Abstract
The nasopharynx is the primary site of colonization by respiratory pathogen that constitutes the port of entrance in the respiratory tract. The role of mucosal respiratory microbiota in infection has been recently emphasized; therefore, we aimed to assess if a specific respiratory microbiota profile was associated with symptomatic infection and/or with presence of respiratory viruses. We performed a case-control study to characterize the healthy respiratory microbiota and its alteration during acute viral infections. Next-generation sequencing of the 16S rRNA gene was applied to 225 nasopharyngeal samples from 177 patients with viral respiratory infection and 48 matched healthy controls. We evidenced an important decrease of bacterial alpha-diversity in patients with symptomatic respiratory infection and a loss of the healthy core microbiota, specifically anaerobes and Prevotella spp. Moreover, eight respiratory pathogens were enriched in these patients, including Staphylococcus aureus, Haemophilus influenzae, Streptococcus pneumoniae, Moraxella catarrhalis, Dol osigranulum pigrum and Corynebacterium propinquum/pseudodiphtheriticum, whose role in respiratory infection is unclear. The asymptomatic carrier of influenza harbors a microbiota similar to healthy subjects, suggesting a critical role of microbiota in the clinical expression of viruses. These data suggest that the commensal microbiota plays a significant role in susceptibility to viral infection. The frequent co-detection of virus and bacteria raises the question of a strategy to prevent bacterial disease, focusing on the prevention of nasopharyngeal colonization through effective antibiotic treatment. In addition to antibiotics, further studies should test preventive or therapeutic interventions for maintaining or restoring a healthy nasopharyngeal microbiota.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute viral respiratory infections are very common with a mean of five episodes per person/year [1]. Influenza virus, respiratory syncytial virus, metapneumovirus and rhinovirus are prevalent sources of respiratory illnesses [1], usually resulting in a relatively mild and self-limiting infection, but also leading to severe disease. Secondary bacterial infection can complicate these viral diseases, notably involving Streptococcus pneumoniae, Haemophilus influenza, Moraxella catarrhalis, and Staphylococcus aureus, which are transient colonizers of the healthy nasopharynx and are commonly responsible for pneumonia [2, 3].
These pathogens cohabit with a complex microbial community constituting the respiratory microbiota [3]. Its composition varies throughout the respiratory tract, with a gradual decrease of bacterial density in the lower airways [3]. A high inter-individual variability has been observed [3] and several factors influence its composition, including age, antibiotics, vaccine, smoking, and seasons [3]. Differences in the microbiota between healthy persons and patients with chronic respiratory disease [3] or respiratory infection [3,4,5,6] suggest an important role of these bacterial communities in the development of these diseases.
The pathophysiology of respiratory infection is complex. A substantial proportion of asymptomatic carriers of viruses [7] as high as 68% in children [7] and the presence of a significant number of respiratory viruses in the environment outside epidemic periods [8] suggest that the nasopharyngeal microbiota may play a critical role in the genesis and clinical expression of viral respiratory infection, challenging Koch’s postulates [9]. The hypothesis that respiratory infections are linked to an imbalance of the nasopharyngeal microbiota has recently emerged [3,4,5,6]. Few studies have investigated the link between the respiratory microbiota and viral disease [4, 6, 10, 11] but all suggest that a disruption of the airways microbiota may be linked to the occurrence and severity of viral infection [6, 10,11,12]. To characterize the healthy microbiota and its alteration during acute viral respiratory infections, we performed a case-control study using 16S DNA sequencing.
Materials and methods
Patients
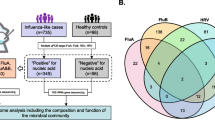
In February–March 2015, we included symptomatic patients with a respiratory infection (cough, fever, sore throat, coryza, chills) for whom a viral analysis was requested in our microbiology department (Hopital de la Timone, Marseille, France). We included five groups, each of approximately 25 individuals positive for only one tested virus (influenza A, influenza B, rhinovirus, metapneumovirus, and respiratory syncytial virus) and approximately 50 virus-negative symptomatic individuals. An age and sex group-matching was applied (Fig. 1) and the samples included were randomly selected to obtain six groups with similar baseline characteristics. We also recruited 48 asymptomatic healthy individuals through a snowball approach. This design was chosen to obtain a 2:1 ratio between cases and controls. The study was approved by the local Ethics Committee (number 2016–016). Informed consent was obtained for all the controls. Each cases and controls provided one nasopharyngeal swab, which was analyzed for both viral qPCR [13] and 16S DNA large-scale sequencing.
