Abstract
Due to low consumption and high efficiency, in situ microbial remediation of petroleum hydrocarbons (PHs)-contaminated sites in in-service petrochemical enterprises has attracted more and more attention. In this study, a degrading strain was isolated from oil depot–contaminated soil with soil extract (PHs) as the sole carbon source, identified and named Rhodococcus sp. OBD-3. Strain OBD-3 exhibited wide adaptability and degradability over a wide range of temperatures (15–37 °C), pH (6.0–9.0), and salinities (1–7% NaCl) to degrade 60.6–86.6% of PHs. Under extreme conditions (15 °C and 3–7% salinity), PHs were degraded by 60.6 ± 8.2% and more than 82.1% respectively. In OBD-3, the alkane monooxygenase genes alkB1 and alkB2 (GenBank accession numbers: MZ688386 and MZ688387) were found, which belonged to Rhodococcus by sequence alignment. Moreover, strain OBD-3 was used in lab scale remediation in which the contaminated soil with OBD-3 was isolated as the remediation object. The PHs were removed at 2,809 ± 597 mg/kg within 2 months, and the relative abundances of Sphingobium and Pseudomonas in soil increased more than fivefold. This study not only established a system for the isolation and identification of indigenous degrading strains that could efficiently degrade pollutants in the isolated environment but also enabled the isolated degrading strains to have potential application prospects in the in situ bioremediation of PHs-contaminated soils.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Global oil consumption is increasing every year (Bharti et al. 2021), and the USA and China are among the top countries worldwide in terms of oil refining and oil consumption (Al-Fattah and Aramco 2021). In 2020, the USA had 135 operable refineries and approximately 115,000 gas stations (EIA 2021; Patel 2021). China also had 28 10-million-ton refining and chemical enterprises and 119,000 gas stations (Liu 2022). However, oil leaks inevitably occur during processing, storage, and use, causing serious pollution to the local or surrounding soil of in-service enterprises such as refining and chemical enterprises, oil depots, and gas stations (Liu et al. 2015; Wu et al. 2022). Petroleum hydrocarbons (PHs) are typical pollutants in oil-contaminated soils. In the Dagang Oilfield of China, the concentrations of total petroleum hydrocarbons (TPHs) in soils collected from oil refineries and transportation zones ranged from (2.0 ± 0.5) × 104 to (2.3 ± 0.2) × 104 mg kg−1 dm (Liu et al. 2015). At a gas station in Nanjing, the C10–C40 hydrocarbons concentrations were high in most samples, especially at shallow depths, reaching 3680 mg/kg (Wu et al. 2022). In addition, when a sufficient amount of PHs was released on the (underground) surface, they migrated vertically downward until reaching the groundwater and then spread laterally (Banerji et al. 1995), causing serious pollution to groundwater and even surrounding rivers or waters. At the abovementioned Nanjing gas station, the maximum concentration of TPHs in groundwater was 13.1 mg/L (Wu et al. 2022). In the shallow groundwater around the gas station in Chongqing, the detection rate of PHs was 96.3% (Zhao et al. 2016). PHs pose high potential risks and harmful effects to humans and other organisms surrounding contaminated aquatic and terrestrial ecosystems (Haider et al. 2021). Therefore, the PHs pollution of in-service petrochemical enterprises urgently needs to be remediated.
The contaminated sites of in-service enterprises could be remediated by in situ remediation, mainly due to it was deeply repaired without excavation (Kuppusamy et al. 2016), to avoid affecting normal production. However, the safety requirements are high in the remediation process, and pollution easily rebounds (O'Connor et al. 2018), so it is necessary to choose a safe, efficient, and continuous in situ remediation method. Microbial remediation is a better choice due to its flexibility, efficiency, effectiveness, economy, and eco-friendliness (Azubuike et al. 2020). Bioaugmentation is a type of microbial remediation that degrades pollutants by added indigenous or allochthonous degrading microbial agents (Yu et al. 2014). The microbial agents used in remediation need to have efficient degradation activity, good environmental adaptability, and no biological risk. Therefore, the isolation of high-efficiency degrading microorganisms from contaminated soil has been the focus of research (Song et al. 2021).
At present, there are many strains that can degrade PHs, including Pseudomonas (Xie et al. 2011), Rhodococcus (Huang et al. 2008; Takei et al. 2008; Li et al. 2013; Liu et al. 2016; Hu et al. 2020), Bacillus (Wang et al. 2020), Acinetobacter (Lal and Khanna 1996), Alcanivorax (Hara et al. 2004), and Sphingomonas (Li et al. 2013; Wang et al. 2020). However, some pathogenic or opportunistic pathogens among these degrading strains were not safe for soil remediation. In addition, non-indigenous degrading strains were used to remediate soil, which might be difficult to adapt to the environment due to temperature, salinity, and other factors, resulting in no survival. Therefore, it was particularly important to isolate indigenous strains that were highly adaptable, and could be efficiently degraded and engineered.
