Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression via the recognition of their target messenger RNAs. MiR-10a-3p plays an important role in the process of ossification. In this study, we obtained the precursor sequence of miR-10a-3p in the pearl oyster Pinctada fucata martensii (Pm-miR-10a-3p) and verified its sequence by miR-RACE technology, and detected its expression level in the mantle tissues of the pearl oyster P. f. martensii. Pm-nAChRsα and Pm-NPY were identified as the potential target genes of Pm-miR-10a-3p. After the over-expression of Pm-miR-10a-3p, the target genes Pm-nAChRsα and Pm-NPY were downregulated, and the nacre microstructure became disordered. The Pm-miR-10a-3p mimic obviously inhibited the luciferase activity of the 3′ untranslated region of the Pm-NPY gene. When the interaction site was mutated, the inhibitory effect disappeared. Our results suggested that Pm-miR-10a-3p participates in nacre formation in P. f. martensii by targeting Pm-NPY. This study can expand our understanding of the mechanism of biomineralization in pearl oysters.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs), the noncoding single-stranded RNA molecules encoded by endogenous genes that are approximately 20–24 nucleotides in length, participate in post-transcriptional gene expression regulation (Khan et al. 2009). They downregulate their target genes mainly through complementary pairing with the 3′ untranslated regions (UTRs) of messenger RNAs (mRNAs). MiR-1990c-3p and miR-9a-3p in pearl oyster may influence shell formation by controlling matrix proteins or mineralized cells (Huang et al. 2019). MiR-29a regulates the immune response and nacreous formation of pearl oysters by regulating Y2R (Tian et al. 2015). Previous studies have shown that miRNAs are associated with biological mineralization (Jiao et al. 2014). A growing number of scholars are paying attention to the miR-10 family because of its protective effect and role in the Hox cluster of developmental regulators (Lund 2010). Decreased expression of miR-10a-3p can promote osteogenic differentiation of BMSC to improve osteoporosis (Liu et al. 2021). The misregulation of miR-10a-3p may cause acute myeloid leukemia (Yu et al. 2016; Li et al. 2013), and participates in ossification, and promotes osteogenic differentiation and mineral deposition of the posterior ligament (Xu et al. 2018, 2019). MiR-10a-3p has also been reported to be associated with the development of adult acute myeloid leukemia by targeting SLC14A1, ARHGAP5, and PIK3CA (Chen et al. 2020), and it also involved in cholesterol metabolism, extracellular matrix degradation, and primary chondrocyte inflammation (Li et al. 2021). The downregulation of miR-10a-3p may be associated with bone degeneration (Kmetzsch et al. 2021).
The pearl oyster Pinctada fucata martensii is an economically important shellfish. Its natural population is mainly distributed in southern China and Japan. Shell formation by mollusks is a typical biological mineralization process. Studying the mechanism of biological mineralization helps understand the pearl-producing mechanism of pearl shellfish. Another important reason to study shell mineralization is that biological minerals have incomparable material advantages over inorganic minerals. The mantle is the main organ of shell formation, the protein produced and secreted, and the free calcium carbonate ions constantly combine to form the nacreous layer and prism layer. The nacre layer of the pearl shell, a product of biomineralization, is composed of aragonite crystals and organic matrix (including chitin and proteins) that are arranged in an orderly staggered manner (Addadi and Weiner 1997). The formation of molluskan shells is generally believed to be related to the mantle, and the biomineralization abilities of different areas of the mantle differ (Zhang et al. 2020). The mantle central (MC) plays a crucial part in the formation of the nacre layer, while the mantle edge (ME) is mainly connected to the formation of the prism layer (Marin et al. 2012). The mechanism of biomineralization is essential for the adaptation of P. f. martensii (He et al. 2021), and many biomineralization-related proteins, such as nacrein, MSI-60, Pif, dermatopontin, BMP7, and PmCHST1b, were detected and functionally analyzed (Zheng et al. 2017; Sudo et al. 1997; Jiao et al. 2012; Suzuki et al. 2009; Yan et al. 2014; Hao et al. 2018). From 2012, we began to focus on miRNAs associated with mineralization and found that miR-2305, miR-29a, Pm-miR-124, Pm-miR-9a-5p, and miR-29b were involved in nacre formation (Jiao et al. 2015; Tian et al. 2015; Zhang et al. 2021). Our previous study also showed that miR-10a-3p of P. f. martensii (Pm-miR-10a-3p) is highly expressed in the MC compared with ME (Zhang et al. 2021). Therefore, we speculated that Pm-miR-10a-3p may have potential functions in nacre formation. This study aims to verify the role and mechanism of Pm-miR-10a-3p in the formation of nacre in the pearl oyster P. f. martensii.
