Abstract
Besides phenotypic antimicrobial susceptibility testing (AST), whole genome sequencing (WGS) is a promising alternative approach for detection of resistance phenotypes. The aim of this study was to investigate the concordance between WGS-based resistance prediction and phenotypic AST results for enterococcal clinical isolates using a user-friendly online tools and databases. A total of 172 clinical isolates (34 E. faecalis, 138 E. faecium) received at the French National Reference Center for enterococci from 2017 to 2020 were included. AST was performed by disc diffusion or MIC determination for 14 antibiotics according to CA-SFM/EUCAST guidelines. The genome of all strains was sequenced using the Illumina technology (MiSeq) with bioinformatic analysis from raw reads using online tools ResFinder 4.1 and LRE-finder 1.0. For both E. faecalis and E. faecium, performances of WGS-based genotype to predict resistant phenotypes were excellent (concordance > 90%), particularly for antibiotics commonly used for treatment of enterococcal infections such as ampicillin, gentamicin, vancomycin, teicoplanin, and linezolid. Note that 100% very major errors were found for quinupristin-dalfopristin, tigecycline, and rifampicin for which resistance mutations are not included in databases. Also, it was not possible to predict phenotype from genotype for daptomycin for the same reason. WGS combined with online tools could be easily used by non-expert clinical microbiologists as a rapid and reliable tool for prediction of phenotypic resistance to first-line antibiotics among enterococci. Nonetheless, some improvements should be made such as the implementation of resistance mutations in the database for some antibiotics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Enterococci are long-standing members of the gastrointestinal tract (GIT) of humans and many animals, and they are also ubiquitously distributed in natural environments [1]. Classically considered as harmless bacteria, two main species (namely Enterococcus faecalis and Enterococcus faecium) have become major human opportunistic pathogens, especially in hospital settings [2]. Of the greatest concern, there is a worldwide dissemination of multidrug-resistant (MDR) enterococci, especially vancomycin-resistant enterococci (VRE) and more recently linezolid-resistant enterococci (LRE), for which limited therapeutic options remain [3, 4].
Antimicrobial susceptibility testing (AST) is a key element for an effective antibiotic treatment and allows epidemiological surveillance of antibiotic resistance. Because they are simple and cost-effective, phenotypic AST methods (especially MIC determination and disk diffusion) remain commonly used in clinical laboratories [5]. However, they cannot be applied to all bacteria (such as non-cultivable and difficult-to-grow bacteria) and they suffer from reproducibility issues due to technical limitations even if large efforts have been made into standardizing and improving quality of AST (methodology, growth conditions, and interpretation criteria) [6].
Due to the advent of next-generation sequencing (NGS) technologies, whole-genome sequencing (WGS) has become a powerful tool for the detection of resistance mechanisms in bacterial pathogens, particularly for Staphylococcus aureus, Enterobacterales, Pseudomonas aeruginosa, Campylobacter, or Mycobacterium tuberculosis [7,8,9,10,11,12,13,14]. In theory, the use of WGS could potentially replace phenotypic AST methods but the current state of the art is partially achieved [15]. In addition, several obstacles remain to be lifted, particularly the possibility of all clinical microbiologists (expert or not in bioinformatics) to rapidly provide reliable information to clinicians in a format as easy-to-understand as the current AST report [16].
Many different open-access bioinformatics pipelines for detecting AMR mechanisms from NGS data have been published, such as ResFinder, CARD, ARG-ANNOT, and NDARO [15, 17]. However, most of these databases are not user friendly for clinical microbiologists and does not provide translation of genotypes into predicted phenotypes directly usable for clinicians. Currently, there is no consensus on which database should be used for inferring profiles of antibiotic resistance from WGS data. Nonetheless, ResFinder, launched in 2012, is the first web-based, open, and easily accessible tool for users with no bioinformatics expertise and then seems to be the more appropriate for clinical microbiologists [18]. Available from the portal of the Center for Genomic Epidemiology (CGE) (https://cge.cbs.dtu.dk), this service is highly solicited from all over the world and around 15,000 analyses are processed per month [19]. The last version ResFinder 4.0 contains four databases including AMR genes (ResFinder), chromosomal mutations (PointFinder), translation into phenotypes, and species-specific panels for in silico antibiograms [20]. For enterococci, PointFinder database contains only chromosomal mutations conferring resistance to ampicillin and to ciprofloxacin [21]. Interestingly, ResFinder 4.0 can be used with raw reads and with assembled sequences as inputs, and it performs database searches using kmer-based (KMA) and BLAST, respectively [22]. ResFinder 4.0 performs poorly in prediction of linezolid resistance (only acquired genes are detected) and then another CGE tool, called LREfinder 1.0, can be used for detecting resistance mutations in enterococci [23].