In this study, the indigenous PHs degrading strain OBD-3 was isolated from the oil depot contaminated soil with soil extract (PHs) as the sole carbon source. OBD-3 was identified by bacterial morphology and 16S rRNA gene sequence analysis. Its degradation characteristics were investigated by degrading PHs at different medium pH, culture temperatures, and salinities. The PHs degradation genes of OBD-3 were detected by PCR amplification, and their amino acid sequences were analyzed. Thus, an isolation and identification system of indigenous degrading strains that could efficiently degrade pollutants in the isolated environment was formed. In addition, OBD-3 was utilized to remediate the contaminated soil from which it was isolated, and the effects of the remediation on the microbial community structure in the in situ soil were investigated. These findings provide strain resources and theoretical support for the in-situ bioremediation of PHs-contaminated soil.
Materials and methods
Chemicals and culture media
The PHs used in all experiments were extracted with dichloromethane, concentrated and finally dissolved in n-hexane to form a liquid with a density of 0.8 g/cm3. All other reagents and solvents used were of analytical grade and the highest purity available. 1% salinity mineral salt medium (MSM) was used to isolate strains and degrade PHs, which contained the following: 0.2 g/L NH4Cl, 7.95 g/L NaCl, 0.77 g/L MgCl2·6H2O, 1.05 g/L MgSO4·7H2O, 0.076 g/L CaCl2, 0.22 g/L KCl, 0.01 g/L NaHCO3, 0.026 g/L NaBr, 0.25 g/L K2HPO4, and trace element solution (1 mL/L) (Feng et al. 2012). Other salinity MSM were prepared in proportion. Luria–Bertani (LB) medium (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl) was used for the isolation and cultivation of strains. Solid agar plates were prepared with the addition of 1.5% (w/v) agar to the LB liquid medium.
Isolation and identification of degrading strains
The soil samples for isolation of potential PHs degrading strains were collected from the contaminated soil of an oil depot in Shanghai, and were transported aseptically at 4 °C to the laboratory. A soil sample (5 g) was used to inoculate a 250-mL flask with 100 mL of MSM containing 800 mg/L PHs as the sole carbon source (Sood and Lal 2008). The flask was shaken at 28 °C, 180 r/min, in the dark and transferred every 4 days, and the inoculation volume was 10% (v/v). After multiple transfers, the remaining PHs content was determined, and the microbial community with PHs degradation ability was selected to isolate single strain. The microbial community was diluted and spread on LB solid medium and incubated at 28 °C for 2 days. Single bacterial colonies were selected to be streaked and separated on the LB agar plates, and finally, the PHs degrading single strain was obtained.
The genomic DNA of strain OBD-3 was extracted by using Fast DNA™ Spin DNA extraction kit (MP Biomedical, USA). The 16S rRNA gene was amplified from genomic DNA using the universal primers 27F and 1492R (STable 1). The gyrB gene was amplified from genomic DNA using gyrB-F and gyrB-R (STable 1) (Táncsics et al. 2014). The amplifications were performed using Taq DNA polymerase under standard reaction conditions, and sequenced by Sangon Biotech Co., Ltd. Sequences were aligned and analyzed by NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast) server. The phylogenetic analysis was performed using the neighbor-joining method by MEGA X (version 10.2.6).
Degradation of PHs by degrading strain
To study the degradation characteristics of strain OBD-3, the effects of pH, temperature, and salinity on degradation were investigated. Different pH values (pH 6.0, pH 7.0, pH 8.0, pH 9.0) were tested. Strain OBD-3 was cultured in LB medium containing 1000 mg/L PHs, and shaken at 28 °C for 1 day. The cells were centrifuged at 10,000 × g for 5 min, washed with PBS twice, and transferred to 50 mL of MSM containing 1000 mg/L PHs to make the initial OD600 = 0.2. The degradation system was shaken at 180 r/min for 7 days in the dark. Similarly, the effects of culture temperature (15 °C, 28 °C, 37 °C) and salinity (1%, 3%, 5%, 7%) in MSM were also tested for strain OBD-3. Three parallel experiments were performed in each group, and blank control (degradation system without strain OBD-3) was performed. The residual PHs in MSM were extracted with an equal volume of n-hexane, and the extract was filtered through 0.22 μm millipore filter to prepare the samples for subsequent analysis by gas chromatography (GC).
