Abstract
Nrf2 is an important transcription factor involved in the antioxidant response and is widely expressed in animal tissues. The function of Nrf2 is regulated by its negative regulator Keap1 by inducing its cytoplasmic degradation. Recent studies have suggested that Nrf2 is also regulated post-transcriptionally via miRNAs. However, to date, how miRNAs regulate Nrf2 in fish skeletal muscles is unknown. In this study, the full-length cDNAs with 2398 bp of the Nrf2 was firstly cloned by SMART RACE amplification tools from Chinese perch. The Nrf2 gene structure and its 3’-UTR region for possible miRNA binding sites, as well as its spatial expression profile were assayed. Then, we employed TargetScan Fish tool MiRNAnome to predict putative sites for five miRNAs including miR-181a-5p, MiR-194a, MiR-216a, miR-459-5p, and miR-724. Using qRT-PCR assay, we found that Nrf2 mRNA levels have negative correlation with all five miRNAs expression in muscle of nutritionally deprived fish, and that ectopic expression of miR-181a-5p alone reduces Nrf2 mRNA levels. Luciferase reporter assay in a heterologous cell system revealed that each of the five miRNAs reduced Nrf2 expression, suggesting a direct regulatory mechanism. Moreover, the miR-181a-5p suppression using specific antagomir led to a significant increase in Nrf2 expression in vivo. At the same time, the expression levels of the antioxidant enzymes CAT, ZnSOD, GPx, GSTA, and GSTA genes increased significantly after injecting miR-181a-5p antagomir. Taken together, these findings provide evidence that miRNAs are involved in the Nrf2 signaling networks in regulation of oxidative stress in fish, at least in Chinese perch muscle.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nrf2, an important transcription factor involved in the antioxidative response, is widely expressed in animal tissues, but is only activated through a series of oxidative and electrophilic stimuli (including ROS, some antioxidants, heavy metals, and certain disease processes) (Ma 2008, Kensler et al. 2007). Excessive accumulation of ROS caused by oxidative stress increases cellular autophagy levels, including the antioxidant signaling pathway Nrf2-Keap1. In its static form, Nrf2 protein is localized in the cytoplasm and rapidly transferred through a specific ubiquitin-26S proteasome pathway controlled by Keap1/Cul3-dependent ubiquitin ligase (E3) (Kobayashi et al. 2004, He et al. 2006, Kusik et al. 2008). A chemical-protein thiol interaction between an inducer and the Keap1 protein causes signal transduction, leading to the stabilization and activation of the Nrf2 protein (Dinkova-Kostova et al. 2002). Nrf2 binds to antioxidant response elements (AREs) in the promoter of the target antioxidant genes, such as NAD (P) H-quinone oxidoreductase 1 (NQO1), and strictly regulates their transcription (Ma et al. 2004, He et al. 2006, Nguyen et al. 2003, Itoh et al. 1997). Other antioxidant enzymes included blood oxygenase-1, SOD, catalase, reduced glutathione (GSH), and NADPH-quinone oxidoreductase (Liu et al. 2015). Nrf2 contains a conserved cysteine in the DNA-binding domain (Cys-514) (Wang et al. 2013), and it is necessary to protect the heart from glucose-induced oxidative stress and cardiomyopathy in rat. Loss of Nrf2 function in rat cardiac muscle cells significantly increased high glucose-induced oxidative stress and apoptosis, and in turn reduced contractility (He et al. 2009). Mechanistically, Nrf2 regulates basal expression and induction of regulated cytoprotective genes in primary cardiomyocytes, in vivo cardiomyocytes, and in vitro in cardiomyoblast cell line H9C2 (He et al. 2009). Muthusamyet et al. reported that Nrf2-antioxidant response signals in the myocardium can be activated by continuous exercise for 2 days, which may be a non-pharmacological method of activating the antioxidant pathway (Muthusamy et al. 2012). Previous research found that glucagon-like peptide 1 (GLP-1) can enhance Nrf2 activation in endothelial cells (Jaiswal 2004), suggesting that there may be a correlation between dietary protein levels and fish muscle antioxidant enzymes and related signaling pathways. However, the relationship between starvation-induced H2O2 production and Nrf2 signaling/antioxidant enzymes in fish skeletal muscle has not been studied.