Material and Methods
Experimental Samples and RNA Extraction
In this experiment, adult P. f. martensii (approximately 2 years old) pearl oysters were collected from Xuwen County, Zhanjiang City, Guangdong Province, and pretreated in circulating water (25–30 ℃) for 2 days. The ME and MC of the pearl oysters were stored in liquid nitrogen immediately after collection. Small RNA was extracted from the mantle tissues by using RNAiso for small RNA (Takara, Dalian, China), and total RNA was extracted from all previously treated tissues by using TRIzol (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions.
miR-RACE and qRT-PCR
The mature Pm-miR-10a-3p sequence was obtained from the miRNA transcriptome database of P. f. martensii and has been uploaded to SAR (Accession no. PRJNA628844). Poly(A) polymerase (Takara) and 5′ adaptors (CGACUGGAGCACGAGGACACUGAAAA) were used to tail and adapt small RNA. After reverse transcription, 3′ and 5′ RACE-specific primers for PCR amplification were designed. All fragments were cloned into pMD-19 T and sequenced. Complementary DNA (cDNA) was specifically reverse transcribed by using stem-loop primers in accordance with the instructions of the M-MLV kit (Takara). Then, the cDNA template was diluted 100-fold, and stem-loop qRT-PCR was utilized with U6 snRNA as the internal reference gene to evaluate the differential expression of Pm-miR-10a-3p in the ME and MC. The expression of the target genes was quantified by using qRT-PCR with GAPDH as the internal reference. Thermo Scientific DyNAmo Flash SYBR Green qPCR Kit (Thermo Scientific, Shanghai, China) was applied by following the manufacturer’s protocol. Each sample was run in triplicate along with the reference gene. The primers were designed as previously described (Caifu et al. 2005) and are listed in Table 1.
miR-10a-3p Sequence Analysis
M-FOLD program (http://unafold.rna.albany.edu/?q=mfold/DNA-Folding-Form) was used to predict the secondary structure of Pm-miR-10a-3p. The miR-10a-3p sequence of Sus scrofa (MIMAT0022954), Capra hircus (MIMAT0035912), Gallus gallus (MIMAT0007732), Macaca mulatta (MIMAT0026797), Mus musculus (MIMAT0004659), Columba livia (MIMAT0038405), Homo sapiens (MIMAT0004555), Danio rerio (MIMAT0003391), Branchiostoma floridae (MIMAT0009467), Branchiostoma belcheri (MIMAT0031573), and Melibe leonine (MIMAT0048709) was downloaded from miRBase (http://www.mirbase.org/), and CLUSTALW (https://www.genome.jp/tools-bin/clustalw) was utilized for multiple sequence alignment. In accordance with the transcriptome database, mRNAs related to biomineralization or immunity were selected as the targets for analysis (miRanda and RNAhybrid). A low free (< − 10 kcal/mol) energy between mRNA and miRNA was considered to be indicative of potential interactions between the mRNA and miRNA. ClustalW was used for multiple sequence alignment.
Over-expression of miR-10a-3pIn Vivo
The blank control group (RNase-free water), the negative control group (negative control mimic), and experimental group (Pm-miR-10a-3p mimic) were set up. The mimics required for the experiment were synthesized by GenePharma, Shanghai, China. One-year-old pearl oysters with similar sizes were randomly divided into three injection groups, thirty in each group. The blank control group was injected with 100 μL RNase-free water, and the experimental group and NC group were injected with 10 μg Pm-miR-10a-3p and negative control mimic (100 μL) each time. Injection was conducted twice, and the interval between the first and second injection was 4 days. Four days after the second injection, the mantle tissues of the samples were cut into small pieces and quickly frozen in a liquid nitrogen tank. After the shell was cut, its internal surface was observed with a FEIQuanta200 scanning electron microscope.