The GC analysis was carried out on GC-2014 (Shimadzu company, Japan), equipped with flame ionization detector (FID), and the injection chromatographic column was HP-5 capillary column (30 m × 0.25 mm × 0.25 μm. Agilent Technology Co., Ltd.). The conditions were based on the China HJ 1021–2019 methods, as follows: inlet temperature 300 °C and detector temperature 325 °C. The program was as follows: initial temperature 50 °C for 2 min, heating at 40 °C/min to 230 °C; then, the temperature was increased at the rate of 20 °C/min up to 320 °C, with a hold time of 20 min. The column flow rate was 1.5 mL/min. The biodegradation efficiency (%) was calculated as follows: biodegradation efficiency (%) = (C0-Ct-CL)/(C0-CL) × 100%, where C0 is the initial concentration of PHs in the culture medium, Ct is the concentration of PHs in the culture medium after degradation for a certain time, and CL is the concentration of volatilization loss.
The detection of degradation genes
The degradation genes were amplified from genomic DNA using the primers of alkane monooxygenase genes alkB1 and alkB2 (Yang et al. 2015), as shown in STable 1. The amplifications were performed using Taq DNA polymerase. The annealing temperature was 62 °C, and the extension time was 1 min. The PCR products were detected by DNA gel electrophoresis and sequenced by Sangon Biotech Co., Ltd. The analysis of sequences was described in “Isolation and identification of degrading strains.” Motif searches were performed using the Vector NTI AlignX software (version 11.0).
Remediation of contaminated soil
The tested soil was the contaminated soil used to isolate the degrading strain and was collected after grinding and screening (30 mesh). Strain OBD-3 was cultured as described above. The cells were centrifuged and washed to form resting cells (OD600 = 10.0) in 50 mM phosphate buffer (pH 7.4). The resting cells were evenly sprayed into 500 g of polluted soil according to the water soil ratio of 1:10. The experimental soil was placed in a constant-temperature incubator (25 °C) for cultivation, and a moisture content of 20–25% was maintained. The samples were taken every 30 days. The contaminated soil with deionized water was used as the control group. Three parallel experiments were performed in each group.
For the extraction of PHs in soil, 5 g of dry weight soil, dichloromethane (v:w = 4:1) as extractant, ultrasonic extraction for 10 min, centrifugation at 5000 × g for 10 min, was repeated 4 times (Guo et al. 2017). The extract was collected and concentrate it to about 1 mL, added 10 mL of n-hexane to concentrate to 1 mL, repeat for 2 times, and finally diluted to 1 mL with n-hexane. The prepared samples were diluted 100 times with n-hexane and passed through 0.22 μm millipore filter, and the above method was used to detect the content of PHs by GC.
Microbial community structure analysis
The initial contaminated soil (CK 0 M) and remediated soil after 2 months (E 2 M) were collected for MiSeq sequencing. DNA was extracted according to a Power Soil DNA extraction kit (MoBio Laboratories, USA). For microbial community structure analysis, primers 341F and 805R were used to amplify the V3–V4 region as reported (Qu et al. 2016). The PCR products were loaded on Illumina-MiSeq device according to the manufacturer’s protocols (Wu et al. 2019). After sequencing, data were collected and processed by Sangon Biotech Co., Ltd. The operational taxonomic unit (OTU) clustering was performed using Usearch (version 8.1.1831) at 97% sequence similarity threshold.
Results
Isolation and identification of degrading strains
After multiple transfers, the remaining PHs content was determined, and the microbial community with PHs degradation ability was selected to isolate single strain. In the present study, 6 strains were isolated from LB solid medium, and only 2 strains (OBD-1 and OBD-3) showed obvious degradation ability (SFig. 1). Strain OBD-1 and strain OBD-3 could degrade 63% and 82% of PHs in 7 days, respectively. Strain OBD-3 had higher degradation effect than strain OBD-1 under the same conditions. Therefore, strain OBD-3 with higher degradation effect on PHs degradation was selected for identification.
The strain OBD-3 cultivated on LB solid medium showed a cheese colored, round, and glossy after cultivation for 3 days (SFig. 2a). The strain morphology of OBD-3 was rod-shaped (SFig. 2b). Based on alignment of the partial 16S rRNA gene sequence (Fig. 1a), the sequence of OBD-3 (GenBank accession No.: MW 404441) showed 100% identity with model strain Rhodococcus qingshengii djl-6 (Chuang et al. 2021), 99% identity with Rhodococcus erythropolis T7-2 (Huang et al. 2008), and 97% identity with model strain Rhodococcus opacus DSM 43205 T. On the other hand, the gyrB gene of OBD-3 revealed 100% identity with Rhodococcus qingshengii PT3-14, 99% identity with Rhodococcus thermopolis JCM 2892, and 87% with Rhodococcus globerulus ATCC 19370 (Fig. 1b). Thus, strain OBD-3 was Rhodococcus qingshengii, named Rhodococcus sp. OBD-3, which was stored in the China Center for Type Culture Collection (CCTCC M 2020978).