A few studies have been conducted on enterococci but several limits can be raised from a clinical point of view: isolates from a single center, isolates of animal origin, surveillance of antimicrobial resistance, small collections of clinical isolates, limited number of tested antibiotics, use of complex home-made bioinformatics pipelines, or low prevalence of resistance to major antibiotics (e.g., vancomycin, linezolid) [20, 24,25,26,27]. The aim of this study was then to evaluate the performances of WGS for predicting antimicrobial resistance profiles of a large collection of well-characterized enterococcal clinical isolates by using conventional phenotypic AST methods as the gold standard.
Materials and methods
Bacterial isolates
A total of 172 epidemiologically unrelated enterococcal clinical isolates (34 E. faecalis, 138 E. faecium) received at the National Reference Center for Enterococci (NRC-Enc) from 81 different cities all over France between 2017 and 2020 were included in this study. Strains were identified by MALDI-TOF mass spectrometry (Microflex; Brucker Daltonics, Bremen, Germany).
Phenotypic antimicrobial susceptibility testing
Routine antimicrobial susceptibility testing (AST) was performed in vitro by the disk diffusion method (ampicillin, gentamicin, erythromycin, quinupristin-dalfopristin, levofloxacin, tetracycline, chloramphenicol, cotrimoxazole, rifampicin) or by MIC determination by the broth microdilution reference method (Sensititre; Thermo Fisher Scientific, Courtaboeuf, France) for vancomycin, teicoplanin, linezolid, daptomycin, and tigecycline. Results were interpreted according to the Comité de l’Antibiogramme de la Société Française de Microbiologie (CASFM, 2020) (www.sfm-microbiologie.org).
WGS and bioinformatic analysis
Genomic DNA was isolated with the Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research, Irvine, CA, USA). DNA libraries were prepared with the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) and sequenced as paired-end reads (2 × 300 bp) on an Illumina MiSeq platform with the MiSeq Reagent Kit version 3.
In order to check the quality of WGS data, the Illumina reads were de novo assembled into a draft genome with SPAdes v3.14.0 software [28], while contigs below 500 bp were discarded. QC parameters used as guidelines were as follows: read depth > 60 × , N50 > 30,000 bp, number of contigs < 300. Then, a phylogenetic analysis based on SNPs (single-nucleotide polymorphisms) was performed with Snippy (https://github.com/tseemann/snippy) on a reference core genome MLST of E. faecalis (1972 genes) [29] or E. faecium (1423 genes) [30] available at “cgmlst.org” website (https://www.cgmlst.org/ncs/schema/3887469/ and https://www.cgmlst.org/ncs/schema/991893/). The resulting filtered SNPs were transformed into a distance matrix for tree construction with the NJ (neighbour-joining) algorithm using Python implementation in “Scikit-bio” package (http://scikit-bio.org/docs/0.5.2/generated/skbio.tree.nj.html). For E. faecalis and E. faecium, the sum of nucleotides considerate from cgMLST after filtering step was 699.839 nt and 520.407 nt, respectively. For E. faecalis and E. faecium, the distance matrix used for the phylogenetic reconstruction is based on SNP difference between strains. The total of these unique SNPs for each species was 14.247 nt and 3.877 nt, respectively. The phylogenetic tree and a heatmap for resistance mechanisms were generated with iTOL v5 (https://itol.embl.de/) [31].
Raw reads (in FastQ format) were submitted to the Center for Genomic Epidemiology (https://cge.cbs.dtu.dk). MLST was performed using the MLST 2.0 server, while identification of acquired resistance genes and chromosomal resistance mutations was carried out using ResFinder v4.1 (with default parameters: 90% identity threshold and 60% minimum length) and LRE-Finder v1.0 (with default parameter: 80% identity threshold) services [20, 23].
Statistical analysis
Concordance between WGS-predicted resistance and phenotypic susceptibility was determined and was defined as the presence or the absence of a genetic resistance determinant to a specific antibiotic in phenotypically-categorized resistant and susceptible isolates, respectively. The sensitivity and specificity (and their corresponding 95% CI) were calculated for each antibiotic/organism as well as rates of very major errors (VME, i.e., susceptible genotype with resistance phenotype) and major errors (ME, i.e., resistant genotype with susceptible phenotype). Phenotypic results (disk diffusion, BMD) were considered as the gold standard. Statistical calculations were performed using GrapPad Prism v.5.01 (GraphPad Software Inc., San Diego, CA, USA).