Nrf2 expression can be regulated by two mechanisms, Keap1-dependent and Keap1-independent. It has been determined that the negative regulator Keap1 binds and isolates Nrf2, resulting in ubiquitination and proteasome degradation of Nrf2 (Wakabayashi et al. 2004, Kobayashi and Yamamoto 2006). Inactivation of Keap1 releases Nrf2, leading to nuclear translocation of Nrf2 and subsequent activation of Nrf2-dependent gene transcription (Niture et al. 2010). Nrf2 is also regulated by aromatic hydrocarbon receptor (AhR) transcription (Miao et al. 2005). In addition, previous studies have shown that ectodermal cortex 1 (ENC1) inhibits Nrf2 expression in a Keap1-independent manner by reducing Nrf2 protein synthesis without affecting Nrf2 transcription or ubiquitination of Nrf2 protein (Wang and Zhang 2009). In a mouse prostate transgenic adenocarcinoma (TRAMP) model, epigenetic mechanisms (DNA methylation and histone deacetylation) contribute to Nrf2 gene silencing (Yu et al. 2010). These studies indicated that in addition to Keap1 post-translationally regulating Nrf2 expression, Nrf2 is also regulated at the level of transcription and translation. The function of Nrf2 is regulated by its negative regulator Keap1 after translation. This protein binds to Nrf2 and induces the degradation of cytoplasmic Nrf2. Nrf2 is a key transcription factor that regulates the expression of several detoxifying enzymes by binding to ARE in the gene promoter. These Nrf2-dependent detoxifying enzymes, including glutathione S-transferase (GST), NQO1, γ-glutamyl cysteine synthetase (GCL), and glucuronyl transferase (UDP), protect cells against carcinogen-induced DNA damage and cytotoxic effects (Fields et al., 1999). Other studies have shown that Nrf2 expression can inhibit oxidative stress and induce lung and prostate canceration (Harvey et al. 2009, Barve et al. 2009). Breast cancer research has found that Nrf2 can inhibit H2O2-induced oxidative stress by activating Nrf2-dependent genes in breast cancer cells (Hsieh et al. 2010). Under copper exposure treatment, fish brain Nrf2 nuclear accumulation can increase its ability of binding to ARE (CuZnSOD), which improves its antioxidant capacity (Jiang et al. 2014). The Nrf2 pathway plays a protective role against cellular oxidative damage by regulating the activation of oxidase and is important in maintaining zebrafish olfactory function in the Cd exposure environment (Wang and Gallagher 2013).
MicroRNAs (miRNAs) are a class of endogenous non-coding small-molecule RNAs with a length of approximately 22 to 25 nucleotides, regulating approximately 30% of human gene expression at post-transcriptional and translational levels (Bartel 2004) involved in a variety of biological processes, including programmed cell death (apoptosis and autophagy), muscle development, and many human diseases (Lima et al. 2011, Cho 2010, Guo et al. 2015), as well as regulating the response to oxidative cell stress (Mateescu et al. 2011, Padgett et al. 2009). Increasing evidence shows that many miRNAs can regulate Nrf2 directly and indirectly by targeting Nrf2 or by targeting the expression of proteins involved in the Nrf2 pathway and participate in oxidative stress processes. Bach1, a negative regulator of Nrf2 activity, has been shown to be a target of miR-196b, miR-155, miRNA-let-7b, miRNA-let-7c, miR-98, and miR-122 (Sajadimajd and Khazaei 2018, Zhang et al. 2015). In addition, another negative regulator of Nrf2, c-Myc is also regulated by miRNA, including miR-520D-3p, miRNA-135b, miR-34b/c, miR-744, miR-184, miR-135a, miR-145, MiR-126, miR-449c, miR-196b, miR-34a, miR-33b, miR-98, miR-let-7, and miR-185-3p (Zhang et al. 2015, Sajadimajd and Khazaei 2018). The Nrf2 chaperone c-Jun is also targeted by miR-125b and miR-155. Keap1 is a Nrf2 inhibitor and a target of miR-200a and miR-141 (Sajadimajd and Khazaei 2018, Zhang et al. 2015, Lin 2019). In order to obtain the complete picture of Nrf2 regulation and better understand the extent of its regulation by miRNAs, it is therefore necessary to find other miRNAs involved in regulating Nrf2 signaling by targeting Nrf2 or its related proteins.
In this study, we cloned the full-length sequences of Nrf2 of the Chinese perch and analyzed the 3’-UTR for possible miRNAs binding sites. We predicted miR-216a, miR-724, miR-459-5p, miR-194a, and miR-181a-5p were potential regulators of Nrf2. We then verified that miR-216a, miR-724, miR-459-5p, miR-194a, and miR-181a-5p could inhibit Nrf2 mRNA expression through direct interaction between miRNAs and the Nrf2. These results highlight the miR-216a, miR-724, miR-459-5p, miR-194a, and miR-181a-5p have possible important roles in the oxidative stress mediated by the antioxidant factor Nrf2.
Materials and Methods
Ethics Approval and Consent for Participation
This study was conducted following the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health of Hunan University, China. All procedures for fish handling were performed after fish were anesthetized using MS-222 (3-aminobenzoic acid ethyl ester methane sulfonate).
Fish Fasting Treatment
Juvenile Chinese perch were obtained from the Hunan Fisheries Science Institute (Changsha, China). The fish were adapted to the experimental environment at least for 2 weeks. A total of 60 similarly sized Chinese perch (10 g ± 2 g) were randomly assigned to 6 concrete tanks (3-m diameter and 1.5-m water depth), resulting in 10 fish per tank. All fish in each tank were fed live bait once a day (07:00). Rearing water temperature was 25 ± 0.8 °C, and dissolved oxygen was 85 ± 2%. Fish were consistently maintained under a natural light and dark cycle during the experiment. When the normally fed samples (fasting 0 day) were obtained, the remaining fish were circulated to 6 new tanks to ensure that the starvation experiment and the culture conditions remained unchanged.
