Abstract
Staphylococcus aureus (SA) is the leading cause of bloodstream infection (BSI). The incidence of methicillin-resistant SA (MRSA) has decreased in France and Europe since one decade. Early and precise prediction of methicillin susceptibility is needed to improve probabilistic antibiotic therapy of MRSA-BSI. The aim of this study was to identify MRSA-BSI risk factors at admission and evaluate which patients need costly rapid diagnostic tests. A single-center retrospective descriptive study of all diagnosed SA-BSI was conducted in a French University Hospital between January 2015 and December 2016. All medical charts were reviewed. Univariate and multivariate analyses by a logistic regression model were performed on the data. We then build a prediction score of MRSA-BSI by assigning one point for each of the risk factor identified. During the study period, 151 SA-BSI were identified including 32 (21%) MRSA-BSI. In multivariate analysis, three factors were associated with MRSA-BSI: coming from long-term care facility, known previous MRSA colonization and/or infection, and chronic renal disease. Among our population, respectively, 5% and 100% had a MRSA-BSI when no or three risk factors were identified. Therefore, among the PCR performed, 43 (96%) could be avoided according to our clinical score. In our study, methicillin-susceptible SA and MRSA-BSI can be predictable by counting MRSA risk factors. This prediction rule could avoid the use of expensive rapid diagnostic tests. Prospective studies and prediction rules could help physicians to predict SA-BSI susceptibility to improve appropriate empiric therapy choice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Staphylococcus aureus (SA) is one of the most frequent causes of community- and healthcare-associated bloodstream infection (BSI). SA-BSI is associated with a high morbidity and mortality [1]. Appropriate antimicrobial therapy (AAT) seems to be the most important factor associated with a better outcome. Indeed, several studies [2,3,4,5,6,7] suggested that inappropriate antibiotic therapy was associated with a higher risk of mortality and extended hospital length of stays. Moreover, recent studies highlighted a higher risk of secondary localization and mortality among methicillin-susceptible SA (MSSA) BSI treated with glycopeptides compared with those treated by β-lactam such as cloxacillin or cefazolin [8,9,10,11,12,13]. Also, guidelines for multi-drug-resistant bacteria control include prudent use of vancomycin that appears to be a risk factor for vancomycin-resistant Enterococcus acquisition. The choice of empiric antimicrobial therapy for suspected SA-BSI is usually based on local or national prevalence of methicillin-resistant SA (MRSA). In France, during the last decade, a sustained reduction of MRSA had been obtained and MRSA prevalence is now estimated to be 16.4% of SA in healthcare institutions [14] and 26.5% in hospital-acquired infection [15]. Despite this reduction, MRSA active drug prescription is increasing [16]. Moreover, as early AAT is associated with a better outcome, early and precise prediction of methicillin susceptibility is needed to improve the first-line empirical therapy choices and avoid superfluous use of MRSA-active antibiotics.
Recently, new technologies can inform on bacterial identification and antibiotic susceptibilities sooner such as matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS identification combined to PBP2a detection test [17], SA immunochromatographic test combined to PBP2a detection test [18], and real-time polymerase chain reaction (PCR) detecting both S. aureus species and methicillin susceptibility. Xpert MRSA PCR is widely used because of its high sensibility, specificity [19], efficiency, and rapidity. MRSA PCR performed on blood cultures appears to reduce mortality, duration of antimicrobial administration, and length of stay [20, 21]. However, because of their cost, MRSA PCR utilization should be delimited. Decision models based on MRSA risk factors are needed to determine if costly specific tests are required.
Considering the epidemiological changes and the need to improve diagnosis, we conducted a retrospective study with the aim to identify risk factors associated with MRSA bloodstream infections and to evaluate which patient needs rapid diagnostic test.