Degradation of PHs by strain OBD-3
The effect of conditions (pH, temperature, and salinity) on the biodegradation of PHs by strain OBD-3 was investigated as shown in Fig. 2. Strain OBD-3 biodegraded 75.3–86.6% of PHs at pH 6.0–9.0 (Fig. 2a), and more than 82.0% in neutral and alkaline environments (pH 7.0–9.0). Under different temperature conditions (Fig. 2b), PHs were degraded by more than 83.4% at 28–37 °C, and 60.6% ± 8.2% at low temperature (15 °C). In addition, strain OBD-3 has good degradation effect on PHs when the total salinity was 1–7% (Fig. 2c), which degraded 81.1–84.8%.
Under relatively optimal conditions (28 °C, pH 8.0, total salinity 1%), strain OBD-3 rapidly degraded 75.8 ± 4.7% of the PHs within 3 days, and 89.1 ± 1.8% in 7 days (Fig. 3c). During the degradation process, the contents of short-chain and medium-chain alkanes were significantly reduced (Fig. 3a and b).
The detection of degradation genes in strain OBD-3
The alkB1 and alkB2 are the most common genes of alkane monooxygenase gene (Whyte et al. 2002), and their primers were used for PCR amplification with the OBD-3 genome as a template. The products were 634 bp and 500 bp, respectively (SFig. 3). Through the recovery and sequencing of products, the partial amino acid sequences were aligned (Fig. 4). Both AlkB1 and AlkB2 of strain OBD-3 have EHN(V)R(K)GHH and NYXEHYGL motifs that are highly conserved among all bacterial alkane monooxygenases (Whyte et al. 2002). The AlkB1 in strain OBD-3 showed 100% identity with AlkB1 from the low-temperature PHs degrading strain Rhodococcus sp. TMP2 (Takei et al. 2008), 100% identity with AlkB1 from another low-temperature PHs degrading strain Rhodococcus sp. Q15 (Whyte et al. 2002), and 69% identity with AlkB2 in strain OBD-3 itself. The AlkB2 in strain OBD-3 revealed a 100% identity with AlkB2 from Rhodococcus sp. NRRL b-16531 and 99% identity with AlkB2 from Rhodococcus sp. Q15 (Whyte et al. 2002). AlkB1 and AlkB2 in strain OBD-3 were 50% and 50% identical to AlkB in the PHs degrading strain Pseudomonas putida GPo1 (Xie et al. 2011), respectively. Moreover, they showed 44–53% identity with AlkB1 and AlkB2 from Pseudomonas aeruginosa and Alcanivorax borcumensis. Therefore, AlkB1 and AlkB2 expressed by the alkB1 (MZ688386) and alkB2 (MZ688387) genes in strain OBD-3 all belonged to Rhodococcus.
The remediation of contaminated soil by strain OBD-3
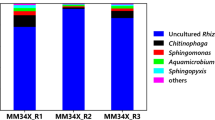
The concentration of PHs in the test soil was 8556 ± 803 mg/kg, as determined by GC detection. After 2 months of remediation by strain OBD-3, the PHs in the soil were removed at 2049 ± 347 mg/kg in 1 month, and 2809 ± 597 mg/kg in 2 months. The microbial community structure in the soil also changed. The top 25 predominant genera in each sample and their relative abundances were shown in Fig. 5. In the initial contaminated soil (CK 0 M), unclassified Gammaproteobacteria, Escherichia Shigella and unclassified Pseudonocardineae were the dominant genera with relative abundances of 8.6%, 5.5%, and 5.1%, respectively. The relative abundances of Sphingobium, Rhodococcus, and Pseudomonas in the remediated soil (E 2 M) were 25.2%, 12.1%, and 10.3%, respectively. In addition, Achromobacter (0.27% in CK 0 M vs. 5.20% in the E 2 M), Olivibacter (0.01% in CK 0 M vs. 1.80% in the E 2 M), Pandoraea (0.02% in CK 0 M vs. 1.04% in the E 2 M), Brevibacillus (0.01% in CK 0 M vs. 0.62% in the E 2 M), and Azospirillum (0.03% in CK 0 M vs. 0.58% in the E 2 M) were significantly increased in relative abundance level of soil.
Discussion
In this study, the substrate (PHs) extracted from contaminated soil was used as the sole carbon source to isolate and identify high-efficiency degrading indigenous strains from the contaminated soil of oil depots. The reason for using the substrate extracted from the soil was to isolate indigenous degrading strains that were more suitable for the soil (Zhao et al. 2017). After the degrading strains were isolated, the degradation performance of the strains was studied by changing the pH, temperature, and salinity, and the degradation function genes of the strains were detected. The above process formed an isolation and identification system of indigenous degrading strains. In addition, the degrading strains were applied to the remediation of in situ contaminated soil, and the remediation effect of the strains was measured from the degradation effect and changes in microbial community structure. Finally, the isolation and application system of indigenous degrading strains was formed (Fig. 6).