DNA extraction and sequencing
The samples were extracted using the NucleoSpin Tissue kit (Macherey Nagel, Hoerdt, France) and with a second method using a deglycosylation step and the EZ1 Advanced XL device (Qiagen, Courtaboeuf, France) [14]. The V3-V4 region of the 16S ribosomal RNA (rRNA) gene was amplified by PCR and a subsequent limited cycle amplification step was performed to add multiplexing indices and Illumina sequencing adapters according to the Nextera XT library kit (Illumina). Libraries were sequenced by MiSeq technology (Illumina, Inc., San Diego, CA 92121, USA) in 500 cycles. The raw data were configured in fastq files for R1 and R2 reads.
Metagenomic analyses
The paired-end reads of corresponding raw fastq files were assembled into longer joined sequences by using FLASH with a quality score cutoff of 33 [15]. The high quality sequences were then filtered using QIIME [16]. Primers were removed. Sequences shorter than 200 nts and greater than 1000 nts were also discarded. ChimeraSlayer was used to remove chimeric sequences. These sequences were clustered de novo into OTUs using UCLUST at 97% similarity [16]. The OTUs were searched against the Silva SSU and LSU reference database [17] of release 123 using blast [18]. The best matches, ≥ 80% identity and 100% coverage, were selected from the reference database, and taxonomy was assigned by applying majority voting [19].
Statistical analysis
For qualitative variables, Chi-square (when applicable) or Fisher’s exact test were used. When the sample size was very low, the two-sided Barnard’s test was used, as it is more powerful than Fisher’s exact test. For quantitative variables, statistical analyses were performed with GraphPad Prism, version 6.0 (La Jolla, CA). Normal distribution was assessed by visual exam and the Kolmogorov-Smirnov test. Student’s t test or the Mann-Whitney test was used. When three or more groups were compared, Dunn’s multiple comparison test was used. Linear discriminant analysis effect size (LEfSe) was performed as previously described [20]. To focus on the main differentially abundant species, only bacterial species identified with an absolute LDA score ≥ 3.6 were discussed. Bacterial species were defined as a “frequent respiratory pathogen” if present in the top 15 bacterial species most frequently isolated from respiratory samples and as a “frequent human pathogen” if present in the top 15 species most frequently isolated among all human samples in our lab in the 2014–2015 period (Supplementary Table 1, Table 2, Fig. 4 and Fig. 5). QIIME was used for rarefaction analysis and PCoA analysis [16]. For PcoA analysis, we rarefied the data to 20,000 reads per sample and used a weighted unifrac distance of QIIME [16]. The Adonis test (weighted unifrac distance) was used to test whether our seven groups were significantly different [21]. The Shannon indices were calculated using the formula H = −∑pi* ln(pi), where pi is the proportion of species relative to the total number of species. The AOR was calculated as previously described [19]. All tests were two-sided. A p value < 0.05 was considered significant.
Data availability
All the raw sequences of fastq files have been submitted to EMBL-EBI (http://www.ebi.ac.uk/) with the accession number PRJEB14780.
Results
Subjects’ characteristics and nasopharyngeal microbiota composition
We characterized the nasopharyngeal microbiota of 225 patients by metagenomics, including 26 symptomatic patients positive for influenza A, 27 for influenza B, 26 for respiratory syncytial virus, 28 for metapneumovirus, 23 for rhinovirus, and 47 virus-negative symptomatic patients (Fig. 1). We also enrolled 48 asymptomatic controls. An age and sex group-matching was applied to obtain seven groups with similar baseline characteristics (Table 1).