Methods
Design, setting, and population
This study was conducted at the Avicenne Hospital, France, a 500-bed adult teaching hospital, including surgical, medical, and intensive care unit (ICU) wards. From our database, we retrospectively extracted all the blood cultures with an identified S. aureus, occurring between the first of January 2015 and the last of December 2016. Data were extracted from available medical records and included the following variables: age, demographic characteristics, presence of underlying disease (chronic cardiac failure, chronic pulmonary disease, chronic renal disease, diabetes, hematologic malignancies, inflammatory disease, solid cancer), Charlson Comorbidity Index [22], origin of onset (community or hospital acquired), prior known MRSA colonization or infection (indefinite period of time), prior use of antibiotics (during the last 3 months), prior surgery (within the last 3 months), prior hospitalization (within the last 12 months), site of infection, presence of central venous catheter at time of BSI, ward of admission (surgical, medical, or intensive care). Diseases that caused chronic renal disease included type 1 or type 2 diabetes, high blood pressure, glomerulonephritis, and interstitial nephritis. Outcome data that were collected included duration of hospital stay, secondary transfer to ICU, deterioration of clinical status within 48 h, severity of infection at time of diagnosis (sepsis, severe sepsis, or septic shock), and in-hospital mortality. Antibiotics susceptibility tests were extracted from microbiologic laboratory database.
Definition
An episode of S. aureus bacteremia was defined as a positive culture in at least one or more set of blood cultures. If a patient had two separate episodes of SA-BSI during the study period (30 or more days after the initial positive blood culture), both episodes were separately included. We divided the origin of bacteremia in three categories: community-acquired (CAI), hospital-acquired (HAI) and healthcare-associated infection (HCAI). Hospital-acquired infection was defined as a SA-BSI episode occurring 48 h or longer after hospital admission. Healthcare-associated infection was defined as a SA-BSI present at hospital admission or within 48 h of admission in patients that fulfilled one or more Friedman’s criteria [23]. The primary source of bacteremia was determined according to CDC criteria [24]. Otherwise, it was defined as primary bacteremia with no determined portal of entry. The severity of illness was defined as severe sepsis or septic shock according to Bone’s criteria [25].
Laboratory methods
For all positive blood culture with a Gram positive suggesting staphylococci, a direct tube coagulase test (rabbit plasma for coagulase test; Bio-Rad) was performed to detect free coagulase produced by S. aureus. 0.5 mL of blood culture broth was mixed to 0.5 mL reconstituted rabbit plasma in hemolysis tubes and incubated at 37 °C. The tubes were examined after 2, 4, and 6 h of aerobic incubation. Results were transmitted to the physician in charge. Xpert MRSA PCR (Xpert® MRSA/SA blood culture; Cepheid) was performed directly on positive blood culture before or after tube coagulase test at the practitioner’s discretion. S. aureus species were identified on subculture with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry ([MALDI-TOF MS] Microflex LT; Bruker Daltonics). Susceptibility testing and reporting were performed according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guideline. MRSA were identified by cefoxitin resistance testing on subculture.
Statistics
Statistics were performed on R software (version 3.3.2). We first used a univariable logistic regression model in order to describe the relationship between each study covariate and the occurrence of MRSA bloodstream infection. With this, we obtained odds ratios and corresponding confidence intervals. Next, we fitted a multivariable logistic regression model on the data. In a first step, using stepwise backward variable selection, we selected the covariates that led to the model with the lowest Bayesian information criterion (BIC). We then incorporated pairwise interaction effects among the retained covariates, and by the same BIC, we further suppressed non-significant interactions. We tested differences in SA-BSI outcome using the χ2 test. Taking into account the several risk factors identified in our study, we tried to build a prediction score of MRSA-BSI. We assigned one point for each of the risk factor identified in multivariate analysis.
Ethics
Ethical approval was not required as all procedures were part of our routine care. All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki Declaration and its later amendments.
Results
Study population and patients’ characteristics
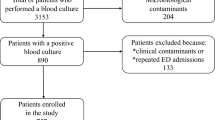
From January 2015 to December 2016, around 6000 blood culture remained positive at the laboratory for all bacteria species. Among them, a total of 162 SA-BSI episodes were identified. Eleven (7%) episodes were excluded because of lack of sufficient data, resulting in the final 151 SA-BSI episodes included in this study.