Nucleotide sequence accession numbers
The genomic sequences of the 172 strains generated in this study were deposited in GenBank as bioproject PRJNA875074.
Results
Diversity of E. faecalis and E. faecium populations
Assemblies for E. faecalis isolates (n = 34) had a median of 45 contigs (range 14–85), N50 of 188,904 bp (range 105,380–479,327 bp), coverage of 133 × (range 62–336). For the 138 E. faecium isolates, the corresponding medians were 172 contigs (range 68–259) and 43,716 bp (range 19,451–136,307 bp), coverage of 142 × (range: 63–296).
Phylogenetic analysis illustrated that the population of both E. faecalis and E. faecium was heterogeneous (Figures S1 and S2). Indeed, a high genetic diversity was documented in E. faecalis with up to 20 sequence types (STs) among the 34 isolates studied. The E. faecium population seemed to be less diverse in terms of STs (25 different STs among 138 isolates) but was representative of vancomycin-resistant E. faecium clinical isolates circulating in France since the majority of them belonged to the CC17 with the most common STs being ST80 (n = 58, 42.0%) and ST117 (n = 21, 15.2%). Using the higher-resolution core genome SNP analysis, it was confirmed that the E. faecalis population was genetically diverse with only a few strains potentially clonally related. For E. faecium, clinical isolates were distributed over the entire phylogenetic tree and covered almost every branch, indicating that the strain collection reflects an extensive and diverse selection of the E. faecium population structure.
Phenotypic resistance overview
Of the 34 E. faecalis clinical isolates selected for the study, none was resistant to ampicillin, vancomycin, and teicoplanin. Ten (29.4%) displayed a high-level resistance to gentamicin. A majority was resistant to erythromycin (n = 25, 73.5%), tetracycline (n = 27, 79.4%), chloramphenicol (n = 24, 70.6%), and linezolid (n = 67.6%). A smaller proportion of isolates was resistant to ciprofloxacin (n = 13, 38.2%), cotrimoxazole (n = 16, 47.1%), and rifampicin (n = 12, 35.3%). Only one strain (2.9%) was phenotypically resistant to tigecycline, while two (5.9%) were categorized as resistant to daptomycin. As expected, all isolates were resistant to quinupristin-dalfopristin since it is an intrinsic resistance.
Of the 138 E. faecium clinical isolates, all exhibited resistance to at least one antibiotic and most of them were resistant to different antimicrobial classes. All except one were highly resistant to ampicillin (n = 137, 99.3%), while the large majority was resistant to vancomycin (n = 118, 85.5%) and teicoplanin (n = 102, 73.9%). Eighty-six (62.3%) displayed a high-level resistance to gentamicin. More than 60% of isolates were resistant to erythromycin (n = 131, 94.9%), rifampicin (n = 126, 91.3%), ciprofloxacin (n = 121, 87.7%), tetracycline (n = 90, 65.2%), and cotrimoxazole (n = 84, 60.9%). Around 40% of isolates were resistant to quinupristin-dalfopristin (n = 57, 41.3%), while less than 20% of isolates were categorized as resistant to linezolid (n = 24, 17.4%), chloramphenicol (n = 18, 13.0%), and tigecycline (n = 10, 7.2%). For daptomycin, 14 strains (10.1%) were categorized as resistant.
Concordance between phenotypes and genotypes in E. faecalis
The analysis of genomic sequences of E. faecalis revealed the presence of 184 resistance genes or mutations within 23 different loci (Table 1). Of 408 isolate/antibiotic combinations (34 strains × 12 antibiotics), concordance between the two methods was 94.9%, but increased to 97.6% by omitting rifampicin (see below) (Table 2).
No E. faecalis isolates were resistant to ampicillin, vancomycin, or teicoplanin and as expected, none harboured pbp5 mutations or van operons. For gentamicin, all predictions were correct with 14 resistant isolates carrying the aac(6′)-aph(2″) gene and 24 susceptible isolates with no other gentamicin-resistance genes. Concordance was also 100% for chloramphenicol.