Overall, we obtained 14 million 16S rRNA gene sequence reads, classified into 2023 OTUs (operational taxonomic units), corresponding to 24 bacterial phyla. Although a high inter-individual variation was observed, the 3 dominant phyla were Proteobacteria, Firmicutes, and Actinobacteria for most subjects, 1000 different bacterial and archaeal species were identified in all the individuals tested, 303 were common to all groups, and 201 were specifically found only in virus-positive cases, 105 in virus-negative cases, and 77 in controls (Supplementary Figs. 1 and 2).
Decrease of global bacterial abundance and diversity in patients with respiratory infection
The number of reads by sample was significantly lower in virus-positive and virus-negative cases compared to controls (Fig. 2a). The Adonis test gave a p value of 0.001 and R2 value of 24, indicating that at least 24% of the variation in distances was explained by our grouping. The rarefaction curves confirmed a decreased global bacterial abundance in cases (Fig. 2b). The lower number of species per samples in cases compared to controls showed a decrease of biodiversity (Fig. 2c). The alpha-diversity, as measured by the Shannon index, was significantly lower in virus-positive (mean 1.9) than that in virus-negative patients (2.3) compared to controls (3.1, two-sided Mann-Whitney test with Dunn’s multiple correction test, p < 0.0005, Fig. 2d). A gradient was observed from controls to virus-negative and virus-positive cases.
Impoverished nasopharyngeal microbiota during viral respiratory tract infection. a While using identical metagenomic methods, the number of reads by sample was decreased in cases. b α-diversity estimated using rarefaction curves among patients with influenza A virus (n = 26), influenza B virus (27), respiratory syncytial virus (26), metapneumovirus (28), rhinovirus (23), patients with negative qPCR for virus (47), and controls (48). The vertical axis shows the number of OTUs that would be expected to be found after sampling the number of reads shown in the horizontal axis. c The decreased number of species by sample in cases confirmed the lower diversity. d α-diversity (Shannon diversity index) among virus-positive cases (n = 130), virus-negative cases (47) and controls (48). For a, c, and d: Two-sided Mann-Whitney test with Dunn’s multiple comparison test. Median and interquartile range are represented. A gradient was observed from asymptomatic controls to virus-negative and virus-positive cases
Possible correlation between the microbiota and the clinical expression of viral invasion
A principal co-ordinate analysis (PCoA) showed a distinct cluster grouping the controls, whereas patients were heterogeneously and indistinguishably distributed (Fig. 3). Among the healthy subjects, we found one asymptomatic carrier for influenza A. Interestingly, it clusterized with the controls, suggesting a correlation between the microbiota and the clinical expression of viral invasion. A restriction of heterogeneity of the nasopharyngeal microbiota was observed in controls and confirmed by the comparison of the Euclidian distance to the centroid of each group (Supplementary Fig. 3), suggesting the existence of a healthy core microbiota. Among the cases, patient groups by virus were indistinguishable from each other by principal component analysis. This suggest that the microbiota alteration in viral symptomatic infection is not specific of each virus but results from a common mechanism.
Principal co-ordinate analysis showing the alteration of the nasopharyngeal microbiota during viral respiratory tract infection. Principal component analysis (PCoA) was performed on species raw data (number of reads) using the unweighted UniFrac distance matrix. Each sample is represented by a point, with patients with influenza A virus in black (n = 26), patients with influenza B virus in green (27), patients with RSV in blue (26), patients with rhinovirus in yellow (23), patients with metapneumovirus in purple (28), symptomatic qPCR negative patients in red (47), and asymptomatic controls in dark blue (48)
Enrichment of aerotolerant pathogenic bacteria in cases
Enrichment of aerotolerant pathogenic bacteria in patients with respiratory infection
At the species level, LEfSe analysis showed an enrichment of aerotolerant pathogenic bacteria in cases versus controls, including 8 of the top 15 respiratory pathogens isolated in our microbiology department, including S. aureus, H. influenzae, Klebsiella pneumoniae, Staphylococcus epidermidis, S. pneumoniae, Serratia marcescens, Moraxella catarrhalis, and Enterobacter aerogenes (Figs. 4a, b; Supplementary Table 1 and Fig. 4). Surprisingly, there was also enrichment in Escherichia coli, which is not usually isolated from respiratory samples but which was the first pathogen isolated from all human samples in our laboratory (Supplementary Table 2 and Fig. 5). Other bacteria infrequently associated with infection were enriched in cases, including Dolosigranulum pigrum, Corynebacterium propinquum/pseudodiphteriticum, and Corynebacterium macginleyi (Fig. 4a, b). The microbiota of controls was enriched with commensal bacteria commonly present in the nasopharynx, including oral Streptococcus and different species of Rothia spp., Veillonella spp., Prevotella spp., and Neisseria spp. LEfSe analysis between positive and negative virus groups showed an enrichment of the same pathogenic bacteria as above in virus-positive cases, except M. catarrhalis, which was significantly associated with the virus-negative patients.