Based on this system, the isolated strain OBD-3 was identified as Rhodococcus. Rhodococcus is a common hydrocarbons degrading bacterial genu that can persist and grow in highly contaminated soils and waters, and even under oxygen- and nutrient-limited conditions (Kuyukina and Ivshina 2010). Rhodococcus sp. HX-2 was a salt tolerant degrading strain screened from the oil field that could degrade more than 50% of 0.4% (v/v) diesel oil at 5% salinity (Hu et al. 2020). Rhodococcus erythropolis T7-2 was isolated from the oil-polluted sea-bed mud of Bohai Sea, which degraded diesel oil at 15 °C (Huang et al. 2008). Rhodococcus sp. JZX-01 decomposed 65.27 ± 5.63% of crude oil in 9 days, and had good oil degradation ability at low temperatures as well as under high salt conditions (Li et al. 2013). Rhodococcus rubber JC-106 could efficiently degrade crude oil at low temperature, which degraded 41.61% and 58.18% with crude oil as the sole carbon source at 15 °C and 35 °C for 15 days (Liu et al. 2016). In this study, strain OBD-3 had high degradation activity at 15 ℃, pH 9.0 and 7% salinity, and degraded most of PHs within 3 days. OBD-3 combined the characteristics of low-temperature resistance, saline alkali resistance, and rapid degradation in a short time. Compared with other Rhodococcus, OBD-3 has strong environmental adaptability, degradation ability, and potential application prospects.
Strain OBD-3 was more likely to degrade short-chain and medium-chain alkanes but not completely degraded PHs. Alkanes are most easily degraded by microorganisms due to their simple structure compared to other hydrocarbons (Verma et al. 2006). However, short-chain alkanes also have the potential for incomplete degradation due to their bioavailability and solubility in cell membranes, and substrate toxicity may be the reason for the limited biodegradation of long-chain alkanes reported by various studies (Lal and Khanna 1996; Deng et al. 2014). In the aerobic degradation of alkanes, alkanes were mainly catalyzed by alkane monooxygenases, which catalyzed terminal methylated carbon oxidation (Ji et al. 2013). The Alk system is one of the major alkane monooxygenases and the most studied hydrocarbon hydroxylation system. The Alk system in Pseudomonas putida GPo1 could oxidize C5–C12 n-alkanes to 1-alkanols, and catalyze a variety of reactions including the hydroxylation of linear and branched aliphatic, cycloaliphatic, and alkylaromatic compounds, branched demethylated methyl ethers and epoxidation of terminal olefins (van Beilen et al. 1994; van Beilen and Funhoff 2005). In Alcanivorax borkumensis SK2, there were two non-heme hydroxylases AlkB1 and AlkB2. AlkB1 preferentially hydroxylated C5–C12 hydrocarbons, while AlkB2 preferred to catalyze C8-C16 hydrocarbons (Hara et al. 2004). The alkB1 and alkB2 genes of Rhodococcus opacus B-4 were heterologously expressed in E. coli JM109, making JM109 degradable to C5-C16 alkanes (Sameshima et al. 2008). Rhodococcus sp. TMP2 contained five alkane-degrading monooxygenase genes alkB1–alkB5, which degraded straight-chain and branched alkanes from C9 to C24, but only the expression of alkB1 and alkB2 was induced by n-alkanes (Takei et al. 2008). Strain OBD-3 possessed alkB1 and alkB2 genes belonging to Rhodococcus. These genes played an important role in the degradation of alkanes, which was consistent with the characteristic that strain OBD-3 rapidly degraded short-chain and medium-chain alkanes.
In the bioremediation process, strain OBD-3 continuously degraded PHs in soil within 2 months. However, the remediation effect of OBD-3 was not particularly good, possibly due to the relatively high concentration of PHs in the soil and the lack of nutrient supplementation (Yuniati 2018). Except for the degradation of PHs, the microbial community structure also changed with the addition of OBD-3. Rhodococcus, Sphingobium and Pseudomonas became the dominant bacteria. The relative abundance of Rhodococcus was upregulated 388.5-fold compared with the initial contaminated soil. The other two dominant genera, Sphingobium and Pseudomonas, were upregulated 5.0-fold and 13.1-fold, respectively. These genera were frequently present in PHs-contaminated soils (Wang et al. 2020; Rodríguez-Uribe et al. 2021). Sphingobium was known for its natural ability to adjust to contaminated environments and use contaminants as a growth and energy source (Waigi et al. 2015). Pseudomonas was a common bacterium capable of degrading hydrocarbons, which could degrade crude oil, diesel oil and gasoline (Wongsa et al. 2004; Xie et al. 2011; Li et al. 2013). In addition, Achromobacter (upregulated 19-fold) and Olivibacter (upregulated 138-fold), which did not account for a large proportion but increased by a large number, were also to degrade PHs (Deng et al. 2014; Szabo et al. 2011). In microbial communities that grew and degraded under PHs, the amplifiers that originated from the six major bacterial genera (Pseudomonas, Sphingobium, Ochrobactrum, Achromobacter, Cupriavidus, and Parvibaculum) accounted for more than 97% of the total amplifiers sequenced from diesel fuel cultures. Among them, Pseudomonas was the most important (relative abundance was 70.7%), followed by Sphingobium (12.3%), Ochrobactrum (6.1%), Achromobacter (4.6%), Cupriavidus (2.2%), Parvibaculum (1.1%), and Olivibacter (1.0%). This result was also similar to the microbial community structure in soil after adding strain OBD-3. The addition of strain OBD-3 not only degraded PHs in the soil but also promoted the growth of other PHs degrading strain, to a obtain better remediation effect.