Sample Collection
At the termination of the feeding trial, nine individuals (10 ± 2 g) were randomly sampled from each tank as control group (the normally fed samples). The same number of fasting samples (9 individuals) was collected at two time points, the second and fifth days. The tissues including the white muscle, red muscle, liver, spleen, kidney, gut, brain, and heart were collected from the normally fed fish while the white muscle samples were collected from both the treated and control fish, quickly washed with cold sterile × 1 PBS to remove contaminating blood before immersion in liquid nitrogen and storage at − 80 °C until RNA extraction.
Molecular Cloning the Core Autophagy-Related Genes
To amplify cDNA fragments of Nrf2, gene specific primers were designed either from contigs of the assembled expressed sequence tags obtained from the Chinese perch muscle transcriptome database (accession no. SRX1738860) or the genes’ homologous sequences from large yellow croaker and tilapia. Full-length cDNAs of the Nrf2 was obtained using a SMART RACE cDNA amplification kit (Takara, Dalian, China). Specific nested PCR primers were designed based on the RACE cloned partial sequences (Table 1). For the 5’-RACE, amplification was conducted as follows: 39 cycles of 94 °C for 30 s, 60 °C for 90 s, and 72 °C for 2 min. For the 3’-RACE, two sequential amplifications were performed under the same conditions: 39 cycles of 94 °C for 30 s, 58 °C for 90 s, and 72 °C for 2 min.
Prediction of Nrf2-Binding miRNAs
To identify miRNAs that potentially bind 3’-UTR of Nrf2, we utilized the TargetScan Fish 6.2 (http://www.targetscan.org/fish_62/) and MicroCosm targets version 5(http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) for target prediction.
Genes and miRNA Expression Analysis
Total RNA from the prepared muscle tissues was extracted with the TRIzolR Reagent (Monad, China), and treated with RNAse-free DNAse I (Monad Biotech, Wuhan, China) in the presence of RNAse inhibitor (Monad, Wuhan, China) followed by ethanol precipitation. The obtained RNA (1 μg) was reversely transcribed using Superscript III RNase H-reverse transcriptase (Monad, Wuhan, China) and Mir-X™ miRNA first-strand synthesis kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. Nrf2, 7 antioxidant enzymes genes (CAT, MnSOD, ZnSOD, GPx, GSTA, GSTA, and GST4A), and miRNAs primers were designed with the software Primer 5.0 (Table 1). cDNAs of skeletal muscle (prived quantity) samples were used as templates for quantitative RT-PCR assays using a SYBR Green PCR reaction kit (Takara, Dalian, China), and the amplification reaction was carried out using the CFX96TM real-time PCR detection system (Bio-Rad, Hercules, CA, USA). The subsequent protocol used standard cycling for the qPCR. The relative expression ratios (R) of target mRNAs were calculated by R = 2−ΔΔCt, where Ct is the cycle threshold, normalized to rpl13 (Zhu et al. 2015).
Plasmids, Transfection, and Luciferase Assay
Wild-type Nrf2 3’-UTR sequences of (355) nt was cloned into pGL4-CMV-luc-vector to generate pGL4-CMV-luc-Nrf2 3’-UTR reporter plasmid (Promega, USA). 3’-UTR mutant pGL4-CMV-luc-Nrf2 reporter plasmids were generated by amplification of the WT plasmid with primers designed to omit the core-binding sites corresponding to each of the selected miRNAs. The primers for DNA sequence clones are listed in Table 1. Site-directed mutagenesis PCR (Sangon Biotech, Shanghai) was conducted as follows: initial denaturation 95 °C, 3 min; 18 cycles of denaturation 95 °C, 30 s; annealing 55 °C, 1 min; extension 68 °C, 1 min; final extension 68 °C, 10 min. HEK293T cells in 96-well plates were transfected with pGL4-CMV-luc-Nrf2, pGL4-CMV-luc-Nrf2-mutant, pRL-CMV vectors (expressing Renilla luciferase) as a normalization control, and miRNA mimics/negative control (NC) using Lipofectamine™ 3000 Transfection Reagent (Thermo Fisher) following the manufacturer’s instructions. The luciferase activity was performed using the dual-luciferase reporter assay system (Promega; Madison, WI) 48 h after the transfection as described previously. Each sample was measured after adding firefly luciferase substrate (F), followed by measurement after the addition of Renilla luciferase substrate (R). Nrf2 3’ UTR relative expression levels were extrapolated from the F/R ratio, as relative luciferase activity (RLA) compared to the non-miRNA control. Each experiment was set up with 5 parallel groups to eliminate the accidental errors introduced in the operation as much as possible. The mutant group data was pCMV-luc-Nrf2-Mut-sch-miRNAs+pRL-CMV + sch-miRNA mimics/pCMV-luc-Nrf2-Mut + pRL-CMV + negative control. And the WT group data was pCMV-luc-Nrf2 + pRL-CMV + sch-miRNAsmimics/pCMV-luc-Nrf2 + pRL-CMV + negative control.