Alteration of the nasopharyngeal microbiota during viral respiratory tract infection at the species level resolution. Linear discriminant analysis (LDA) effect size (LEfSe) showing the enrichment of bacterial species in a virus-positive cases (n = 130) compared to controls (48) and in b virus-negative cases (47) compared to controls (48). Only bacterial species identified with an LDA score ≥ 3.6 are shown. Red: bacteria found in the top 15 human respiratory pathogens (Table S1 & Fig. S1) and top 15 human pathogens (Table S2 & Fig. S2). No human pathogens were found enriched in asymptomatic controls
Depletion of anaerobic bacteria in patients with respiratory infection
A depletion of anaerobic bacteria in patients with respiratory disease was observed. In fact, 39% (40/103) of the species enriched in controls were strict anaerobes, versus 13% (7/55) in virus-positive patients (aerotolerant odds ratio [AOR] 4.31, 95% CI 1.83–11.2, p < 0.0006). And 70% (40/57) of the enriched species in controls were strictly anaerobic, compared to 26% (23/87) in the virus-negative patients (AOR 6.45, 95% CI 3.1–13.8, p < 10−6). Moreover, Only 6% (1/18) of the species enriched in the virus-positive patients were anaerobic, and this was significantly lower than in virus-negative cases (30% (23/76), AOR 0.13, 95% CI: 0.0–0.8, p = 0.047).
The core respiratory microbiota
Because a high inter-individual variation was observed, we applied a less strict definition to characterize the group of microbes commonly found in each group. Accordingly, we defined the 50%-core as the group of OTUs present in at least 50% of the individuals. We observed a 50%-core microbiota of 13 bacterial species (Supplementary Table 3) among all the subjects. The 50%-core microbiota was composed of 60 species in the controls but only 15 in virus-positive cases and 17 in virus-negative cases. The 50%-core microbiota represented 12% of all species found in controls versus 2.7% in virus-negative and 1.9% in virus-positive cases (two-sided Chi-square test, p < 10−7) (Table 2). Fifty-two species of the healthy 50%-core microbiota were not found in virus-positive or in virus-negative cases, suggesting that these species were missing in all cases; 24 (46%) of them were strict anaerobes. In contrast, the 50%-core microbiota specific for all cases was much poorer (7 species identified, including M. catarrhalis and H. influenzae) and did not contain anaerobes (two-sided Barnard test, p = 0.014) (Supplementary Table 3). Remarkably, the strict anaerobic Prevotella spp. was the most represented genus within the 50%-core microbiota of controls.
Discussion
We showed that the healthy nasopharyngeal microbiota is composed of a diverse well-balanced aero-anaerobic bacterial core consortium which is altered during respiratory infection. Respiratory infections were associated with a predominantly anaerobic depletion and an invasion by aerotolerant frequent respiratory pathogens. To the best of our knowledge, this is the largest metagenomic study exploring the human nasopharyngeal microbiota of patients with viral respiratory illnesses and controls. A high inter-individual heterogeneity in airway microbiota composition was observed [5, 22], therefore a large number of subjects is required to reveal significant differences between the different groups. Identical protocols were performed for sampling, sequencing and data processing for samples from cases and controls. Two different DNA extraction protocols were used to obtain a higher microbial richness and diversity and to reduce variability in deep sequencing, resulting in DNA extraction bias [14].