Vector Construction
The 3′ UTRs of Pm-nAChRsα and Pm-NPY were obtained by using PrimeSTAR HS DNA Polymerase (Takara). The restriction enzymes SpeI and HindIII were used to insert the 3′ UTR sequence into the pMIR-reporter plasmid (Ambion). The Pm-NPY mutant plasmid was constructed by randomly changing the Pm-miR-10a-3p target site ACGAATT in the 3′ UTR of Pm-NPY to TCCATTA. First, the recombinant plasmid (pMIR-reporter-3′ UTR/Pm-NPY) was extracted by using an Endo-Free Plasmid Mini Kit (OMEGA). After obtaining the plasmid template, a mutant target gene fragment was obtained by using PCR. The result of vector construction was determined by sequencing. The primers used in this experiment are listed in Table 1.
Cell Culture and Transfection
HEK-293 T cells were cultured in Dulbecco’s modified Eagle’s medium with 10% peptide bovine serum in a 37 ℃ CO2 incubator (providing 5% CO2) under high-glucose conditions to provide the proper environment for cell culture. Plasmid transfection was carried out by strictly following the manufacturer’s instructions for Lipofectamine 2000. One day before transfection, HEK-293 T cells in the logarithmic growth phase were inoculated into 48-well culture plates, at the density of 5 × 104 per hole with 500 μL of medium. When the cells reached over 70–80% confluence, the previous culture medium was replaced with fresh medium. Then, the constructed pMIR-reporter plasmid was co-transfected with miRNA or negative control mimics. Moreover, each well was transfected with 4 ng of plasmid pRL-TK vector as an internal quality control to correct the differences among the groups accurately. After 48 h of transfection, the cells were lysed, and luciferase activity was detected by using a dual-luciferase detection kit (Promega, Fitchburg, WI, USA) (Jiao et al. 2020).
Statistical Analysis
SPSS software (IBM, Chicago, IL, USA) was utilized for one-way analysis of variance for statistical analysis. All the data were presented as mean ± SD. A difference with p < 0.05 was deemed to be statistically significant by Duncan’s multiple comparison test.
Results
Sequence Analysis of miR-10a-3p
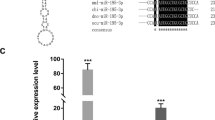
MiR-RACE technology was used to verify the sequence of mature Pm-miR-10a-3p (Fig. 1a). Then, we obtained the secondary stem-loop structure of Pm-miR-10a-3p by using M-fold. Pm-pre-miR-10a-3p had a typical stem-loop structure with the length of 79 bp, and its mature sequence started from the 3′ arm (Fig. 1b). The multiple comparative analysis of the mature Pm-miR-10a-3p sequences and miR-10a-3p of other species suggested that all analyzed miRNAs had the same nucleotide sequences in their seed regions (Fig. 1c).
Sequence analysis of miR-10a-3p. a Pm-miR-10a-3p-RACE sequencing result. Red nucleotides indicate mature sequences. b Secondary structure of Pm-miR-10a-3p was obtained by M-fold, and the annotation is based on either p-num or ss-count information. Red line: most likely to be single stranded; purple line: least likely to be single stranded. Bold bases indicate mature sequences. c Multi-alignment of mature Pm-miR-10a-3p with that from other species (ssc, Sus scrofa; chi, Capra hircus; gga, Gallus gallus; mml, Macaca mulatta; mmu, Mus musculus; cli, Columba livia; has, Homo sapiens; dre, Danio rerio; bfl, Branchiostoma floridae; bbe, Branchiostoma belcheri; mle, Melibe leonine). Conserved nucleotides of all animals are represented on a gray background
Functions of miR-10a-3p in Nacre Layer Formation
Stem-loop qRT-PCR was utilized to detect the expression of Pm-miR-10a-3p in the ME and MC of P. f. martensii. The results showed that Pm-miR-10a-3p was differentially expressed in different tissues of the mantle and was especially highly expressed in the MC compared with ME (Fig. 2a/Supplement Fig. 2). This expression pattern suggested that Pm-miR-10a-3p may be involved in nacre formation. We injected Pm-miR-10a-3p mimics into the adductor muscle of P. f. martensii to verify the mechanism of Pm-miR-10a-3p in pearl oysters. The expression levels of Pm-miR-10a-3p respectively increased by 2.2- and 1.7-fold in the ME (Fig. 2b) and 1.9- and 1.5-fold in the MC (Fig. 2c) in the mimic injection group compared with that in the blank and NC groups. Then, we took the vigorously growing part of the junction area between the prismatic layer and the nacre layer of the shell for the observation of inner surface morphology by using SEM (Supplement Fig. 1). In the NC and blank groups, the boundaries between aragonite crystals were clear, and single crystals were nearly hexagonal and uniform in size in the NC and blank groups. However, in the experimental group, the internal surface microstructure of the shell nacre layer exhibited disordered growth, and the crystal shape was fragmented (Fig. 2d). The above results indicated that Pm-miR-10a-3p has a vital influence on nacre layer formation.