Regulation of miR-181a-5p In vivo with Antagomirs
Chemically modified antisense oligonucleotides (antagomir) was synthesized to regulate miR-181a-5p expression, which showed the highest upregulation after 5 days starvation (GenePharma, Jiangsu, China). The 3’-end of the oligonucleotides was conjugated to cholesterol to enable cell permeation, and all the bases were 2’-OMe modified to increase stability and specificity. The antagomir oligonucleotides were HPLC purified. Juvenile Chinese perch weighing about 5 g received muscle injection of saline (contained same doses of antagomir negative control (CAGUACUUUUGUGUAGUACAA)) or antagomir at a dose of 60 mg/kg body weight for twice times (6 h and 24 h). The muscle tissues were collected 24 h after the last injection for experimental analysis.
Statistical Analysis
Nrf2 and miRNAs expression levels were analyzed by one-way ANOVA procedures using SPSS software. Duncan’s multiple range tests were used to compare the difference between the control and experimental groups. The luciferase assay data also were analyzed by one-way ANOVA procedures using SPSS software. Duncan’s multiple range tests were used to compare the difference between the control and experimental groups. The differences were considered to be statistically significant when the P value was less than 0.05. Data are shown as the means ± SEM (n = 5). Correlation between the Nrf2-miRNAs expression was assayed with Pearson’s correlation test (r).
Results
Cloning and the Spatial Expression of Nrf2 in the Chinese Perch
The full-length cDNAs of the Nrf2 (accession number, MT270449) was cloned with SMART RACE amplification tools. As shown in Fig. 1, the Nrf2 cDNAs is 2398 bp long including the 5′-non-coding region (5’-UTR) of 156 bp, an open reading frame (ORF) of 1836 bp and 3′-non-coding region (3’-UTR) of 406 bp. The Chinese perch Nrf2 contained the conserved bZIP_NFE2-like, bZIP_Maf, and BRLZ domains (Fig. 1).
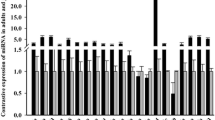
To assay its potential function, the tissue specific patterns of Nrf2 expression were analyzed in Chinese perch by quantitative real-time PCR (qRT-PCR) in 8 different tissues including the fast muscle, slow muscle, heart, liver, spleen, kidney, gut, and brain tissues of normally fed juveniles. As showed in Fig. 2, Nrf2 mRNAs is differentially expressed in the assayed tissues and it is highly expressed in the liver and gut, followed in the fast and slow muscles and the heart tissues with the lowest expression in spleen.
Inverse Expression Patterns of Nrf2 mRNA and miRNAs in the Chinese Perch Muscle After Starvation
We first set to determine the muscular expression level of each miRNAs as well as of Nrf2 under normal feeding and starvation conditions. We found that after 2 days starvation, mRNA level of Nrf2 increased while those of the tested miRNAs decreased. After 5 days starvation however, Nrf2 was downregulated and the miRNAs levels were upregulated. Specifically, the levels of miR-194a and miR-459p were 4 and 8 times upregulated after 5 days starvation. Clearly, Nrf2 transcript level was negatively correlated with miR-181a-5p, miR-194a, miR-216a, miR-459-5p, and miR-724, and the reverse-correlation was highly significant (r < − 0.8) (Figs. 3 and 4 and Table 2).
MiR-181a-5p/194a/216a/459-5p/724 Target the 3’-UTR of Nrf2 mRNA
To determine whether miR-181a-5p/194a/216a/459-5p/724 directly interact with the 3’-UTR of the Nrf2 mRNA to inhibit Nrf2 expression, the luciferase reporter assay were utilized. It is worth noting that there was 9 bp site on the 3’-UTR of Nrf2 and it is predicted complementarily matching miR-181a-5p. Chinese perch wild-type Nrf2 3’-UTR or mutant Nrf2 3’-UTR, lacking the binding site (Fig. 5a) driving firefly luciferase expression constructs were transfected to HEK293T cells along with the Renilla luciferase vector. As demonstrated in Fig. 5b, the luciferase activity was significantly reduced (P < 0.05) when each of the miR-181a-5p/194a/216a/459-5p/724 mimic was added, in the wild-type 3’-UTR Nrf2 wild-type constructs. These results confirm that miR-181a-5p/194a/216a/459-5p/724 are able to negatively regulate Nrf2 expression by targeting the 3’-UTR of Nrf2 mRNA.
