobium sangaii under nitrogen limitation. J Appl Phycol 32:233–242
Wu HJ, Sun LB, Li CB, Li ZZ, Zhang Z, Wen XB, Hu Z, Zhang YL (2014) Enhancement of the immune response and protection against Vibrio parahaemolyticus by indigenous probiotic Bacillus strains in mud crab (Scylla paramamosain). Fish Shellfish Immunol 41:156–162
Xie Y, Khoo KS, Chew KW, Devadas VV, Phang SJ, Lim HR, Rajendran S, Show PL (2022) Advancement of renewable energy technologies via artificial and microalgae photosynthesis. Bioresour Technol 363:127830
Yee CS, Okomoda VT, Hashim F, Waiho K, Sheikh Abdullah SR, Alamanjo C, Abu Hasan H, Muzalina Mustafa E, Kasan NA (2021) Marine microalgae co-cultured with floc-forming bacterium: Insight into growth and lipid productivity. PeerJ 9:e11217
Zheng H, Ge F, Song K, Yang Z, Li J, Yan F, Wu X, Zhang Q, Liu Y, Ruan R (2022) Docosahexaenoic acid production of the marine microalga Isochrysis galbana cultivated on renewable substrates from food processing waste under CO2 enrichment. Sci Total Environ 848:157654
Zhu BH, Tu CC, Shi HP, Yang GP, Pan KH (2017) Overexpression of endogenous delta-6 fatty acid desaturase gene enhances eicosapentaenoic acid accumulation in Phaeodactylum tricornutum. Process Biochem 57:43–49
Funding
This research was supported by Ningbo Science and Technology Research Projects, China (2019B10006), the earmarked fund for CARS-49, Ningbo Public Welfare Science and Technology Program (2023S123), the Natural Science Foundation of Ningbo Government (2023J109), and Zhejiang Provincial Department of Education Scientific Research Project (Y202249030).
Author information
Authors and Affiliations
Contributions
Jiayi Cao, Jilin Xu and Yijun Xu conceived of the study and participated in its design and coordination. Yijun Xu, Minnan Wu, Yingying Wang and Yanrong Li performed the research. Yijun Xu, Minnan Wu, Yingying Wang, Lin Zhang and Jiayi Cao designed and performed the bioinformatics and statistical analysis. Yijun Xu, Minnan Wu and Jiayi Cao wrote the first draft. All authors contributed to interpreting the data and writing the manuscript. The manuscript is approved by all authors for publications.
Corresponding authors
Ethics declarations
Ethics Approval
This article does not involve any studies with human participants or animal-based experiments.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, Y., Wu, M., Cao, J. et al. Enhancement of Docosahexaenoic Acid and Eicosapentaenoic Acid Biosynthesis in Isochrysis galbana by Bacillus jeotgali. Mar Biotechnol (2024). https://doi.org/10.1007/s10126-024-10337-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10126-024-10337-5