Function verification of Pm-miR-10a-3p. a The expression of Pm-miR-10a-3p in ME and MC under normal conditions. b, c The expression of Pm-miR-10a-3p in ME/MC after mimic injection. Different letters mean a significant difference (p < 0.05). Error bars correspond to mean ± SD. d SEM images of the nacre layer in NC, blank, and mimic injection groups, respectively. The bars are 5 μm in the nacre images. The blank control group was injected with 100 μL RNase-free water, and the experimental group and NC group were injected with 10 μg Pm-miR-10a-3p and negative control mimic (100 μL) each time
Target Prediction
Two bioinformatics tools (miRanda and RNAhybrid) were utilized to predict the target genes of Pm-miR-10a-3p in the pearl oyster P. f. martensii. As shown in Fig. 3, Pm-nAChRsα and Pm-NPY were predicted as the potential targets of Pm-miR-10a-3p. Then, the effects of the over-expression of Pm-miR-10a-3p on the potential target genes Pm-nAChRsα and Pm-NPY were detected by using qRT-PCR. The expression levels of the Pm-nAChRsα and Pm-NPY genes in the MC group had reduced by 80% relative to those in the control group (Fig. 4a, b). In the ME, the expression of Pm-nAChRsα did not change significantly, whereas that of Pm-NPY had reduced by 76% (Fig. 4c, d).
Effect of Pm-miR-10a-3p on target genes in MC and ME. a, b The relative expression of Pm-nAChRsα/Pm-NPY gene in MC after Pm-miR-10a-3p mimic injection. c, d The relative expression of Pm-nAChRsα/Pm-NPY gene in ME after Pm-miR-10a-3p mimic injection. GAPDH gene was used as the reference gene. Different letters mean a significant difference (p < 0.05). Error bars correspond to mean ± SD. The blank control group was injected with 100 μL RNase-free water, and the experimental group and NC group were injected with 10 μg Pm-miR-10a-3p and negative control mimic (100 μL) each time
Target Verification
We performed dual-luciferase reports by using a luciferase reporter containing the 3′ UTRs of Pm-nAChRsα and Pm-NPY to verify that the target gene is negatively regulated by Pm-miR-10a-3p. We also constructed the Pm-NPY mutation vector to validate the specificity of miRNA target sites. We randomly changed the sequence ACGAATT at the NPY target of Pm-miR-10a-3p to TCCATTA. Pm-miR-10a-3p mimic or the negative control were transfected into HEK-293 T cells with the reporter plasmids. The transfected cells were treated through incubation for 24 h, and luciferase was detected. The relative luciferase activity of the reporter containing the 3′ UTR of the Pm-NPY had was decreased by approximately 29% (Fig. 5a, b), whereas the Pm-miR-10a-3p mimic had no obvious effect on the Pm-NPY mutation vector. These results showed that Pm-NPY could be regulated by Pm-miR-10a-3p.
Target verification of Pm-miR-10a-3p. a, b Target verification between Pm-miR-10a-3p and target genes. Pm-miR-10a-3p mimic obviously inhibited the luciferase activity of the 3′ UTR of the Pm-NPY gene detected by dual-luciferase analysis. Different letters mean a significant difference (p < 0.05). Error bars correspond to mean ± SD
Discussion
In mammals, miRNA-10a-3p is considered to be a myelopoiesis-associated miRNA, and miR-10a-3p can efficiently inhibit ID3 expression level, thus promoting mineral deposition and the osteogenic differentiation of the posterior ligaments (Xu et al. 2018). In patients with osteoporosis, miR-10a-3p is expressed at levels that are significantly higher than normal (Jin et al. 2018). In consideration of the homology between bone formation and pearl formation, and conservation of miRNA in sequence and function, we hypothesized that miR-10a-3p is associated with shell formation in pearl oysters. We are interested in Pm-miR-10a-3p because it is differentially expressed in the MC and ME of pearl oysters. The high expression of Pm-miR-10a-3p in the MC indicates that Pm-miR-10a-3p is likely to participate in nacre formation. Our research showed that after mimic injection, the nacre microstructure became disordered and the crystal shape became irregular and fragmented. This result demonstrated that Pm-miR-10a-3p can efficiently regulate the formation of nacre.