Among the 151 included BSI episodes, 32 (21%) were due to MRSA and the prevalence increased from community to HAI (Table 1). Only 39 patients had a SA-BSI without any Friedman’s criteria. Among HAI, 47 (59%) patients were in a medical ward, 21 (27%) in an intensive care unit, and 11 (14%) in the surgery department. The mean Charlson score at diagnosis was 3.8, and 136 (90%) patients had at least one comorbidity. The two most frequent primary sources of infections were catheter related with 46 (29%) SA-BSI and skin and soft tissue with 36 (24%) SA-BSI, whereas primary bacteremia was identified for 40 (26%) patients (Table 1). At time of BSI, almost half of patients had intravenous catheter. Among the 40 patients with primary bacteremia, 22 had a catheter and 10 of them were removed. Eighty percent of the studied population has at least one of the following criteria: antibiotics therapy within the previous 3 months (34%), surgery within the previous 3 months (28%), and hospitalization in the last 12 months (70%). Ten patients with MRSA-BSI had a previous MRSA colonization/infection in the last year (from 3 days to 9 months ago) and only one patient with MSSA-BSI had a previous MRSA colonization/infection 14 months ago. MRSA and MSSA antibiotics susceptibility is listed in Table 2.
MRSA risk factors
By univariate analysis, six factors were significantly associated with higher risk of MRSA bloodstream infection (Table 1): long-term care facility or home medical care (OR = 7.2, 95% CI = [2.6, 20.6], p < 0.01), hospitalization in the previous 12 months (OR = 4.5, 95% CI = [1.4, 19.8], p = 0.02), previous MRSA colonization or infection (OR = 42.9, 95% CI = [7.3, 820.8], p < 0.01), antibiotic therapy in the previous 3 months (OR = 4.3, 95% CI = [1.8, 10.9], p < 0.01), chronic renal disease (OR = 4.1, 95% CI = [1.7, 10.3], p < 0.01), and Charlson score (OR = 1.1, 95% CI = [1.0, 1.3], p = 0.04).
By multivariate analysis, only three factors remained associated with higher risk of MRSA-BSI (model with Bayesian information criterion, 41.1 and Akaike information criterion, 25.4): long-term care facility, known previous MRSA colonization or infection, and chronic renal disease (Table 1).
Prediction rule
To establish a prediction score, we assigned one point for each of the risk factor identified by multivariate analysis. Sensitivity, specificity, negative predictive value, and positive predictive value were calculated and listed in Table 3. MRSA and MSSA risks based on the score’s result are listed in Table 4. Among our population, the risk to be infected with a MRSA strain was 5% when our score was at 0 and increase to 100% when it reaches 3 with a small sensitivity. Focusing on patients with a score at 0, we then calculated this risk according to the onset of infection: 7.3%, 5.3%, and 3.5% for respectively hospital-acquired infection, healthcare-associated infection, and community-acquired SA-BSI.
Independent of the prediction rule, we extracted all Xpert MRSA PCR performed on blood culture during the study period. From January 2015 to December 2016, 171 PCR Xpert MRSA have been recorded: 101 identifying coagulase-negative staphylococci and only 70 S. aureus. Among them, 45 patients had a score at zero predicting MSSA, and methicillin phenotypic concordance was observed in 43 out of the 45 (96%) S. aureus. The combination of rapid coagulase test and the clinical score could have avoided 144 out of the 171 PCR performed (84%).
Outcome
Among our population, the in-hospital rate of mortality was 27% and was significantly associated with MRSA-BSI compared with MSSA-BSI (47% versus 22%, p < 0.01). Also, septic shock and deterioration of clinical status within 48 h were higher with MRSA-BSI (respectively 19% and 28% versus 10% and 22%). In-hospital mortality was surprisingly twofold higher in the subgroup with SA-BSI of unknown origin (45% versus 21%; p = 0.04).
Discussion
The present study highlighted three factors associated with MRSA-BSI: long-term care facility, known previous MRSA colonization and/or infection, and chronic renal disease. These risk factors allow us to establish a simple and friendly use of clinical score at the bedside. Also, it could help in our practice to spare the use of costly tests in 96% for patients with no MRSA risk factors. Indeed, the three MRSA-SI risk factors found are easily identifiable by a physician and help us to exclude MRSA-BSI. Several studies had previously identified these risk factors as associated with a higher risk of MRSA-BSI [26,27,28,29]. However, combination of them differs between studies. In our study, knowing previous MRSA colonization/infection was found as the main risk factor. With a high OR, it seems the best variable to predict MRSA-BSI. Indeed, 10 out of 11 patients with previous MRSA colonization/infection had a MRSA-BSI. The only MRSA carrier with MSSA-BSI has been a carrier for over a year. In contrast with other previous published studies, skin or soft tissue infection, presence of indwelling venous catheter, or even previous surgery was not identified as risk factors associated with MRSA-BSI in our study. This could be explained by a heterogeneous MRSA strain distribution between countries and infection prevention behaviors. Indeed, a 2015 European multicenter study showed that MRSA was involved in only 15.1% of CA-SA skin and soft tissue infection in Europe with a gradient distribution from north to south, and no USA300 CA-MRSA clone was detected [30].