Conclusions
The indigenous PHs degrading strain Rhodococcus sp. OBD-3 was isolated and identified from the oil depot contaminated soil. It could degrade 60.6–86.6% of 1000 mg/L PHs within a broad range of temperatures (15–37 °C), pH (6.0–9.0), and salinities (1–7% NaCl), and could be efficiently degraded under extreme conditions (15 °C and 3–7% salinity), with good environmental adaptability. Strain OBD-3 contained alkane monooxygenase genes alkB1 and alkB2, which belonged to Rhodococcus. An isolation and identification system of indigenous degrading strains was formed from the isolation of indigenous strains, the study of degradation performance to the detection of degradation genes. In addition, strain OBD-3 had a certain effect on the remediation of in situ contaminated soil, and increased the relative abundance of PHs degradation strains in soil. Strain OBD-3 had potential application prospects for the in situ bioremediation of PHs-contaminated soils.
Data Availability
Data available on request from the authors.
References
Al-Fattah SM, Aramco S (2021) Application of the artificial intelligence GANNATS model in forecasting crude oil demand for Saudi Arabia and China. J Petrol Sci Eng 200:108368. https://doi.org/10.1016/j.petrol.2021.108368
Azubuike CC, Chikere CB, Okpokwasili GC (2020) Bioremediation: an eco-friendly sustainable technology for environmental management. In: Saxena G, Bharagava R (eds) Bioremediation of Industrial Waste for Environmental Safety. Springer, Singapore, pp 19–39
Banerji SK, Zappi ME, Teeter CL et al (1995) Bioremediation of soils contaminated with petroleum hydrocarbons using bioslurry reactors. Final report. United States
Bharti MK, Chalia S, Thakur P et al (2021) Nanoferrites heterogeneous catalysts for biodiesel production from soybean and canola oil: a review. Environ Chem Lett 19:3727–3746. https://doi.org/10.1007/s10311-021-01247-2
Chuang S, Yang H, Wang X et al (2021) Potential effects of Rhodococcus qingshengii strain djl-6 on the bioremediation of carbendazim-contaminated soil and the assembly of its microbiome. J Hazard Mater 414:125496. https://doi.org/10.1016/j.jhazmat.2021.125496
Deng MC, Li J, Liang FR et al (2014) Isolation and characterization of a novel hydrocarbon-degrading bacterium Achromobacter sp. HZ01 from the crude oil-contaminated seawater at the Daya Bay, southern China. Mar Pollut Bull 83:79–86. https://doi.org/10.1016/j.marpolbul.2014.04.018
EIA (2021) Refinery closures decreased US refinery capacity during 2020. U.S. Energy Information Administration. https://www.eia.gov/todayinenergy/detail.php?id=48636. Accessed 8 July 2021
Feng T, Cui C, Dong F et al (2012) Phenanthrene biodegradation by halophilic Martelella sp. AD-3. J Appl Microbiol 113:779–789. https://doi.org/10.1111/j.1365-2672.2012.05386.x
Guo J, Chen S, Tan X et al (2017) Simple and efficient extraction method of petroleum hydrocarbon in soil. Environ Eng (Beijing, China) 35:150–154. https://doi.org/10.13205/j.hjgc.201709030
Haider FU, Ejaz M, Cheema SA et al (2021) Phytotoxicity of petroleum hydrocarbons: sources, impacts and remediation strategies. Environ Res 197:111031. https://doi.org/10.1016/j.envres.2021.111031
Hara A, Baik S, Syutsubo K et al (2004) Cloning and functional analysis of alkB genes in Alcanivorax borkumensis SK2. Environ Microbiol 6:191–197. https://doi.org/10.1111/j.1462-2920.2004.00550.x
Hu X, Li D, Qiao Y et al (2020) Salt tolerance mechanism of a hydrocarbon-degrading strain: salt tolerance mediated by accumulated betaine in cells. J Hazard Mater 392:122326. https://doi.org/10.1016/j.jhazmat.2020.122326
Huang L, Ma T, Li D et al (2008) Optimization of nutrient component for diesel oil degradation by Rhodococcus erythropolis. Mar Pollut Bull 56:1714–1718. https://doi.org/10.1016/j.marpolbul.2008.07.007
Ji Y, Mao G, Wang Y et al (2013) Structural insights into diversity and n-alkane biodegradation mechanisms of alkane hydroxylases. Front Microbiol 4:58. https://doi.org/10.3389/fmicb.2013.00058
Kuppusamy S, Thavamani P, Megharaj M et al (2016) In-situ remediation approaches for the management of contaminated sites: a comprehensive overview. In: de Voogt P (ed) Reviews of Environmental Contamination and Toxicology, vol 236. Springer, Cham, pp 1–115