As an important pathway in neuroimmune regulation, cholinergic anti-inflammatory pathway can inhibit the release of proinflammatory factors by macrophages and inhibit inflammatory response. NAChRs are a signaling ion channel protein between cholinergic synapses, which is activated after binding to acetylcholine or nicotine and inhibits macrophages form producing TNF, IL and other inflammatory factors through cell signal transduction to prevent excessive inflammatory responses in the body. Acetylcholine plays a crucial regulatory role in bone turnover through the nervous system. The nAChR subunit has been found in primary human osteocyte cultures and human bone biopsy sites (Rothem et al. 2009). NAChR subunit α9 mRNA was highly expressed in bone marrow cells of developing juvenile and adult rats (Baumann et al. 2019). The lack of α7 nAChR in mice can lead to increased bone mass and reduced osteoclast production (Mandl et al. 2013). In addition, in mammals, nAChR is mainly distributed in the nervous system and participates in regulating cell excitation, neurotransmitter release, and neuronal integration. This role is essential for regulating networks and also have effects on anxiety, food intake, memory attention, and stress response (Changeux and Edelstein 2001). A growing body of research has shown that nAChR has immune suppression effects in mollusks (Yu et al. 2021). Pm-nAChRsα is one of Pm-miR-10a-3p’s target genes as predicted by miRanda and RNAhybrid. When Pm-miR-10a-3p was highly expressed in P. f. martensii, the expression of Pm-nAChRsα in the MC obviously reduced significantly. However, in the dual-luciferase experiment, Pm-miR-10a-3p could not inhibit the 3′ UTR of Pm-nAChRsα. We hypothesized that Pm-miR-10a-3p may indirectly target Pm-nAChRsα expression and participate in the regulation of the choline anti-inflammatory pathway.
NPY plays an important role as a transmitter in regulating inflammatory response and maintaining homeostasis in the neuroimmune regulatory system. NPY not only participates in nerve and immune activities, but also participates in the regulation of bone formation in human body, affecting the active bone metabolism, such as periosteum surface and bone marrow, and participating in bone formation and injury repair. NPY regulates the transport of bone nutrients and bone formation matrix by regulating blood flow within blood vessels on the periosteum surface (Yi et al. 2018). It can also bind to the surface parathyroid hormone receptor of osteoclasts to inhibit osteoclast activity and promote bone formation. NPY has an important role in some physiological functions, such as circadian rhythm, food intake, energy homeostasis, and cognition, and is also considered to be an important component of the stress response (Reichmann and Holzer 2016). In addition, NPY is a critical integrator of bone homeostatic signals, and when hypothalamus NPY expression is low, mice adjust their bone mass in accordance with “starvation” conditions (Baldock et al. 2009). The NPY receptor system plays a decisive role in bone regulation (Allison et al. 2007) and shields against excessive stress‐induced bone loss (Baldock et al. 2014). NPY not only regulates bone formation and absorption directly, but also affects bone formation through intestinal microbiota (Chen and Zhang 2022). In general, the biological mineralization of NPY does not occur independently, but is jointly regulated by the nervous and immune systems, which influence each other. Although shellfish have a simpler nervous and immune system than mammals, they also have a complete processing pathway for stimulus signals. Double luciferase experiments and miRNA overexpression analysis proved that Pm-miR-10a-3p can directly regulate the expression of Pm-NPY in the mantle of pearl oysters. When Pm-miR-10a-3p was highly expressed in pearl oysters, the expression level of Pm-NPY in the mantle decreased, and the nacre became disordered. The Pm-miR-10a-3p mimics apparently hampered the luciferase activity of the 3′ UTR of the Pm-NPY gene. On the basis of these data, we can speculate that in pearl oysters, Pm-miR-10a-3p participates in shell biomineralization by regulating the expression of Pm-NPY.