One of the interesting points of our clinical score lies in the fact that when none of the identified factors was present at admission, it rules out the diagnosis of MRSA with a high predictive negative value (95%). Also, although only 3 patients presented cumulative associated risk factors, all of them had a MRSA-BSI. For these two situations (none or 3 MRSA risk factors), our clinical score could help physicians for empiric therapeutic choice and help us to avoid a large number of unnecessary costly tests. Indeed, using retrospectively the clinical score could help avoid 43 MRSA PCR out of 45 done for patients with a score at zero. In contrast, S. aureus susceptibility seems difficult to predict for patients with one or two risk factors and MRSA PCR could be useful for these patients. Several authors have studied the cost-effectiveness of MRSA PCR to detect MRSA carriers and concluded that rapid technologies are probably not leading to overall cost savings [31, 32]. Using a pharmacoeconomic methodology, Brown et al. [20] compared PCR testing to empiric use of vancomycin or semi-synthetic penicillin cost-effectiveness. Their study supports the supposition that methicillin susceptibility information by PCR testing available at the time of initial antibacterial prescribing could be cost-effective, with several limitations. The use of PCR could be more efficient if targeted. Several microbiologists or clinicians use the PCR before identification of S. aureus while direct tube coagulase test could rapidly and easily detect it from positive blood culture because of its high sensitivity and specificity [33, 34]. Indeed, in our study, direct coagulase test result could avoid 101 out of 171 MRSA PCR. The microbiology laboratory should be able to keep control on the indications in which performance of a PCR assay for MRSA would be allowed (only after and not before identification of S. aureus by coagulase testing). Combination of direct coagulase test and clinical score could avoid up to 84% of all PCR performed.
The MRSA incidence is decreasing in France and Europe. In our study, from 2015 to 2016, MRSA were involved in 21% of all SA-BSI, which complies with national French epidemiology. SA antibiotics susceptibility confirms epidemiology is changing: only few MRSA were resistant to gentamicin and 1 out of 3 was susceptible to ofloxacin. Even though we have not tested all fluoroquinolones’ susceptibility, they have been described as a strong co-resistant marker of MRSA. Walter et al. [35] had observed the same change in the co-resistance profiles of S. aureus in Germany. In our population, we found a considerable part of avoidable SA-BSI. First, healthcare-associated and hospital-acquired infections represented the majority of MRSA-BSI and MSSA-BSI; catheter-related infection represented the most frequent source of SA-BSI. Moreover, the source of bacteria was not found in a large number of SA-BSI cases. Catheter-related infections could be underestimated among our population leading to a considerable part of avoidable S. aureus infection.
In our population, septic shock and deterioration of clinical status within 48 h were higher among MRSA-BSI than those of MSSA-BSI, and the in-hospital rate of mortality was 27% and was significantly associated with an MRSA infection which has previously been found [36]. These results support the necessity to identify MRSA risk factors to improve appropriate antimicrobial therapy and SA-BSI management. Old studies warned that lack of identifiable focus aggravates and prolongs SA-BSI [37, 38]. In our population, we find that in-hospital mortality was twofold higher in the subgroup with SA-BSI of unknown origin that confirms the identification of the site of infection for bacteremia is primordial.
The major limitation of our study is we obtained data in a limited number of case observations from a small sample size and from a single healthcare center. Like all retrospective studies, we observed a lack of accuracy of the data. For example, only 11 patients were known as MRSA carriers while the MRSA rate is higher in the general population. However, previous MRSA colonization/infection was found as the highest risk factor in our analysis. Another limitation is that no antibiotics data were analyzed which has a significant impact on SA-BSI mortality but it does not affect the principal aim of our study: identify MRSA risk factors. Even though the number of study subjects was small, we achieved to identify several MRSA-BSI risk factors. To build our clinical score, we assigned one point for each risk factor without considering their strength. For example, previous MRSA colonization/infection was the best variable predicting MRSA-BSI in univariate analysis and could have the strongest impact in our clinical score but we choose to build a user-friendly clinical score.