Out of 25 erythromycin-resistant isolates, 24 harbored one or two erm genes, including 16 strains positive for erm(B), 7 for erm(A) + erm(B) and 1 for erm(C), whereas a single VME was observed. Thirteen isolates were resistant to ciprofloxacin and all but one showed mutations in QRDRs of GyrA and/or ParC, the most frequent ones being S83Y in GyrA and S80I in ParC. For cotrimoxazole, 16 isolates were resistant to trimethoprim (in addition to the intrinsic resistance to sulphonamides) but only 15 acquired drfD or dfrG genes, corresponding to a single VME for this antibiotic. MEs were also observed for cotrimoxazole with 3 susceptible strains (2 dfrD + and 1 dfrG +). Note that resistance gene or mutation was found for rifampicin or tigecycline.
Twenty-eight isolates were predicted to be resistant to tetracycline with detection of a tet gene [mostly tet(L) and tet(M)] but only 27 were phenotypically resistant corresponding to one ME for a susceptible tet(L)-positive strain. For linezolid, an optrA-positive strain was categorized as susceptible, whereas all 23 resistant isolates harbored at least one resistance gene or mutation (Table 1).
Note that daptomycin resistance is not predicted by ResFinder since resistance mutations are not included in the database, and then, concordance was not analyzed, while only two daptomycin-resistant strains were studied. Also, since E. faecalis is intrinsically resistant to lincosamides and streptogramins (the so-called LSAP phenotype) [32], results for quinupristin-dalfopristin were not included.
Concordance between phenotypes and genotypes in E. faecium
The analysis of genomic sequences of E. faecium revealed the presence of 997 resistance genes or mutations within 25 different loci (Table 1). Of 1794 isolate/antibiotic combinations (138 strains × 13 antibiotics), concordance between the two methods was 86.0%, but it increased to 95.7% by omitting rifampicin and quinupristin-dalfopristin (see below) (Table 2).
Almost all E. faecium isolates (136/138) were highly resistant to ampicillin and harbored pbp5 mutations, with a concordance of 98.6% concordant with phenotypic susceptibility testing results (1 VME and 1 ME). All VRE isolates (n = 118) were resistant to vancomycin, including 100 vanA-, 16 vanB-, and 4 vanD-positive strains. All vanA-positive strains were resistant to both vancomycin and teicoplanin except two susceptible strains that harbored the vanA operon. As expected, the 16 vanB-positive strains were predicted resistant to vancomycin only since vanB does not confer to teicoplanin.
For gentamicin, all resistant strains harbored the aac(6′)-aph(2″) gene but 10 of the 52 susceptible strains were falsely predicted as resistant by WGS, corresponding to 19.2% of MEs. Note that aph(3′)-Ia, aph(3′)-IIIa, aac(6′)-Ii, and ant(6′)-Ia were commonly found but were not associated with high-level gentamicin resistance.
A large majority of strains (n = 131, 94.9%) were resistant to erythromycin, and 130 of them (99.2%) carried at least one erm gene, predominantly erm(B) and erm(T), corresponding to a single VME (0.8%). All 57 isolates resistant to quinupristin-dalfopristin were not detected The same observation was made for both tigecycline and rifampicin, as for E. faecalis, for which resistance was not predicted for 10 and 126 isolates, respectively (100% VME).
A large proportion of isolates (n = 123, 89.1%) were resistant to ciprofloxacin, and all showed mutations in QRDRs of GyrA and/or ParC, whereas two were falsely predicted as susceptible. For cotrimoxazole, 14 resistant strains were predicted as susceptible with no dfr genes detected (16.7% VME) and three dfrG-positive strains were susceptible in vitro. Discordances were also observed for chloramphenicol (1 VME and 13 MEs) for which concordance was below 90%.
For tetracycline, concordance was 93.5% with 2 VMEs and 7 MEs including 3 tet(M)-, 1 tet(L), 1 tet(M) + tet(L)-, and 1 poxtA-positive strains. Twenty-eight isolates were predicted to be resistant to tetracycline with detection of a tet gene [mostly tet(L) and tet(M)] but only 27 were phenotypically resistant corresponding to one ME for a susceptible tet(L)-positive strain.
Interestingly, concordance was 100% for linezolid susceptibility prediction with 24 phenotypically resistant strains harboring different types of resistance mechanisms (optrA, poxtA, and/or 23S rRNA mutations) and 124 susceptible strains with none resistance determinants.
As for E. faecalis, the 14 daptomycin-resistant were not detected by WGS since resistance mutations were not included in the database, and then concordance was not analyzed.