miR-181a-5p/194a/216a/459-5p/724 targets the 3’-UTR of Nrf2 mRNA. a Schematics of Nrf2 mRNA 3′UTR and its potential mi181a-5p/194a/216a/459-5p/724 binding site. Nrf2 3’-UTR mutant was generated with point mutations in the miR181a-5p/194a/216a/459-5p/724 binding site. b HEK293T cells transfected with wild-type (WT) or mutant Nrf2 mRNA 3’-UTR reporter plasmids with vehicle control or miRNA mimics. The relative luciferase activities were calculated by normalizing to that of vehicle controls. Error bars indicate the mean ± SEM (n = 5). A single asterisk above each bar indicates statistical difference (P < 0.05)
The Role of miR-181a-5p Regulating Nrf2 Expression in Chinese Perch Skeletal Muscle
Among the predicted miRNA targeted to Nrf2, we chose one microRNA, miR-181a-5p to determine its role in regulating Nrf2-Keap1 pathway in vivo. We designed a specific miRNA antagomir to knock down the selected miRNA function. When the miR-181a-5p antagomir was injected into juvenile Chinese perch muscle tissues, the relative expression level of miR-181a-5p was significantly decreased. However, when treatment with miR-181a-5p antagomir, the mRNA level of the Nrf2 and Keap1 were significantly increased compared with the control (Fig. 6). The effects of miR-181a-5p silencing on antioxidant parameters in the muscle of juvenile Chinese perch are summarized in Fig. 6. The expression levels of the antioxidant enzymes CAT, ZnSOD, GPx, GSTA, and GSTA genes also increased significantly (P < 0.05). The injection of miR-181a-5p antagomir significantly increased the expression of the antioxidant signal molecule Nrf2. These results indicate that miR-181a-5p silencing could affect juvenile Chinese perch in vivo, implying that miR-181a-5p interaction likely play a role in modulating transcription levels of Nrf2 in a Keap1-independent pathway.
Discussion
In this study, we cloned the full-length cDNA sequences of Nrf2 from Chinese perch (Sinipercachuatsi) and investigated the expression profiles in various tissues. Nrf2 was highly expressed in liver and gut followed with higher expression in both fast and slow muscles, but the lowest expression in spleen. The 3’-UTR of the Nrf2 sequence in Chinese perch is 406 bp in length and bioinformatic predicted binding sites for miR-181a-5p/194a/216a/459-5p/724. Based on real-time PCR analysis, the miR-181a-5p/194a/216a/459-5p/724 and Nrf2 mRNA exhibited an opposite pattern of expression in the normally fed and starvation Chinese perch muscle. The luciferase reporter assay and the miR-181a-5p/194a/216a/459-5p/724 suppression experiment further verified the direct interaction between the miR-181a-5p/194a/216a/459-5p/724 and Nrf2 in vitro. The miR-181a-5p silencing results in an increase in Nrf2 gene expression.
Nrf2 is a key transcription factor that regulates the expression of several antioxidant enzymes by binding to AREs in the gene promoter (Yu and Kong 2007). Nrf2 can inhibit H2O2-induced oxidative stress by activating Nrf2-dependent genes in the liver and intestine of grass carp (Zhao et al. 2015). According to earlier reports, it showed that the highly expression of Nrf2 in grass carp intestine and liver (Zhao et al. 2015, Wu et al. 2018) and Chinese perch skeletal muscle (Wu et al. 2020) can effectively offset the oxidative stress caused by ROS under nutrient stress environment. Our result of Nrf2 high expression levels in the liver, gut, and skeletal muscle supports the notion that it could plays an important role in antioxidant signaling pathway to defend tissues against oxidative insults. When exposed to external stress, Nrf2 levels also increased in some tissues. In our early report, the effect of long-term and short-term starvation on antioxidative signal molecules (Nrf2, Keap1, mTOR, and S6K1) in the muscle of juvenile Chinese perch can be confirmed that Nrf2 likely play an active role in oxidative stress (Wu et al. 2020). Interestingly, the mRNA levels of Nrf2 in Chinese perch skeletal muscle and cardiac muscle were not as prominent in the liver and intestine compared to adult muscles. One possible explanation that antioxidative response in juvenile muscle is minor during the growth phase (Wu et al. 2018). This result is in agreement with the expression patterns of Nrf2 in muscle of another fish species, the grass carp during different growth stages (Wu et al. 2018).