As previously reported, we found a reduction of diversity and/or richness in patients with respiratory disease, including viral infection [4, 5, 11, 23], and patients with respiratory diseases presented an increased abundance of pathogenic bacteria, including S. aureus, S. pneumoniae, M. catarrhalis and H. influenzae [2]. Other metagenomic studies have reported a transient incursion of potentially pathogenic bacteria in viral diseases, but not all identified bacteria have risen to the rank of species [4, 6, 10, 11, 23, 24]. However, species resolution is mandatory, as different species may have a different effect on infection risk or resilience, as found here between H. influenzae (enriched in cases) and Haemophilus parainfluenzae (enriched in controls). Most studies have reported that Moraxella spp. and Haemophilus spp. were associated with viral infection and severity of illness, although Streptococcus and Staphylococcus gave inconsistent results. Higher nasopharyngeal carriage of pathogenic bacteria was reported by qPCR in patients with a concurrent viral respiratory infection, ranging from 23 to 83% of subjects [2, 10] and showed an impact on the clinical severity [25].
We did not observe any difference in microbiota profiles with regard to the causal virus type. Yi et al. analyzed the oropharyngeal microbiota of 59 patients with 7 diverse viral respiratory illnesses and came to the same conclusion [4]. The only asymptomatic patient carrying the influenza A virus in our study harbors a microbiota exactly comparable to healthy subjects, suggesting that the nasopharyngeal microbiota may play a critical role in the clinical expression of viruses. In fact, interactions between pathogens and microbiota may play a role in disease pathogenesis, and the commensal microbiota can be manipulated to cure some infections [9]. The airways microbiota seems to locally influence the immune system, and bacteria or bacterial ligands may enhance or reduce viral infection in multiple pathways [26]. As in the gut, we hypothesized that the nasopharyngeal microbiota may play a living instrumental role, sensitive to several environmental and host factors, and may be able to modify resilience or risk of infection [19]. As in Alice’s living croquet theory [27], causality is not so simple when the mechanism is composed of different living organisms. Moreover, the stability of the respiratory microbiota remains unclear. Longitudinal studies and animal models are required in order to establish if imbalance of the nasopharyngeal microbiota is a cause and/or a consequence of viral infection, and to elucidate the mechanism of susceptibility to viral infection.
Depletion in anaerobes appears to be linked to the onset of viral respiratory disease, suggesting the protective role of commensal anaerobic bacteria against these infections. Remarkably, the Prevotella genus was the most represented gender within the 50%-core microbiota specific to healthy controls [5]. It was shown that, contrary to H. influenzae and M. catarrhalis, Prevotella spp. have no pro-inflammatory effects on human dendritic cells and may have a protective role, by reducing secretion of pro-inflammatory cytokines induced by H. influenzae [28]. Prevotella spp. was also found to decrease significantly in patients with H1N1 flu, pneumonia and sinusitis [5, 29, 30]. Preventive and/or therapeutic intervention may aim at maintaining or restoring the healthy aero-anaerobic nasopharyngeal microbiota. We previously showed that antioxidants, including ascorbate, enabled the aerobic culture of anaerobes including Prevotella spp. [31]. On the contrary, we observed that C. propinquum/pseudodiphteriticum and D. nigrum were found to be associated with susceptibility to viral infection. However, the role of these two bacteria in respiratory infection is unclear. In fact, very few cases of opportunistic respiratory infection are reported for these two bacteria. They are known to be positively associated with breastfeeding and colonize the nasopharynx during the first year of life, then persist in adults, with lower relative abundance compared to children [32]. They are often cultured together from respiratory samples, suggesting a mutualistic relationship [33]. They are rather found to be associated with lower rates of respiratory infection in young children [34]. Enrichment of the upper respiratory tract with C. pseudodiphteriticum is associated with resistance to RSV and secondary pneumococcal pneumonia compared with controls in mice and could be an interesting candidate for enhancing respiratory immunity [35]. However, some of bacterial properties could be strains specific.