Shellfish biomineralization and crystallization are complex biological processes involving the synergy of multiple systems. Previous studies have shown that Pm-miR-2305 participates in nacre Formation by targeting pearlin in the pearl oyster Pinctada fucata martensii (Jiao et al. 2015). Pm-miR-29a overexpression leads to downregulation of Y2R and affects the nacre formation (Tian et al. 2015). Pm-miR-124 involved in nacre formation by targeting Pm-nAChRsα and PfCHS (Zhang et al. 2021). In this study, Pm-NPY was evidenced as one target of Pm-miR-10a-3p. NPY can control the transport of bone nutrients and the formation of the bone matrix by regulating blood flow in the blood vessels on the surface of the periosteum or by binding to the parathyroid hormone receptor on the surface of osteoclasts to inhibit osteoclast activity and promote bone formation (Lindblad et al. 1994). These results showed that biomineralization requires the participation of multiple systems. The immunoregulatory role of miRNAs in pearl production is also worthy of further study.
In conclusion, the mature sequence of Pm-miR-10a-3p was verified by miR-RACE technology, and its expression levels in the mantle tissues of the pearl oyster P. f. martensii were detected. The results of functional analysis revealed that Pm-miR-10a-3p engages in nacre formation in the pearl oyster P. f. martensii. All the results demonstrated that Pm-miR-10a-3p participates in nacre formation in P. f. martensii by targeting Pm-NPY.
Data Availability
Methods, materials, and data used in this study are fully delineated in the text.
References
Addadi L, Weiner S (1997) A pavement of pearl. Nature 389:912–913
Allison SJ, Baldock PA, Herzog H (2007) The control of bone remodeling by neuropeptide Y receptors. Peptides 28:320–325
Baldock PA, Lee NJ, Driessler F, Lin S, Allison S, Stehrer B, Lin EJ, Zhang L, Enriquez RF, Wong IP, McDonald MM, During M, Pierroz DD, Slack K, Shi YC, Yulyaningsih E, Aljanova A, Little DG, Ferrari SL, Sainsbury A, Eisman JA, Herzog H (2009) Neuropeptide Y knockout mice reveal a central role of NPY in the coordination of bone mass to body weight. PloS one 4:e8415
Baldock PA, Lin S, Zhang L, Karl T, Shi Y, Driessler F, Zengin A, Hörmer B, Lee NJ, Wong IP, Lin EJ, Enriquez RF, Stehrer B, During MJ, Yulyaningsih E, Zolotukhin S, Ruohonen ST, Savontaus E, Sainsbury A, Herzog H (2014) Neuropeptide Y attenuates stress-induced bone loss through suppression of noradrenaline circuits. J Bone Miner Res 29:2238–2249
Baumann L, Kauschke V, Vikman A, Dürselen L, Krasteva-Christ G, Kampschulte M, Heiss C, Yee KT, Vetter DE, Lips KS (2019) Deletion of nicotinic acetylcholine receptor alpha9 in mice resulted in altered bone structure. Bone 120:285–296
Changeux J, Edelstein SJ (2001) Allosteric mechanisms in normal and pathological nicotinic acetylcholine receptors. Curr Opin Neurobiol 11:369–377
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33:e179
Chen QC, Zhang Y (2022) The role of NPY in the regulation of bone metabolism. Front Endocrinol 13:833485
Chen S, Chen Y, Zhu Z, Tan H, Lu J, Qin P, Xu L (2020) Identification of the key genes and microRNAs in adult acute myeloid leukemia with FLT3 mutation by bioinformatics analysis. Int J Med Sci 17:1269–1280
Hao R, Zheng Z, Wang Q, Du X, Deng Y, Huang R (2018) Molecular and functional analysis of PmCHST1b in nacre formation of Pinctada fucata martensii. Comp Biochem Physiol B Biochem Mol Biol 225:13–20
He G, Liu X, Xu Y, Liang J, Deng Y, Zhang Y, Zhao L (2021) Repeated exposure to simulated marine heatwaves enhances the thermal tolerance in pearl oysters. Aquat Toxicol 239:105959