The dynamic functions of Nrf2 in Chinese perch, grass carp, and soft-shelled turtle in the oxidative stress response have been previously reported (Wu et al. 2020, Wen et al. 2015, Wang et al. 2020). As the typical Nrf2-target genes, glutathione S-transferases (GSTs), glutathione (GSH), and superoxide dismutase (SOD) can avoid oxidative damage when oxidative stress occurs in the organism (Kobayashi and Yamamoto 2005, Jaiswal 2004). However, the mechanism by which upstream genes regulate Nrf2 expression needs to be further elucidated. Increasing evidence shows that many miRNAs can regulate Nrf2 activity by targeting the expression of proteins involved in the Nrf2 pathway and participated in oxidative stress processes. Some miRNAs regulate the Nrf2 pathway by directly targeting Nrf2. For example, miR-101, miR-93, miR-28, miR-129, miR-450-5p, and miR-153 can target Nrf2 mRNA and downregulate Nrf2 protein expression (Yang et al. 2011, Dong et al. 2019, Sajadimajd and Khazaei 2018). Analysis of miRNAs in erythrocytes shows that miR-144 inhibits Nrf2 expression, leading to reduced levels of antioxidant targets, which is associated with severe anemia (Sangokoya et al. 2010). Overall, it looks like miRNAs targeting Nrf2 are tissue- and species-specific. Since the role of miRNAs in the regulation of Nrf2 expression in Chinese perch remains largely unclear, we set decipher this aspect. Through the prediction of http://www.targetscan.org/fish_62/ in zebrafish, we found that miRNA-216a/b and miRNA-181 have a matching relationship with the zebrafish Nrf2b (Accessionnumber:ENSDARG00000089697) gene. Our study is the first to propose that miR-181a-5p/194a/216a/459-5p/724 regulates Nrf2 expression at the post-transcriptional level by binding to the 3’-UTR of Nrf2 mRNA and resulting in Nrf2 mRNA decline in Chinese perch muscle after starvation. In the 2 days short-term starvation, the upstream inducer of miRNAs were silencing, while the target gene Nrf2 was significantly expressed. However, Nrf2 is inhibited by the upstream miRNAs, making the antioxidant effect in the oxidative stress response ineffective after the long-term starvation in juvenile Chinese perch. Meanwhile, after miR-181a-5p antagomir injection, the increasing of Nrf2 gene expression level directly leads to the activation of downstream antioxidant genes, and the ability to resist oxidative injury is significantly improved. This is consistent with the results reported earlier on zebrafish and fish brain, excessive levels of metal ions (Cd and Cu) binding to the AREs sites of Nrf2 led to the activation of downstream antioxidant genes (GSH, GST, and SOD) and enhanced the ability to antioxidant capacity. Especially in the highly metal contents, the metal ions (Cd and Cu) can increase the expression of antioxidant genes by combining with the AREs of Nrf2, thereby enhancing the ability to decrease oxidative injury in zebrafish and fish brain (Jiang et al. 2014, Wang and Gallagher 2013). Our research further suggested a mechanism by which miRNAs can regulate the degradation of Nrf2 in Chinese perch muscle induced by oxidative autophagy under starvation conditions.
Nrf2 expression can be regulated by Keap1-dependent and Keap1-independent mechanisms. Nrf2 is regulated by aryl hydrocarbon receptor (AHR) transcription (Kwak et al. 2002). In addition, studies have shown that ectoderm Eurasian cortex 1 (ENC1) inhibits Nrf2 expression in a manner independent of Keap1 by reducing Nrf2 protein synthesis without affecting Nrf2 transcription or ubiquitination of Nrf2 protein (Wang et al. 2009). Prior to this, studies have shown that miR-28 targets the 3’-UTR of Nrf2 mRNA and causes the degradation of Nrf2 mRNA, which affects normal human breast epithelial cells and breast cancer cells through a Keap1-independentmechanism (Yang et al. 2011). Meanwhile, there are some researchers reported that miR-181a-5p plays a role in Wnt/β-catenin signaling for muscle growth (Sun et al. 2019, Waki et al. 2016). In our research, the upregulation of Nrf2 mRNA in response to miRNA antagomirs suggested that miR-181a-5p may be also direct, independent of changes in Keap1 expression or Keap1/Nrf2 interaction in Chinese perch skeletal muscle. Our future studies will aim at determining the exact mechanism by which miR-181a-5p mediates Nrf2 ubiquitination and degradation pathway.
Conclusions
In summary, our studies found that miR-181a-5p/194a/216a/459-5p/724 may regulate Nrf2 expression by targeting the Nrf2 3’-UTR region and through miR-181a-5p suppression with specific antagomir treatment could increase Nrf2 and antioxidant genes expression. Our results provide new insights into the role of miR-181a-5p/194a/216a/459-5p/724 in the regulation of Chinese perch muscle genes (Fig. 7) and revealed new mechanisms in regulating Nrf2 expression, which may provide implications for oxidative stress-mediated autophagy and may help develop a potential application in fish aquaculture.