In the literature, most of the studies focus only on young children [10, 22, 32]. In our work, the studied population includes infants to elderly persons. They were enrolled during a short period, excluding seasonal variations. However, numerous potential confounding variables could modify the respiratory microbiota including the use of antibiotics that may be correlated with decrease of diversity and changing abundance of some bacterial species [36, 37]. An important limitation of our study is the lack of information about previous and recent antibiotic exposure (including over-the-counter and self-administered antimicrobial agents) of the patients before the sampling. However, most of the samples included were collected from patients visiting the emergency unit, so it is unlikely that many patients received antibiotics prescription before the collection the nasopharyngeal swab. Among the controls, only those that have not received antibiotics within 2 weeks of taking the sample were enrolled in our study. Moreover, we found an increase of pathogenic bacteria in symptomatic patients versus controls which is rather against an important antibiotic consumption in this group. Secondly, antibiotic treatment was related to a reduction in Moraxellaceae, Corynebacterium, and Dolosigranulum although we found an increase of these bacteria in patients compared to the controls [38]. Other limitations of our study is that we have no information about others potential confounding variables such as vaccination, asthma, allergy, and smoking which could also have an influence on microbiota composition. However, the aim of our study is purely descriptive and does not test a causality between the presence of a pathogen and the occurrence of the disease but only describes the observation of the modification of the microbiota, which does not introduce any bias for our results and does not reconsider our conclusions. Moreover, our study is based only on metagenomics approach generating extraction, sequencing, and depth bias [39].
In conclusion, we showed that the healthy nasopharyngeal microbiota is composed of a diverse and well-balanced aero-anaerobic bacterial core consortium, which is altered during viral respiratory infection. A depletion of anaerobic bacteria in the nasopharyngeal microbiota was observed concomitantly to an invasion of aerotolerant and potentially pathogenic microbes. We accurately described the healthy nasopharyngeal microbiota repertoire at the species level and confirmed M. catarrhalis, H. influenzae, S. pneumoniae, and S. aureus among the leaders of the respiratory pathogenic consortia associated with infection. The frequent co-detection of virus and bacteria raises the question of the strategy to prevent bacterial disease, focusing on the prevention of nasopharyngeal colonization through effective antibiotic treatment for these patients. Depletion of anaerobes appears to be linked to the onset of viral respiratory disease, suggesting the protective role of commensal anaerobic bacteria. In addition to antibiotics, further studies should test preventive or therapeutic interventions, including antioxidant-based approach, for maintaining or restoring a healthy nasopharyngeal microbiota.
References
Byington CL, Ampofo K, Stockmann C, Adler FR, Herbener A, Miller T, Sheng X, Blaschke AJ, Crisp R, Pavia AT (2015) Community surveillance of respiratory viruses among families in the Utah better identification of germs-longitudinal viral epidemiology (BIG-LoVE) study. Clin Infect Dis 61(8):1217–1224
Skevaki CL, Tsialta P, Trochoutsou AI, Logotheti I, Makrinioti H, Taka S, Lebessi E, Paraskakis I, Papadopoulos NG, Tsolia MN (2015) Associations between viral and bacterial potential pathogens in the nasopharynx of children with and without respiratory symptoms. Pediatr Infect Dis J 34(12):1296–1301
Man WH, de Steenhuijsen Piters WA, Bogaert D (2017) The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol 15(5):259–270
Yi H, Yong D, Lee K, Cho YJ, Chun J (2014) Profiling bacterial community in upper respiratory tracts. BMC Infect Dis 14:583