To our knowledge, this is the first study evaluating prediction score compared with rapid diagnostic tests in real life. We propose a decision tree to improve SA-BSI management based on our clinical score and rapid diagnostics (Fig. 1). For every positive blood culture suggesting staphylococci, a rapid direct coagulase test should be performed before PCR. Then, with a positive coagulase, MRSA risk factors should be researched to calculate clinical score even though medical information may not necessarily be present in the file or available to the physician at the time the patient develops its infection. MRSA PCR could be avoided for patients with no or three risk factors suggesting strongly MSSA and MRSA-BSI respectively and appropriate empiric therapy should be initiated. MRSA PCR should be performed for patients with one or two risk factors to determine methicillin sensibility. A prospective multicentric study with a bigger sample size is needed to confirm the clinical MRSA score established in this study. Our score should not be transposed without previous local reassessment of the risk factors as these may vary depending on the epidemiology.
Conclusion
Even though MRSA epidemiology varies between countries, our study suggests MRSA risk factors are easily identifiable and should be researched before costly rapid diagnostic tests are executed. Prediction rules could help physicians to predict SA-BSI susceptibility in order to improve appropriate antibiotherapy choice.
References
Keynan Y, Rubinstein E (2013) Staphylococcus aureus bacteremia, risk factors, complications, and management. Crit Care Clin 29:547–562. https://doi.org/10.1016/j.ccc.2013.03.008
Lodise TP, McKinnon PS, Swiderski L, Rybak MJ (2003) Outcomes analysis of delayed antibiotic treatment for hospital-acquired Staphylococcus aureus bacteremia. Clin Infect Dis 36:1418–1423. https://doi.org/10.1086/375057
Gómez J, García-Vázquez E, Baños R, Canteras M, Ruiz J, Baños V et al (2007) Predictors of mortality in patients with methicillin-resistant Staphylococcus aureus (MRSA) bacteraemia: the role of empiric antibiotic therapy. Eur J Clin Microbiol Infect Dis 26:239–245. https://doi.org/10.1007/s10096-007-0272-x
Soriano A, Marco F, Martinez JA, Pisos E, Almela M, Dimova VP et al (2008) Influence of vancomycin minimum inhibitory concentration on the treatment of methicillin-resistant Staphylococcus aureus bacteremia. Clin Infect Dis 46:193–200. https://doi.org/10.1086/524667
Rodríguez-Baño J, Millán AB, Domínguez MA, Borraz C, González MP, Almirante B et al (2009) Impact of inappropriate empirical therapy for sepsis due to health care-associated methicillin-resistant Staphylococcus aureus. J Inf Secur 58:131–137. https://doi.org/10.1016/j.jinf.2008.11.003
Paul M, Kariv G, Goldberg E, Raskin M, Shaked H, Hazzan R et al (2010) Importance of appropriate empirical antibiotic therapy for methicillin-resistant Staphylococcus aureus bacteraemia. J Antimicrob Chemother 65:2658–2665. https://doi.org/10.1093/jac/dkq373
Bassetti M, Trecarichi EM, Mesini A, Spanu T, Giacobbe DR, Rossi M et al (2012) Risk factors and mortality of healthcare-associated and community-acquired Staphylococcus aureus bacteraemia. Clin Microbiol Infect 18:862–869. https://doi.org/10.1111/j.1469-0691.2011.03679.x
Khatib R, Saeed S, Sharma M, Riederer K, Fakih MG, Johnson LB (2006) Impact of initial antibiotic choice and delayed appropriate treatment on the outcome of Staphylococcus aureus bacteremia. Eur J Clin Microbiol Infect Dis 25:181–185. https://doi.org/10.1007/s10096-006-0096-0
Stryjewski ME, Szczech LA, Benjamin DK, Inrig JK, Kanafani ZA, Engemann JJ et al (2007 Jan 15) Use of vancomycin or first-generation cephalosporins for the treatment of hemodialysis- dependent patients with methicillin-susceptible Staphylococcus aureus bacteremia 2018:7. Clin Infect Dis 44(2):190–196
Schweizer ML, Furuno JP, Harris AD, Johnson JK, Shardell MD, McGregor JC et al (2011) Comparative effectiveness of nafcillin or cefazolin versus vancomycin in methicillin-susceptible Staphylococcus aureus bacteremia. BMC Infect Dis 11. https://doi.org/10.1186/1471-2334-11-279