References
Bartel, D.P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Barve A, Khor TO, Nair S, Reuhl K, Suh N, Reddy B, Newmark H, Kong AN (2009) γ-Tocopherol-enriched mixed tocopherol diet inhibits prostate carcinogenesis in TRAMP mice. Int J Cancer 124:1693–1699
Cho, W.C. (2010). MicroRNAs in cancer—from research to therapy. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 1805:209–217
Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P (2002) Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proceedings of the National Academy of Sciences 99:11908–11913
Dong X, Zhang Y, Shang X, Zeng Y (2019) Effects of miR-101 on the proliferation and apoptosis of gastric mucosal epithelial cells via Nrf2/ARE signaling pathway. Eur Rev Med Pharmacol Sci 23:5187–5194
Fields WR, Morrow CS, Doehmer J, Townsend AJ (1999) Expression of stably transfected murine glutathione S-transferase A3-3 protects against nucleic acid alkylation and cytotoxicity by aflatoxin B1 in hamster V79 cells expressing rat cytochrome P450-2B1. Carcinogenesis 20:1121–1125
Guo X, Connick MC, Vanderhoof J, Ishak M-A, Hartley RS (2015) MicroRNA-16 modulates HuR regulation of cyclin E1 in breast cancer cells. Int J Mol Sci 16:7112–7132
Harvey C, Thimmulappa R, Singh A, Blake D, Ling G, Wakabayashi N, Fujii J, Myers A, Biswal S (2009) Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radic Biol Med 46:443–453
He X, Chen MG, Lin GX, Ma Q (2006) Arsenic induces NAD (P) H-quinone oxidoreductase I by disrupting the Nrf2· Keap1· Cul3 complex and recruiting Nrf2· Maf to the antioxidant response element enhancer. J Biol Chem 281:23620–23631
He X, Kan H, Cai L, Ma Q (2009) Nrf2 is critical in defense against high glucose-induced oxidative damage in cardiomyocytes. J Mol Cell Cardiol 46:47–58
Hsieh T-C, Elangovan S, Wu JM (2010) Differential suppression of proliferation in MCF-7 and MDA-MB-231 breast cancer cells exposed to α-, γ-and δ-tocotrienols is accompanied by altered expression of oxidative stress modulatory enzymes. Anticancer Res 30:4169–4176
Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I (1997) An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236:313–322
Jaiswal AK (2004) Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med 36:1199–1207
Jiang W-D, Liu Y, Hu K, Jiang J, Li S-H, Feng L, Zhou X-Q (2014) Copper exposure induces oxidative injury, disturbs the antioxidant system and changes the Nrf2/ARE (CuZnSOD) signaling in the fish brain: protective effects of myo-inositol. Aquat Toxicol 155:301–313
Kensler TW, Wakabayashi N, Biswal S (2007) Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47:89–116
Kobayashi A, Kang M-I, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M (2004) Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24:7130–7139
Kobayashi M, Yamamoto M (2005) Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid Redox Signal 7:385–394
Kobayashi M, Yamamoto M (2006) Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzym Regul 46:113–140
Kusik BW, Carvan Iii MJ, Udvadia AJ (2008) Detection of mercury in aquatic environments using EPRE reporter zebrafish. Mar Biotechnol 10:750–757
Kwak MK, Itoh K, Yamamoto M, Kensler TW (2002) Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol Cell Biol 22:2883–2892
Lima RT, Busacca S, Almeida GM, Gaudino G, Fennell DA, Vasconcelos MH (2011) MicroRNA regulation of core apoptosis pathways in cancer. Eur J Cancer 47:163–174
Lin Y-H (2019) MicroRNA networks modulate oxidative stress in cancer. Int J Mol Sci 20:4497
Liu Y, Zhang L, Liang J (2015) Activation of the Nrf2 defense pathway contributes to neuroprotective effects of phloretin on oxidative stress injury after cerebral ischemia/reperfusion in rats. J Neurol Sci 351:88–92
Ma Q (2008) Xenobiotic-activated receptors: from transcription to drug metabolism to disease. Chem Res Toxicol 21:1651–1671
Ma Q, Kinneer K, Bi Y, Chan JY, Kan YW (2004) Induction of murine NAD (P) H: quinone oxidoreductase by 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin requires the CNC (cap n collar) basic leucine zipper transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2): cross-interaction between AhR (aryl hydrocarbon receptor) and Nrf2 signal transduction. Biochem J 377:205–213
Mateescu B, Batista L, Cardon M, Gruosso T, De Feraudy Y, Mariani O, Nicolas A, Meyniel J-P, Cottu P, Sastre-Garau X (2011) miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med 17:1627
Miao W, Hu L, Scrivens PJ, Batist G (2005) Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway direct cross-talk between phase I And II drug-metabolizing enzymes. J Biol Chem 280:20340–20348
Muthusamy VR, Kannan S, Sadhaasivam K, Gounder SS, Davidson CJ, Boeheme C, Hoidal JR, Wang L, Rajasekaran NS (2012) Acute exercise stress activates Nrf2/ARE signaling and promotes antioxidant mechanisms in the myocardium. Free Radic Biol Med 52:366–376