de Steenhuijsen Piters WA, Huijskens EG, Wyllie AL, Biesbroek G, van den Bergh MR, Veenhoven RH, Wang X, Trzciński K, Bonten MJ, Rossen JW, Sanders EA, Bogaert D (2016) Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. ISME J 10(1):97–108
de Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, Chaussabel D, Cohen DM, Sanders EA, Ramilo O, Bogaert D, Mejias A (2016) Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med 194(9):1104–1115
Jartti T, Jartti L, Peltola V, Waris M, Ruuskanen O (2008) Identification of respiratory viruses in asymptomatic subjects: asymptomatic respiratory viral infections. Pediatr Infect Dis J 27(12):1103–1107
Prussin AJ, Vikram A, Bibby KJ, Marr LC (2016) Seasonal dynamics of the airborne bacterial community and selected viruses in a children’s daycare center. PLoS One 11(3):e0151004
Byrd Allyson L, Segre Julia A (2016) Adapting Koch’s postulates. Science 351(6270):224–226
Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, Holt BJ, Hales BJ, Walker ML, Hollams E, Bochkov YA, Grindle K, Johnston SL, Gern JE, Sly PD, Holt PG, Holt KE, Inouye M (2015) The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17(5):704–715
Korten I, Mika M, Klenja S, Mack I, Barbani MT, Gorgievski M, Frey U, Hilty M, Latzin P (2016) Interactions of respiratory viruses and the nasal microbiota during the first year of life in healthy infants. mSphere 1(6)
Langevin S, Pichon M, Smith E, Morrison J, Bent Z, Green R, Baker K, Solberg O, Gillet Y, Javouhey E, Lina B, Katze MG, Josset L (2017) Early nasopharyngeal microbial signature associated with severe influenza in children: a retrospective pilot study. J Gen Virol. Sep 8
Benkouiten S, Charrel R, Belhouchat K, Drali T, Nougairede A, Salez N, Memish ZA, Al Masri M, Fournier PE, Raoult D, Brouqui P, Parola P, Gautret P (2014) Respiratory viruses and bacteria among pilgrims during the 2013 Hajj. Emerg Infect Dis 20(11):1821–1827
Angelakis E, Bachar D, Henrissat B, Armougom F, Audoly G, Lagier JC, Robert C, Raoult D (2016) Glycans affect DNA extraction and induce substantial differences in gut metagenomic studies. Sci Rep 6:26276
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963
Caporaso JG, Kuczynski J, Stombaugh J et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41(Database issue):D590–D596
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Million M, Tidjani AM, Khelaifia S, Bachar D, Lagier JC, Dione N, Brah S, Hugon P, Lombard V, Armougom F, Fromonot J, Robert C, Michelle C, Diallo A, Fabre A, Guieu R, Sokhna C, Henrissat B, Parola P, Raoult D (2016) Increased gut redox and depletion of anaerobic and methanogenic prokaryotes in severe acute malnutrition. Sci Rep 6:26051
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12(6):R60
Anderson MJ (2001) A new method for non parametric multivariate analysis of variance. Austral Ecology 26:32–46
Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, Bruin J, Montijn R, Bonten M, Sanders E (2011) Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One 6(2):e17035
Ederveen THA, Ferwerda G, Ahout IM, Vissers M, de Groot R, Boekhorst J, Timmerman HM, Huynen MA, van Hijum SAFT, de Jonge MI (2018) Haemophilus is overrepresented in the nasopharynx of infants hospitalized with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses. Microbiome 6(1):10
Chonmaitree T, Jennings K, Golovko G, Khanipov K, Pimenova M, Patel JA, McCormick DP, Loeffelholz MJ, Fofanov Y (2017) Nasopharyngeal microbiota in infants and changes during viral upper respiratory tract infection and acute otitis media. PLoS One 12(7):e0180630
Damasio GA, Pereira LA, Moreira SD, Duarte dos Santos CN, Dalla-Costa LM, Raboni SM (2015) Does virus-bacteria coinfection increase the clinical severity of acute respiratory infection? J Med Virol 87(9):1456–1461
Bosch AA, Biesbroek G, Trzcinski K, Sanders EA, Bogaert D (2013) Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog 9(1):e1003057
Raoult D (2016) Alice’s living croquet theory. Int J Antimicrob Agents 47(4):249