Kuyukina MS, Ivshina IB (2010) Application of Rhodococcus in bioremediation of contaminated environments. In: Alvarez H (eds) Biology of Rhodococcus. Microbiology Monographs, vol 16. Springer, Berlin. https://doi.org/10.1007/978-3-642-12937-7_9
Lal B, Khanna S (1996) Degradation of crude oil by Acinetobacter calcoaceticus and Alcaligenes odorans. J Appl Bacteriol 81:355–362. https://doi.org/10.1111/j.1365-2672.1996.tb03519.x
Li C, Zhou ZX, Jia XQ et al (2013) Biodegradation of crude oil by a newly isolated strain Rhodococcus sp. JZX-01. Appl Biochem Biotechnol 171:1715–1725. https://doi.org/10.1007/s12010-013-0451-4
Liu M, Chen J, Zhou Y et al (2016) Characterization and application of an oil-degrading Rhodococcus rubber for oily wastewater treatment. Acta Sci Circumstant 36:3651–3657. https://doi.org/10.13671/j.hjkxxb.2016.0108
Liu Q, Tang J, Bai Z et al (2015) Distribution of petroleum degrading genes and factor analysis of petroleum contaminated soil from the Dagang Oilfield, China. Sci Rep 5:11068. https://doi.org/10.1038/srep11068
Liu Y (2022) The feasibility of hybrid vehicles in Chinese transportation networks. Third Int Conf Electron Commun Netw Comput Technol (ECNCT 2021) 12167:514–518. https://doi.org/10.1117/12.2628418
Mori JF, Kanaly RA (2020) Multispecies diesel fuel biodegradation and niche formation are ignited by pioneer hydrocarbon-utilizing proteobacteria in a soil bacterial consortium. Appl Environ Microbiol 87:e02268-e2320. https://doi.org/10.1128/AEM.02268-20
O’Connor D, Hou D, Ok YS et al (2018) Sustainable in situ remediation of recalcitrant organic pollutants in groundwater with controlled release materials: a review. J Control Release 283:200–213. https://doi.org/10.1016/j.jconrel.2018.06.007
Patel S (2021) Electric vehicle limbo: The need for charging incentives. Emory Corp Gov Accountability Rev 8:75
Qu Y, Zhang X, Shen W et al (2016) Illumina MiSeq sequencing reveals long-term impacts of single-walled carbon nanotubes on microbial communities of wastewater treatment systems. Bioresour Technol 211:209–215. https://doi.org/10.1016/j.biortech.2016.03.043
Rodríguez-Uribe ML, Peña-Cabriales JJ, del Carmen R-C et al (2021) Native bacteria isolated from weathered petroleum oil-contaminated soils in Tabasco, Mexico, accelerate the degradation petroleum hydrocarbons in saline soil microcosms. Environ Technol Innov 23:101781. https://doi.org/10.1016/j.eti.2021.101781
Sameshima Y, Honda K, Kato J et al (2008) Expression of Rhodococcus opacus alkB genes in anhydrous organic solvents. J Biosci Bioeng 106:199–203. https://doi.org/10.1263/jbb.106.199
Song Y, Li R, Chen G et al (2021) Bibliometric analysis of current status on bioremediation of petroleum contaminated soils during 2000–2019. Int J Environ Res Public Health 18:8859. https://doi.org/10.3390/ijerph18168859
Sood N, Lal B (2008) Isolation and characterization of a potential paraffin-wax degrading thermophilic bacterial strain Geobacillus kaustophilus TERI NSM for application in oil wells with paraffin deposition problems. Chemosphere 70:1445–1451. https://doi.org/10.1016/j.chemosphere.2007.08.071
Szabo I, Szoboszlay S, Kriszt B et al (2011) Olivibacter oleidegradans sp. nov., a hydrocarbon-degrading bacterium isolated from a biofilter clean-up facility on a hydrocarbon-contaminated site. Int J Syst Evol Microbiol 61:2861–2865. https://doi.org/10.1099/ijs.0.026641-0
Takei D, Washio K, Morikawa M (2008) Identification of alkane hydroxylase genes in Rhodococcus sp. strain TMP2 that degrades a branched alkane. Biotechnol Lett 30:1447–1452. https://doi.org/10.1007/s10529-008-9710-9
Táncsics A, Benedek T, Farkas M et al (2014) Sequence analysis of 16S rRNA, gyrB and catA genes and DNA-DNA hybridization reveal that Rhodococcus jialingiae is a later synonym of Rhodococcus qingshengii. Int J Syst Evol Microbiol 64:298–301. https://doi.org/10.1099/ijs.0.059097-0
van Beilen JB, Kingma J, Witholt B (1994) Substrate specificity of the alkane hydroxylase system of Pseudomonas oleovorans GPo1. Enzyme Microb Technol 16:904–911. https://doi.org/10.1016/0141-0229(94)90066-3