Discussion
The study reports an overall high level of concordance (96.1%) between WGS-predicted resistance and phenotypic AST using the ResFinder both for E. faecalis and E. faecium, when omitting antibiotics for which resistance mutations are not included in the ResFinder database. Indeed, the vast majority of VMEs occurred for quinupristin-dalfopristin, tigecycline, and rifampicin. This is the main caveat related to genotypic AST using the ResFinder database since only known AMR mechanisms can be detected whereas resistance due to new mechanisms and/or modulation of gene expression might also be overlooked. Indeed, most of discrepancies for erythromycin, ciprofloxacin, and cotrimoxazole might be explained by resistance mediated by unknown or not detected resistance mechanisms (Table 3). While the poor sensitivity to predict rifampicin resistance was due to the fact that resistance mutations (within the rpoB gene) are not included in the database of ResFinder. It was the same for the lack of detection of the single tigecycline-resistant isolate due to the absence of resistance mutations in rpsJ coding for the S10 ribosomal protein [33] (Table 3). In E. faecium, resistance to quinupristin-dalfopristin were also not detected (100% VME). We hypothesize that it is related since the resistance mutation in the eat(A) gene, main molecular mechanism responsible for the LSA phenotype, is not included in the database of ResFinder [34]. Moreover, several discrepancies for tetracycline, chloramphenicol, gentamicin, linezolid, and vancomycin were probably due to genes weakly or not expressed (Table 3). For instance, both strains harboring a vanA operon with vancomycin sensitive phenotype (classified as MEs) correspond to vancomycin-variable enterococci (VVE). This discrepancies might be seen like as an advantage of WGS. At last, some discrepancies for gentamicin could be explained by a truncated gene which became non-functional but identified by WGS probably because of the default setting (90% identity threshold and 60% minimum length).
To our knowledge, validation of WGS for AMR prediction in enterococci has been only poorly studied and previous studies present one or several limitations [20, 24,25,26,27].
In the first study, the authors used the first version of the ResFinder web server to predict antimicrobial susceptibility to 14–15 antibiotics from WGS data for a collection of 100 enterococcal isolates originating from Danish pigs in 2011 (50 E. faecalis, 50 E. faecium) [24]. The overall concordance was very high (> 99%) but a major limit of the study was the very low frequency of acquired resistance to clinically relevant antibiotics. For example, only two E. faecium strains were resistant to ampicillin, while only one VRE (vanA-positive E. faecium) and no LRE were included.
A second study evaluating WGS in routine resistance surveillance (2014 NARMS program) was conducted on a set of 197 enterococci from various animal and food sources [25]. The authors reported a high overall concordance (96.5%) but a large proportion of studied strains was pan-susceptible (n = 46), while neither VRE nor LRE were included. Note that the authors analyzed WGS data by using different databases combined with a manual approach to detect resistance genes or mutations.
In a monocentric study, Babiker et al. studied the concordance between WGS-based resistance prediction using online tools (downloaded ResFinder and CARD databases) and phenotypic AST results for 100 E. faecium clinical isolates (all VRE) collected at the university hospital of Pittsburgh between 2016 and 2017 [26]. After repeating AST to resolve discordances, the authors observed a high level of concordance (98.7%) with a sensitivity and a specificity of 99.0% (95% CI, 96.4–99.9%) and 98.0% (95% CI, 93.0–99.8%), respectively. However, only four antibiotics (vancomycin, linezolid, erythromycin, and tetracycline) were tested. For both vancomycin and erythromycin, 100% concordance was found, whereas 2 MEs were evidenced for tetracycline (concordance of 95.8%) and the two LRE (with 23S rRNA mutations) were not detected (concordance of 97.9%).