Huang S, Ichikawa Y, Yoshitake K, Kinoshita S, Igarashi Y, Omori F, Maeyama K, Nagai K, Watabe S, Asakawa S (2019) Identification and characterization of microRNAs and their predicted functions in biomineralization in the pearl oyster (Pinctada fucata). Biology 8:47
Jiao Y, Zheng Z, Tian R, Du X, Wang Q, Huang R (2015) MicroRNA, Pm-miR-2305, participates in nacre formation by targeting pearlin in pearl oyster Pinctada martensii. Int J Mol Sci 16:21442–21453
Jiao Y, Gu Z, Luo S, Deng Y (2020) Evolutionary and functional analysis of MyD88 genes in pearl oyster Pinctada fucata martensii. Fish Shellfish Immunol 99:322–330
Jiao Y, Zheng Z, Du X, Wang Q, Huang R, Deng Y, Shi S, Zhao X (2014) Identification and characterization of microRNAs in pearl oyster Pinctada martensii by Solexa deep sequencing. Mar Biotechnol (NY) 16:54–62
Jiao Y, Wang H, Du X, Zhao X, Wang Q, Huang R, Deng Y (2012) Dermatopontin, a shell matrix protein gene from pearl oyster Pinctada martensii, participates in nacre formation. Biochem Biophys Res Commun 425:679–83
Jin D, Wu X, Yu H, Jiang L, Zhou P, Yao X, Meng J, Wang L, Zhang M, Zhang Y (2018) Systematic analysis of lncRNAs, mRNAs, circRNAs and miRNAs in patients with postmenopausal osteoporosis. Am J Transl Res 10:1498–1510
Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS (2009) Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol 27:549–555
Kmetzsch V, Anquetil V, Saracino D, Rinaldi D, Camuzat A, Gareau T, Jornea L, Forlani S, Couratier P, Wallon D, Pasquier F, Robil N, de la Grange P, Moszer I, Le Ber I, Colliot O, Becker E (2021) Plasma microRNA signature in presymptomatic and symptomatic subjects with C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 92:485–493
Li J, Dong J, Zhang Zh, Zhang DC, You XY, Zhong Y, Yun Z, Chen MS, Liu SM (2013) miR-10a restores human mesenchymal stem cell differentiation by repressing KLF4. J Cell Physiol 228:2324–2336
Li W, Ding X, Zhao R, Xiong D, Xie Z, Xu J, Tan M, Li C, Yang C (2021) The role of targeted regulation of COX11 by miR-10a-3p in the development and progression of paediatric Mycoplasma pneumoniae pneumonia. J Thorac Dis 13:5409–5418
Lindblad BE, Nielsen LB, Jespersen SM, Bjurholm A, Bünger C, Hansen ES (1994) Vasoconstrictive action of neuropeptide Y in bone: the porcine tibia perfused in vivo. Acta Orthop Scand 65:629–634
Liu H, Yi X, Tu S, Cheng C, Luo J (2021) Kaempferol promotes BMSC osteogenic differentiation and improves osteoporosis by downregulating miR-10a-3p and upregulating CXCL12. Mol Cell Endocrinol 520:111074
Lund AH (2010) miR-10 in development and cancer. Cell Death Differ 17:209–214
Mandl P, Hayer S, Huck S, Niederreiter B, Scholze P, Sykoutri D, Smolen JS, Redlich K (2013) AB0138 mice lacking the alpha 7 nicotinic acetylcholine receptor have increased bone mass. Ann Rheum Dis 71:645–645
Marin F, Le Roy N, Marie B (2012) The formation and mineralization of mollusk shell. Front Biosci 4:125
Reichmann F, Holzer P (2016) Neuropeptide Y: a stressful review. Neuropeptides 55:99–109
Rothem DE, Rothem L, Soudry M, Dahan A, Eliakim R (2009) Nicotine modulates bone metabolism-associated gene expression in osteoblast cells. J Bone Miner Metab 27:555–561
Sudo S, Fujikawa T, Nagakura T, Ohkubo T, Sakaguchi K, Tanaka M, Nagashima K, Takahashi T (1997) Structure of mollusk shell framework proteins. Nature 387:563–564
Suzuki M, Saruwatari K, Kogure T, Yamamoto Y, Nishimura T, Kato T, Nagasawa H (2009) An acidic matrix protein, Pif, is a key macromolecule for nacre formation. Science 325:1388–2139
Tian R, Zheng Z, Huang R, Jiao Y, Du X (2015) miR-29a participated in nacre formation and immune response by targeting Y2R in Pinctada martensii. Int J Mol Sci 16:29436–29445