van Beilen JB, Funhoff EG (2005) Expanding the alkane oxygenase toolbox: New enzymes and applications. Curr Opin Biotechnol 16:308–314. https://doi.org/10.1016/j.copbio.2005.04.005
Waigi MG, Kang F, Goikavi C et al (2015) Phenanthrene biodegradation by Sphingomonads and its application in the contaminated soils and sediments: a review. Int Biodeterior Biodegrad 104:333–349. https://doi.org/10.1016/j.ibiod.2015.06.008
Wang R, Wu B, Zheng J et al (2020) Biodegradation of total petroleum hydrocarbons in soil: isolation and characterization of bacterial strains from oil contaminated soil. Appl Sci 10:4173. https://doi.org/10.3390/app10124173
Whyte LG, Smits THM, Labbe D et al (2002) Gene cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus strains Q15 and NRRL B-16531. Appl Environ Microbiol 68:5933–5942. https://doi.org/10.1128/AEM.68.12.5933-5942.2002
Wongsa P, Tanaka M, Ueno A et al (2004) Isolation and characterization of novel strains of Pseudomonas aeruginosa and Serratia marcescens possessing high efficiency to degrade gasoline, kerosene, diesel oil, and lubricating oil. Curr Microbiol 49:415–422. https://doi.org/10.1007/s00284-004-4347-y
Wu M, Wu J, Zhang X et al (2019) Effect of bioaugmentation and biostimulation on hydrocarbon degradation and microbial community composition in petroleum-contaminated loessal soil. Chemosphere 237:124456. https://doi.org/10.1016/j.chemosphere.2019.124456
Wu M, Zhao Z, Cai G et al (2022) In situ evaluation of soil contaminated by total petroleum hydrocarbons using membrane interface probe: a case study from Nanjing, China. Bull Eng Geol Environ 81:1–20. https://doi.org/10.1007/s10064-022-02639-6
Verma S, Bhargava R, Pruthi V (2006) Oily sludge degradation by bacteria from Ankleshwar, India. Int Biodeterior Biodegrad 57:207–213. https://doi.org/10.1016/j.ibiod.2006.02.004
Xie M, Alonso H, Roujeinikova A (2011) An improved procedure for the purification of catalytically active alkane hydroxylase from Pseudomonas putida GPo1. Appl Biochem Biotechnol 165:823–831. https://doi.org/10.1007/s12010-011-9300-5
Yang Z, Chen JX, Qin B et al (2015) Characterization and catabolic gene detection of three oil-degrading Rhodococcus spp. Chin J Appl Environ Biol 21:805–812. https://doi.org/10.3724/SP.J.1145.2015.01063
Yu Y, Zhang W, Chen G et al (2014) Preparation of petroleum-degrading bacterial agent and its application in remediation of contaminated soil in Shengli Oil Field, China. Environ Sci Pollut Res 21:7929–7937. https://doi.org/10.1007/s11356-014-2707-0
Yuniati MD (2018) Bioremediation of petroleum-contaminated soil: a review. IOP Conf Ser: Earth Environ Sci 118:012063. https://doi.org/10.1088/1755-1315/118/1/012063
Zhao H, Zhang Y, Xiao X et al (2017) Different phenanthrene-degrading bacteria cultured by in situ soil substrate membrane system and traditional cultivation. Int Biodeterior Biodegrad 117:269–277. https://doi.org/10.1016/j.ibiod.2016.12.016
Zhao L, Zhang Y, Guo J et al (2016) Investigation on pollution characteristics of petroleum hydrocarbon in shallow groundwater around gas stations. Chin J Environ Eng 10:131–136. https://doi.org/10.12030/j.cjee.20160121
Funding
This work was supported by the National Key Research and Development Program of China (No. 2018YFC1803300) and the National Natural Science Foundation of China (No. 41877129).
Author information
Authors and Affiliations
Contributions
CC and YL conceived the idea and designed the experiments. XC conducted the experiments, analyzed the data, and prepared first draft of the manuscript. JS, GS, FZ, and CC reviewed and revised the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This article does not contain any studies with human or animal subjects performed by any of the authors.
Consent for publication
The author agrees to publish.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, X., Shan, G., Shen, J. et al. In situ bioremediation of petroleum hydrocarbon–contaminated soil: isolation and application of a Rhodococcus strain. Int Microbiol 26, 411–421 (2023). https://doi.org/10.1007/s10123-022-00305-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-022-00305-1