In a recent study, several collections of enterococci (2 E. faecium datasets of human clinical isolates from Germany [n = 50] and Belgium [n = 56]; 1 E. faecalis dataset of human and animal origin from Germany [n = 50]) were used for the development and the evaluation of the more advanced online tool, ResFinder 4.0, which allows the generation of in silico antibiograms [20]. For E. faecium isolates from Germany (363 observations, 8 antibiotics tested), overall genotype–phenotype concordance was 92.8% ranging from 64% for chloramphenicol to 100% for ampicillin, ciprofloxacin, erythromycin, gentamicin, and vancomycin. Most discordances were due to the detection of poxtA, cat, and fexB genes with < 100% identity to the database and often with low read depth, while additional discordances were due to the lack of detection of tetracycline and linezolid resistance determinants. For E. faecium isolates from Belgium (159 observations, 5 antibiotics tested), overall genotype–phenotype concordance was 96.2% ranging from 75% for gentamicin to 100% for ampicillin, ciprofloaxin. Discordances were due to the detection of gentamicin resistance genes in gentamicin-susceptible isolates (n = 2) (low read depth), vancomycin resistance genes with reliable coverage (n = 2) with different vancomycin MICs, and to the lack of detection of AMR determinants in isolates with linezolid MICs one step dilution above ECOFF values used as cut-offs in the study to discriminate between wild-type and non-wild-type strains. For the collection of 50 E. faecalis from Germany, overall genotype–phenotype concordance was 97.0% ranging from 96% for erythromycin and linezolid to 98% for tetracycline and vancomycin. Discordances were represented by isolates possessing erm(B) (n = 1) and tet(M) (n = 1) with 100% and < 100% identity, and an MIC distant from ECOFF knowing that tet(M) was detected with a very low read depth. Additional discordances were caused by the lack of detection of determinants of resistance to erythromycin, gentamicin, linezolid, and vancomycin in some isolates with obvious non-wild-type phenotype. Note that genes were scored as “low read depth” with (1) a “depth < 10” or (2) a “depth < 1/10 compared to that of other resistance genes in the same isolate and query coverage < 100%.” Interestingly, the authors also compared performances between input file formats (FASTQ or FASTA) and demonstrated that there was only 0.2% difference between results obtained with raw reads and assembled sequences. Then, they concluded that FASTQ files should be considered as the preferable input for ResFinder 4.0 since they are processed considerably faster than FASTA files.
Finally, a last study was performed using a collection of E. faecium clinical isolates from a single large US center (Massachusetts General Hospital, Boston) temporally divided into derivation (n = 177, 2016–2017) and validation (n = 205, 2018–2019) sets, while they also included dataset from Germany (n = 50) for external validation [27]. Initial susceptibility testing was carried out with Vitek2 for eight antibiotics (ampicillin, gentamicin, ciprofloxacin, levofloxacin, linezolid, vancomycin, tetracycline, and doxycycline), while confirmatory testing was done with BMD (Sensititre) and gradient diffusion (Etest) when needed. Categorical interpretations were done according to CLSI guidelines (document M100). For the derivation set, a rule-based approach (using different database and a home-made approach) was used and different resistance mechanisms were detected: pbp-5 mutations (most resistant alleles were found with a mutation in codon 485: Met485 to Ala or Thr), vanA and vanB vancocymin resistance genes, high-level resistance to gentamicin associated with aac(6′)-aph(2″), mutations in gyrA and parC, and numerous tetracycline resistance genes including tet(M), tet(L) and tet(S). Unfortunately, no rule-based method was possible for linezolid because of the paucity of LRE isolates (< 1%). Using the validation set, the categorical agreement (CA) was generally excellent with an average of 98%, while all drugs achieved a CA > 89.9%: 99% for ampicillin (2 VMEs, other mutations?), vancomycin (1 VVE that lacked vanRS and a silent vanA); 100% for gentamicin, ciprofloxacin, levofloxacin, linezolid (but only 2 LRE included); 97% for tetracycline; and 91% for doxycycline. The proportion of VMEs was 1.4% and that of MEs < 3% for antibiotics tested except tetracycline (14%) and doxycycline (27%) due to the presence of tet genes in phenotypically susceptible isolates. Considering the external German set (n = 50), an average CA of 97% was demonstrated with a CA of 100% that was observed for ampicillin, vancomycin, and ciprofloxacin. For gentamicin, CA was 69% due to 1 false-negative and 3 false positives with aac(6′)-aph(2″). For tetracycline, CA was 94% (2 mE and 1 ME with tet(M). A CA of 100% was found for linezolid but once again the number of LRE was very low (n = 2).
As compared to previous studies, our study has several strengths since we tested a large collection of human clinical isolates representative of the French epidemiology, we included numerous strains with multiple resistance mechanisms (particularly towards clinically relevant antibiotics such ampicillin, vancomycin, gentamicin, or linezolid), we studied a large panel of antibiotics (12 or 13), and we used a user-friendly online interface for bioinformatics analysis (directly with FASTQ files) like every non-expert clinical microbiologist would.