Horino T, Sato F, Hosaka Y, Hoshina T, Tamura K, Nakaharai K et al (2015) Predictive factors for metastatic infection in patients with bacteremia caused by methicillin-sensitive Staphylococcus aureus. Am J Med Sci 349:24–28. https://doi.org/10.1097/MAJ.0000000000000350
McDanel JS, Perencevich EN, Diekema DJ, Herwaldt LA, Smith TC, Chrischilles EA et al (2015) Comparative effectiveness of beta-lactams versus vancomycin for treatment of methicillin-susceptible Staphylococcus aureus bloodstream infections among 122 hospitals. Clin Infect Dis 61:361–367. https://doi.org/10.1093/cid/civ308
Wong D, Wong T, Romney M, Leung V. Comparative effectiveness of β-lactam versus vancomycin empiric therapy in patients with methicillin-susceptible Staphylococcus aureus (MSSA) bacteremia. Ann Clin Microbiol Antimicrob 2016;15:27. https://doi.org/10.1186/s12941-016-0143-3
Jarlier V, Arnaud I. (2017) Surveillance des bactéries multirésistantes dans les établissements de santé. Réseau BMR-Raisin, France. Résultats 2015. Saint-Maurice : Santé publique France, 112 p
Daniau C, Léon L, Blanchard H, Bernet C, Caillet-Vallet E et al. (2018) Enquête nationale de prévalence des infections nosocomiales et des traitements anti-infectieux en établissements de santé, France, mai-juin 2017. Saint-Maurice : Santé Publique France ; 12 p
Meyer E, Schwab F, Schroeren-Boersch B, Gastmeier P (2011) Increasing consumption of MRSA-active drugs without increasing MRSA in German ICUs. Intensive Care Med 37:1628–1632. https://doi.org/10.1007/s00134-011-2335-9
Delport JA, Mohorovic I, Burn S, McCormick JK, Schaus D, Lannigan R et al (2016) Rapid detection of meticillin-resistant Staphylococcus aureus bacteraemia using combined three-hour short-incubation matrix-assisted laser desorption/ionization time-of-flight MS identification and Alere culture colony PBP2a detection test. J Med Microbiol 65(7):626–631. https://doi.org/10.1099/jmm.0.000285
Heraud S, Freydiere A-M, Doleans-Jordheim A, Bes M, Tristan A, Vandenesch F et al (2015) Direct identification of Staphylococcus aureus and determination of methicillin susceptibility from positive blood-culture bottles in a Bact/ALERT system using Binax Now S. aureus and PBP2a tests. Ann Lab Med 35:454. https://doi.org/10.3343/alm.2015.35.4.454
Wolk DM, Struelens MJ, Pancholi P, Davis T, Della-Latta P, Fuller D et al (2009) Rapid detection of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) in wound specimens and blood cultures: multicenter preclinical evaluation of the Cepheid Xpert MRSA/SA skin and soft tissue and blood culture assays. J Clin Microbiol 47:823–826. https://doi.org/10.1128/JCM.01884-08
Brown J, Paladino JA (2010) Impact of rapid methicillin-resistant Staphylococcus aureus polymerase chain reaction testing on mortality and cost effectiveness in hospitalized patients with bacteraemia: a decision model. PharmacoEconomics 28:567–575. https://doi.org/10.2165/11533020-000000000-00000
Page A, O’Rourke S, Brennan M, Clooney L, Le Blanc D, Griffin J et al (2017 Nov 1) Impact of Xpert MRSA/SA blood culture PCR assay on management of positive blood cultures in obstetric patients: a retrospective audit. Ir J Med Sci 186(4):995–998. https://doi.org/10.1007/s11845-017-1581-4
Charlson ME, Pompei P, Ales KL, MacKenzie CR (1987) A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 40:373–383. https://doi.org/10.1016/0021-9681(87)90171-8
Friedman ND, Kaye KS, Stout JE, McGarry SA, Trivette SL, Briggs JP et al (2002) Health care--associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med 137:791–797
Horan TC, Andrus M, Dudeck MA (2008) CDC/NHSN surveillance definition of health care–associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control 36:309–332. https://doi.org/10.1016/j.ajic.2008.03.002
Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA et al (1992) Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest 101:1644–1655. https://doi.org/10.1378/chest.101.6.1644
Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S et al (2007) Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. https://doi.org/10.1001/jama.298.15.1763
Burkey M, Wilson L, Moore R, Lucas G, Francis J, Gebo K (2008) The incidence of and risk factors for MRSA bacteraemia in an HIV-infected cohort in the HAART era. HIV Med. https://doi.org/10.1111/j.1468-1293.2008.00629.x
Callejo-Torre F, Eiros Bouza JM, Olaechea Astigarraga P, Coma Del Corral MJ, Palomar Martínez M, Alvarez-Lerma F et al (2016 Sep 1) Risk factors for methicillin-resistant Staphylococcus aureus colonisation or infection in intensive care units and their reliability for predicting MRSA on ICU admission. Infez Med 24(3):201–209
Kao K-C, Chen C-B, Hu H-C, Chang H-C, Huang C-C, Huang Y-C. Risk factors of methicillin-resistant Staphylococcus aureus infection and correlation with nasal colonization based on molecular genotyping in medical intensive care units: a prospective observational study. Medicine (Baltimore) 2015;94:e1100. https://doi.org/10.1097/MD.0000000000001100
Bouchiat C, Curtis S, Spiliopoulou I, Bes M, Cocuzza C, Codita I et al (2017) MRSA infections among patients in the emergency department: a European multicentre study. J Antimicrob Chemother 72:372–375. https://doi.org/10.1093/jac/dkw431
Conterno LO, Shymanski J, Ramotar K, Toye B, van Walraven C, Coyle D et al (2007) Real-time polymerase chain reaction detection of methicillin-resistant Staphylococcus aureus: impact on nosocomial transmission and costs. Infect Control Hosp Epidemiol 28:1134–1141. https://doi.org/10.1086/520099
Stürenburg E (2009) Rapid detection of methicillin-resistant Staphylococcus aureus directly from clinical samples: methods, effectiveness and cost considerations. GMS Ger Med Sci 7. https://doi.org/10.3205/000065
Davis TE, Fuller DD, Aeschleman EC (1992) Rapid, direct identification of Staphylococcus aureus and Streptococcus pneumoniae from blood cultures using commercial immunologic kits and modified conventional tests. Diagn Microbiol Infect Dis 15:295–300. https://doi.org/10.1016/0732-8893(92)90014-K
Thirunavukkarasu S, Rathish K.C. Evaluation of direct tube coagulase test in staphylococcal bacteremia. J Clin Diagn Res 2014 May; 8(5): DC19–DC21. https://doi.org/10.7860/JCDR/2014/6687.4371
Walter J, Noll I, Feig M, Weiss B, Claus H, Werner G et al (2017) Decline in the proportion of methicillin resistance among Staphylococcus aureus isolates from non-invasive samples and in outpatient settings, and changes in the co-resistance profiles: an analysis of data collected within the Antimicrobial Resistance Surveillance Network, Germany 2010 to 2015. BMC Infect Dis 17. https://doi.org/10.1186/s12879-017-2271-6
Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y (2003) Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis 36:53–59. https://doi.org/10.1086/345476
Nolan CM, Beaty HN (1976) Staphylococcus aureus bacteremia. Current clinical patterns. Am J Med 60:495–500
Finkelstein R, Sobel JD, Nagler A, Merzbach D (1984) Staphylococcus aureus bacteremia and endocarditis: comparison of nosocomial and community-acquired infection. J Med 15:193–211
Author information
Authors and Affiliations
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
Ethical approval was not required as all procedures were part of our routine care. All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki Declaration and its later amendments. This article does not contain any studies with animals performed by any of the authors.
Informed consent
All data of this retrospective study were fully anonymized before the study authors accessed them.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Delerue, T., Cordel, H., Delerue, T. et al. Prediction of methicillin-resistant Staphylococcus aureus bloodstream infection: do we need rapid diagnostic tests?. Eur J Clin Microbiol Infect Dis 38, 1319–1326 (2019). https://doi.org/10.1007/s10096-019-03556-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-019-03556-5