Nguyen T, Sherratt PJ, Pickett CB (2003) Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol 43:233–260
Niture SK, Kaspar JW, Shen J, Jaiswal AK (2010) Nrf2 signaling and cell survival. Toxicol Appl Pharmacol 244:37–42
Padgett KA, Lan RY, Leung PC, Lleo A, Dawson K, Pfeiff J, Mao TK, Coppel RL, Ansari AA, Gershwin ME (2009) Primary biliary cirrhosis is associated with altered hepatic microRNA expression. J Autoimmun 32:246–253
Sajadimajd S, Khazaei M (2018) Oxidative stress and cancer: the role of Nrf2. Curr Cancer Drug Targets 18:538–557
Sangokoya C, Telen MJ, Chi J-T (2010) microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood, The Journal of the American Society of Hematology 116:4338–4348
Sun L, Li Z, Xue H, Ma T, Ren C, Li M, Lu Y, Sun H, Zhang K (2019) MiR-26a promotes fracture healing of nonunion rats possibly by targeting SOSTDC1 and further activating Wnt/β-catenin signaling pathway. Mol Cell Biochem 460:165–173
Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang M-I, Kobayashi A, Yamamoto M, Kensler TW, Talalay P (2004) Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proceedings of the National Academy of Sciences 101:2040–2045
Waki T, Lee SY, Niikura T, Iwakura T, Dogaki Y, Okumachi E, Oe K, Kuroda R, Kurosaka M (2016) Profiling microRNA expression during fracture healing. BMC Musculoskelet Disord 17:83
Wang K, Zhang T, Dong Q, Nice EC, Huang C, Wei Y (2013) Redox homeostasis: the linchpin in stem cell self-renewal and differentiation. Cell Death Dis 4:e537–e537
Wang L, Gallagher EP (2013) Role of Nrf2 antioxidant defense in mitigating cadmium-induced oxidative stress in the olfactory system of zebrafish. Toxicol Appl Pharmacol 266:177–186
Wang N, Wang W, Sadiq FA, Wang S, Caiqin L, Jianchang J (2020) Involvement of Nrf2 and Keap1 in the activation of antioxidant responsive element (ARE) by chemopreventive agent peptides from soft-shelled turtle. Process Biochem 92:174–181
Wang X-J, Zhang DD (2009) Ectodermal-neural cortex 1 down-regulates Nrf2 at the translational level. PLoS One 4
Wen L-M, Jiang W-D, Liu Y, Wu P, Zhao J, Jiang J, Kuang S-Y, Tang L, Tang W-N, Zhang Y-A (2015) Evaluation the effect of thiamin deficiency on intestinal immunity of young grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol 46:501–515
Wu P, Wang A, Cheng J, Chen L, Pan Y, Li H, Zhang Q, Zhang J, Chu W, Zhang J (2020) Effects of starvation on antioxidant-related signaling molecules, oxidative stress, and autophagy in juvenile Chinese perch skeletal muscle. Mar Biotechnol 22:81–93
Wu P, Zheng X, Zhou X-Q, Jiang W-D, Liu Y, Jiang J, Kuang S-Y, Tang L, Zhang Y-A, Feng L (2018) Deficiency of dietary pyridoxine disturbed the intestinal physical barrier function of young grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol 74:459–473
Yang M, Yao Y, Eades G, Zhang Y, Zhou Q (2011) MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast Cancer Res Treat 129:983–991
Yu S, Khor TO, Cheung K-L, Li W., Wu T-Y, Huang Y, Foste, BA, Kan YW, Kong A-N (2010) Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PloS One 5
Yu S, Kong A-N (2007) Targeting carcinogen metabolism by dietary cancer preventive compounds. Curr Cancer Drug Targets 7:416–424
Zhang H, Davies KJ, Forman HJ (2015) Oxidative stress response and Nrf2 signaling in aging. Free Radic Biol Med 88:314–336
Zhao H-F, Feng L, Jiang W-D, Liu Y, Jiang J, Wu P, Zhao J, Kuang S-Y, Tang L, Tang W-N (2015). Flesh shear force, cooking loss, muscle antioxidant status and relative expression of signaling molecules (Nrf2, Keap1, TOR, and CK2) and their target genes in young grass carp (Ctenopharyngodon idella) muscle fed with graded levels of choline. PloS One 10
Zhu X, Li Y-L, Chen D-X, Wu P, Yi T, Chen T, Zhang J-S, Chu W-Y (2015) Selection of reference genes for microRNA quantitative expression analysis in Chinese perch, Siniperca chuatsi. Int J Mol Sci 16:8310–8323
Acknowledgments
We thank Dr. NilliZmora in the University of Maryland reviewed and revised the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Nos. 31820103016 and 31230076).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
This study was conducted following the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health of Hunan University, China. All procedures for fish handling were performed after fish were anesthetized using MS-222 (3-aminobenzoic acid ethyl ester methane sulfonate).
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, P., Chen, L., Cheng, J. et al. MiRNAs-Modulation of Nrf2 Signaling Networks in Regulation Oxidative Stress of Chinese Perch Skeletal Muscle After Fasting Treatment. Mar Biotechnol 22, 620–630 (2020). https://doi.org/10.1007/s10126-020-09982-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-020-09982-3