There are several limitations of this study. First, ResFinder failed to identify chromosomal point mutations conferring resistance to different antibiotics (i.e., quinupristin-dalfopristin, tigecycline, daptomycin, rifampicin) since they are not included in the database. Therefore, this data base should be implemented with these several resistance mutations, which is quite easy to do. Second, a limited number of E. faecalis isolates (even if the collection was enriched by LRE) as compared to that of E. faecium strains.
Genotypic prediction of a resistance phenotype has the potential to supplant traditional AST for the coming decades. Indeed, because of the reduction in cost and turnaround time, results from WGS-based approaches could be available before those from conventional techniques (48–72 h) in the near future, especially with real-time NGS techniques. Here, we showed good performances of WGS (with no bioinformatics skills) to predict difficult-to-detect resistance phenotypes among multidrug-resistant enterococci (e.g., vanB-mediated vancomycin resistance, plasmid-mediated linezolid resistance).
Conclusion
This study confirms the performances of WGS coupled with online automated biofinformatics analysis for AMR prediction among enterococci. With falling costs, reduced turnaround times, and increased sequence quality, WGS has the potential to become a tool routinely used in clinical microbiology laboratories. Since a major barrier to the widespread adoption of WGS for AMR prediction is the lack of biofinformatics expertise of clinical microbiologists, the use of online-available web-based AMR databases easy to use/user-friendly directly from raw NGS data is of paramount importance. In conclusion, this approach (WGS + web-based bioinformatics analysis) could be used by clinical microbiologists in real life for AST prediction of enterococci. However, AMR databases should be completed for some clinically relevant antibiotics.
Data availability
The genomic datasets generated and analyzed during the current study are available in GenBank as bioproject PRJNA875074.
Code availability
Not applicable.
References
Cattoir V (2022) The multifaceted lifestyle of enterococci: genetic diversity, ecology and risks for public health. Curr Opin Microbiol 65:73–80
Arias CA, Murray BE (2012) The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10(4):266–278
Bender JK, Cattoir V, Hegstad K, Sadowy E, Coque TM, Westh H et al (2018) Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: Towards a common nomenclature. Drug Resist Updat 40:25–39
García-Solache M, Rice LB (2019) The Enterococcus: a model of adaptability to its environment. Clin Microbiol Rev 32(2):e00058-e118
Wheat PF (2001) History and development of antimicrobial susceptibility testing methodology. J Antimicrob Chemother 48(Suppl 1):1–4
Khan ZA, Siddiqui MF, Park S (2019) Current and emerging methods of antibiotic susceptibility testing. Diagnostics 9(2):49
Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, Del Ojo EC et al (2013) Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 68(10):2234–2244
Gordon NC, Price JR, Cole K, Everitt R, Morgan M, Finney J et al (2014) Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 52(4):1182–1191
Tyson GH, McDermott PF, Li C, Chen Y, Tadesse DA, Mukherjee S et al (2015) WGS accurately predicts antimicrobial resistance in Escherichia coli. J Antimicrob Chemother 70(10):2763–2769
Bradley P, Gordon NC, Walker TM, Dunn L, Heys S, Huang B et al (2015) Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun 6(1):10063
McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP et al (2016) Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 60(9):5515–5520
Ruppé E, Cherkaoui A, Charretier Y, Girard M, Schicklin S, Lazarevic V et al (2020) From genotype to antibiotic susceptibility phenotype in the order Enterobacterales: a clinical perspective. Clin Microbiol Infect 26(5):643
Dahl LG, Joensen KG, Østerlund MT, Kiil K, Nielsen EM (2021) Prediction of antimicrobial resistance in clinical Campylobacter jejuni isolates from whole-genome sequencing data. Eur J Clin Microbiol Infect Dis 40(4):673–682
Cortes-Lara S, Barrio-Tofiño ED, López-Causapé C, Oliver A, GEMARA-SEIMC/REIPI Pseudomonas study Group (2021) Predicting Pseudomonas aeruginosa susceptibility phenotypes from whole genome sequence resistome analysis. Clin Microbiol Infect 27(11):1631–1637
Anjum MF, Zankari E, Hasman H (2017) Molecular methods for detection of antimicrobial resistance. Microbiol Spectr 5(6). https://doi.org/10.1128/microbiolspec.ARBA-0011-2017
Su M, Satola SW, Read TD (2019) Genome-based prediction of bacterial antibiotic resistance. J Clin Microbiol 57(3):e01405-e1418