Larsen JM, Steen-Jensen DB, Laursen JM, Søndergaard JN, Musavian HS, Butt TM, Brix S (2012) Divergent pro-inflammatory profile of human dendritic cells in response to commensal and pathogenic bacteria associated with the airway microbiota. PLoS One 7(2):e31976
Leung RK, Zhou JW, Guan W, Li SK, Yang ZF, Tsui SK (2013) Modulation of potential respiratory pathogens by pH1N1 viral infection. Clin Microbiol Infect 19(10):930–935
Santee CA, Nagalingam NA, Faruqi AA, DeMuri GP, Gern JE, Wald ER, Lynch SV (2016) Nasopharyngeal microbiota composition of children is related to the frequency of upper respiratory infection and acute sinusitis. Microbiome 4(1):34
Dione N, Khelaifia S, La Scola B, Lagier JC, Raoult D (2016) A quasi-universal medium to break the aerobic/anaerobic bacterial culture dichotomy in clinical microbiology. Clin Microbiol Infect 22(1):53–58
Bosch AA, Levin E, van Houten MA, Hasrat R, Kalkman G, Biesbroek G, de Steenhuijsen Piters WAA, de Groot PCM, Pernet P, Keijser BJF, Sanders EAM, Bogaert D (2016) Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine
Kaspar U, Kriegeskorte A, Schubert T, Peters G, Rudack C, Pieper DH, Wos-Oxley M, Becker K (2016) The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ Microbiol 18(7):2130–2142
Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, Bogaert D (2014) Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 190(11):1283–1292
Kanmani P, Clua P, Vizoso-Pinto MG, Rodriguez C, Alvarez S, Melnikov V, Takahashi H, Kitazawa H, Villena J (2017) Respiratory commensal bacteria Corynebacterium pseudodiphtheriticum improves resistance of infant mice to respiratory syncytial virus and Streptococcus pneumoniae superinfection. Front Microbiol 8:1613
Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, Engstrand L (2010) Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One 5(3):e9836
Salter SJ, Turner C, Watthanaworawit W, de Goffau MC, Wagner J, Parkhill J, Bentley SD, Goldblatt D, Nosten F, Turner P (2017) A longitudinal study of the infant nasopharyngeal microbiota: the effects of age, illness and antibiotic use in a cohort of South East Asian children. PLoS Negl Trop Dis 11(10):e0005975
Prevaes SM, de Winter-de Groot KM, Janssens HM, de Steenhuijsen Piters WA, Tramper-Stranders GA, Wyllie AL, Hasrat R, Tiddens HA, van Westreenen M, van der Ent CK, Sanders EA, Bogaert D (2016) Development of the nasopharyngeal microbiota in infants with cystic fibrosis. Am J Respir Crit Care Med 193(5):504–515
Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, Trape JF, Koonin EV, La Scola B, Raoult D (2012) Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect 18(12):1185–1193
Funding
This work was supported by the French Government under the “Investissements d’avenir” (Investments for the Future) program managed by the Agence Nationale de la Recherche (ANR, fr: National Agency for Research), (reference: Méditerranée Infection 10-IAHU-03).This work was also supported by Région Provence Alpes Côte d’Azur and European funding FEDER PRIMMI (Fonds Européen de Développement Régional - Plateformes de Recherche et d'Innovation Mutualisées Méditerranée Infection).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by the local Ethics Committee (number 2016-016).
Informed consent
Written informed consent was obtained for all the controls included on a voluntary basis. For cases, metagenomics was performed on excess respiratory sample received in our laboratory for routine diagnosis of viral respiratory infections. Under the French law, patient consent was not required for this type of non-interventional study, provided the patients had received information and retained the right to oppose the use of excess respiratory samples and anonymized medical data (14–15).
Rights and permissions
About this article
Cite this article
Edouard, S., Million, M., Bachar, D. et al. The nasopharyngeal microbiota in patients with viral respiratory tract infections is enriched in bacterial pathogens. Eur J Clin Microbiol Infect Dis 37, 1725–1733 (2018). https://doi.org/10.1007/s10096-018-3305-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-018-3305-8