Xu C, Zhang H, Zhou W, Wu H, Shen X, Chen Y, Liao M, Liu Y, Yuan W (2019) MicroRNA-10a,-210, and-563 as circulating biomarkers for ossification of the posterior longitudinal ligament. Spine Journal 19:735–743
Xu C, Zhang H, Gu W, Wu H, Chen Y, Zhou W, Sun B, Shen X, Zhang Z, Wang Y, Liu Y, Yuan W (2018) The microRNA-10a/ID3/RUNX2 axis modulates the development of ossification of posterior longitudinal ligament. Sci Rep 8:1–13
Yan F, Luo S, Jiao Y, Deng Y, Du X, Huang R, Wang Q, Chen W (2014) Molecular characterization of the BMP7 gene and its potential role in shell formation in Pinctada martensii. Int J Mol Sci 15:21215–21228
Yu J, Lu X, Zhang M, Cao Y, Du X (2021) Metabolomic analyses reveal the crucial metabolites involved in the transplantation response of pearl oyster Pinctada fucata martensii. Aquacul Rep 20:100717
Yi M, Li H, Wu Z, Yan J, Liu Q, Ou C, Chen M (2018) A promising therapeutic target for metabolic diseases: neuropeptide Y receptors in humans. Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology 45:88–107
Yu Z, Li D, Ju XL (2016) CD4+ T cells from patients with acute myeloid leukemia inhibit the proliferation of bone marrow-derived mesenchymal stem cells by secretion of miR-10a. J Cancer Res Clin Oncol 142:733–740
Zhang J, Luo S, Gu Z, Deng Y, Jiao Y (2020) Genome-wide DNA methylation analysis of mantle edge and mantle central from pearl oyster Pinctada fucata martensii. Mar Biotechnol (NY) 22:380–390
Zhang Y, Jiao Y, Li Y, Tian Q, Du X, Deng Y (2021) Comprehensive analysis of microRNAs in the mantle central and mantle edge provide insights into shell formation in pearl oyster Pinctada fucata martensii. Comp Biochem Physiol B Biochem Mol Biol 252:110508
Zheng Z, Du X, Xiong X, Jiao Y, Deng Y, Wang Q, Huang R (2017) PmRunt regulated by Pm-miR-183 participates in nacre formation possibly through promoting the expression of collagen VI-like and nacrein in pearl oyster Pinctada martensii. PLoS One 12:e0178561
Acknowledgements
The research was supported by the Fishery College, Guangdong Ocean University, China.
Funding
This work was supported by the Science and Technology Program of Guangdong Province (Grant Nos. 2021A1515011199 and 2021B0202020003), the Department of Education of Guangdong Province (Grant Nos. 2021KTSCX041, 2020ZDZX1045, and 2021KCXTD026), the Department of Agriculture and Rural Affairs of Guangdong Province (Grant No. 2021KJ146), and the China Agriculture Research System of MOF and MARA.
Author information
Authors and Affiliations
Contributions
Min Yang, Yu Jiao, and Yue wen Deng designed and supervised the project. Min Yang, Xin lei Li, and Yu ting Zhang performed the experiments. Xin lei Li analyzed these data. Min Yang organized the data and wrote the manuscript. Yu Jiao and Yue wen Deng gave valuable advice for the modification of this paper. All authors commented on the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
The animal study was reviewed and approved by the Fishery College, Guangdong Ocean University (Zhanjiang, China).
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10126_2023_10216_MOESM1_ESM.jpg
Supplement Fig 1. The detection position of SEM in the shell. The position of the red frame represents the position observed by the electron microscope (JPG 517 KB)
10126_2023_10216_MOESM2_ESM.jpg
Supplement Fig 2. Expression of Pm-miR-10a-3p in different tissues. GI, gill; HE, hepatopancreas; A, adductor muscle; F, foot; M, mantle (JPG 646 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, M., Li, X.l., Zhang, Y.t. et al. miR-10a-3p Participates in Nacre Formation in the Pearl Oyster Pinctada fucata martensii by Targeting NPY. Mar Biotechnol 25, 428–437 (2023). https://doi.org/10.1007/s10126-023-10216-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-023-10216-5