Papp M, Solymosi N (2022) Review and comparison of antimicrobial resistance gene databases. Antibiot (Basel) 11(3):339
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O et al (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67(11):2640–2644
Florensa AF, Kaas RS, Clausen PTLC, Aytan-Aktug D, Aarestrup FM (2022) ResFinder — an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb Genomics 8(1):000748
Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V et al (2020) ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75(12):3491–3500
Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM (2017) PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 72(10):2764–2768
Clausen PTLC, Aarestrup FM, Lund O (2018) Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics 19(1):307
Hasman H, Clausen PTLC, Kaya H, Hansen F, Knudsen JD, Wang M et al (2019) LRE-Finder, a Web tool for detection of the 23S rRNA mutations and the optrA, cfr, cfr(B) and poxtA genes encoding linezolid resistance in enterococci from whole-genome sequences. J Antimicrob Chemother 74(6):1473–1476
Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agersø Y, Lund O et al (2013) Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother 68(4):771–777
Tyson GH, Sabo JL, Rice-Trujillo C, Hernandez J, McDermott PF (2018) Whole-genome sequencing based characterization of antimicrobial resistance in Enterococcus. Pathog Dis 76(2):fty018
Babiker A, Mustapha MM, Pacey MP, Shutt KA, Ezeonwuka CD, Ohm SL et al (2019) Use of online tools for antimicrobial resistance prediction by whole-genome sequencing in methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE). J Glob Antimicrob Resist 19:136–143
Anahtar MN, Bramante JT, Xu J, Desrosiers LA, Paer JM, Rosenberg ES et al (2022) Prediction of antimicrobial resistance in clinical Enterococcus faecium isolates using a rules-based analysis of whole-genome sequences. Antimicrob Agents Chemother 66(1):e01196-e1221
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol J Comput Mol Cell Biol 19(5):455–477
Neumann B, Prior K, Bender JK, Harmsen D, Klare I, Fuchs S et al (2019) A core genome multilocus sequence typing scheme for Enterococcus faecalis. J Clin Microbiol 57(3):e01686-e1718
de Been M, Pinholt M, Top J, Bletz S, Mellmann A, van Schaik W et al (2015) Core genome multilocus sequence typing scheme for high-resolution typing of Enterococcus faecium. J Clin Microbiol 53(12):3788–3797
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49(W1):W293–W296
Singh KV, Weinstock GM, Murray BE (2002) An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob Agents Chemother 46(6):1845–1850
Cattoir V, Isnard C, Cosquer T, Odhiambo A, Bucquet F, Guérin F et al (2015) Genomic analysis of reduced susceptibility to tigecycline in Enterococcus faecium. Antimicrob Agents Chemother 59(1):239–244
Isnard C, Malbruny B, Leclercq R, Cattoir V (2013) Genetic basis for in vitro and in vivo resistance to lincosamides, streptogramins A, and pleuromutilins (LSAP phenotype) in Enterococcus faecium. Antimicrob Agents Chemother 57(9):4463–4469
Funding
This work was supported by “Santé Publique France,” the French national public health agency.
Author information
Authors and Affiliations
Contributions
Malo Penven and Vincent Cattoir contributed to the study conception and design. Material preparation, data collection, and analysis were performed by all the authors. The first draft of the manuscript was written by Malo Penven and Vincent Cattoir, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
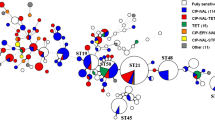
Figure S1.
Genetic relationship and resistance gene content among 34 E. faecalis clinical isolates. Neighbor-joining phylogenetic tree was constructed from aligned core-genome SNPs analysis based on SNPs and visualized together with ST affiliation (colored strips) and a heatmap for resistance mechanisms (black boxes) with iTOL v5. The scale bar represents 100 SNPs. (PNG 405 kb)
Figure S2.
Genetic relationship and resistance gene content among 138 E. faecium clinical isolates. Neighbor-joining phylogenetic tree was constructed from aligned core-genome SNPs analysis based on SNPs and visualized together with ST affiliation (colored strips) and a heatmap for resistance mechanisms (black boxes) with iTOL v5. The scale bar represents 1,000 SNPs. (PNG 914 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Penven, M., Zouari, A., Nogues, S. et al. Web-based prediction of antimicrobial resistance in enterococcal clinical isolates by whole-genome sequencing. Eur J Clin Microbiol Infect Dis 42, 67–76 (2023). https://doi.org/10.1007/s10096-022-04527-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-022-04527-z