Abstract
The aim of this review is to provide an update on the plasmids mediating DHA-1 cephalosporinase in Klebsiella pneumoniae. These plasmids have been mainly found in this bacterium but not only. The first was isolated from Salmonella sp. in France in the early 1990s. They are currently reported worldwide. BlaDHA-1 beta-lactamase gene is usually co-expressed with many other antibiotic resistance genes such as extended-spectrum β-lactamases (blaCTX-M-, bla SHV -types), oxacillinases (blaOXA-1, blaOXA-30), penicillinases (bla TEM -type), carbapenemases (bla OXA48 , blaKPC-2), aminoglycosides (aacA, aadA, armA), fluoroquinolones (qnrB4, aac6′-1b-cr), and sulfonamide (sul1) resistance genes. Plasmids carrying DHA-1 cephalosporinase have different sizes (22 to 313 kb), belong to diverse groups of incompatibility (R, L/M, FII(k), FIB, A/C2, HI2, HIB), and are self-transferable or not. The multidrug resistance region consists of a mosaic structure composed of resistance genes, insertion sequences, composite transposon, and integrons.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Production of β-lactamase is the main mechanism of resistance of Enterobacteriaceae to β-lactam antibiotics. Some Enterobacteriaceae naturally produce chromosome-encoded β-lactamase (penicillinase or cephalosporinase). Oxyimino-β-lactamins, such as cefotaxime or ceftazidime, have been developed to avoid these enzymes. Unfortunately, under antibiotic pressure, over-production of chromosomal AmpC beta-lactamases has occurred by mutation and conferred resistance to these molecules. Similar overproduction has been observed in Enterobacter sp., Citrobacter freundii, Serratia marcescens, Morganella morganii, and Pseudomonas aeruginosa [1]. During the early 1990s, plasmid-encoded AmpC cephalosporinases were reported in species that lack an inducible AmpC enzyme such as Klebsiella pneumoniae, Escherichia coli, and Salmonella sp. [2]. Subsequently, plasmid-mediated AmpC were observed worldwide. They are derivatives of Enterobacter sp. (ACT-1 and MIR-1), C. freundii (CMY-2), M. morganii (DHA type), Hafnia alvei (ACC-1), and Aeromonas sp. (CMY-1, FOX, MOX) [3]. The first DHA-1 cephalosporinase was described in a Salmonella enterica serovar Enteritidis strain isolated in Dhahran, Saudi Arabia, in 1992 [4]. It is the main inducible plasmid-mediated AmpC cephalosporinase (pAmpC).
Detection and susceptibilities
DHA-1 belongs to class C of the Ambler classification and to group 1 of the functional classification of Bush, Jacoby, and Medeiros [5, 6]. pAmpC confers resistance to all penicillins (alone or in combination with β-lactamase inhibitors) and to most cephalosporins (except cefepime), including cephamycin, but is inactive against carbapenems and cefepime. Unlike all other pAmpC, DHA-1 is always inducible because of the systematic presence upstream of blaDHA-1 of the regulatory gene ampR. This regulation explains the antagonism between cefoxitin, clavulanate, and carbapenem, and oxyiminocephalosporins. Such antagonism can be a strong indicator of DHA-1 production in species that do not possess chromosomal-inducible AmpC, such as E. coli or K. pneumoniae. Some phenotypical tests have been developed to detect AmpC production in non-AmpC-producing species, using cephalosporins alone or in combination with cloxacilline or boronic acid, which are inhibitors of AmpC [7,8,9,10]. However, these tests are unable to precisely identify DHA-1 producers and they are unusable on AmpC-producing species such as E. coli, Enterobacter sp., and C. freundii. Molecular methods are the reference strategy for DHA-specific detection, of which Pérez-Pérez multiplex PCR is the most widely used [11].
History (Table 1)
The first plasmid-mediated DHA β-lactamase was identified in a strain of S. enterica serovar Enteritidis from stool samples of a 62-year-old patient with lung carcinoma in Dhahran (Saudi Arabia) in 1992 [4]. A strain of K. pneumoniae-producing DHA-1 was isolated from urine specimens in Los Angeles (USA) in 1994 [12] and another one in Miami (USA) between 1996 and 2000 [13]. Two strains of S. enterica serovar Senftenberg-producing DHA-1 were isolated in London (UK), in 1996 and 1999. They carried plasmids of ca. 98 and 99 kDa that were self-transferable [14]. In France, the first K. pneumoniae-producing DHA-1 was isolated in Paris in 1998 from blood cultures. The plasmid was self-transferable. Nine other K. pneumoniae and one Klebsiella oxytoca-producing DHA-1 strains were later isolated between 1999 and 2003 (five in 2003). Only two were not self-transferable [15]. Fifty-four isolates of K. pneumoniae and 2 isolates of E. coli-producing DHA-1 β-lactamase were identified from blood cultures between 1993 and 2005 in Seoul (Korea). All isolates coproduced qnrB4 (qnr genes are plasmid-mediated quinolone resistance) and some also produced other enzymes (CTX-M types, TEM-52, SHV-12) [16, 17]. Ten isolates of K. pneumoniae-producing DHA-1 β-lactamase were identified between January 1999 and September 2001 in Taiwan. The blaDHA-1 gene was located on 70-kb plasmids that were not self-transferable [18]. A 3-year-old girl with diarrhea and fever was admitted to hospital in October 2002 in Korea. A Salmonella enterica serotype Montevideo DHA-1 positive was isolated from blood and stool specimens. The plasmid was self-transferable [19]. Thirty-six DHA-1-producing K. pneumoniae that co-expressed ESBL enzymes (CTX-M and/or SHV) were collected between January 1999 and June 2002 in Taiwan [20]. From May to July 2004, 11 DHA-1- and SHV-12 ESBL-producing K. pneumoniae were collected in a Korean hospital [21]. Ninety-nine DHA-1-producing K. pneumoniae, 13 DHA-1-producing E. coli, and 1 DHA-1-producing Enterobacter cloacae were collected during 2004 to 2006 in Kyungpook (Korea) [22]. As elsewhere, some of the isolates co-expressed ESBL enzymes (CTX-M and/or SHV). Further, 98/99 isolates of DHA-1-producing K. pneumoniae and 100% isolates of DHA-1-producing E. coli were positive for qnrB4. Then, 89/99 of K. pneumoniae and 7/13 of E. coli isolates were positive for armA, a plasmid-mediated 16S rRNA methylase gene. Conjugation was successful for 38 DHA-1-producing K. pneumoniae, 3 DHA-1-producing E. coli, and 1 DHA-1-producing E. cloacae. Thirty-five transconjugants carried qnrB4, bla DHA , blaSHV-12, and armA; two transconjugants carried qnrB6, blaDHA-1, and armA. Nine plasmids were chosen for further studies, which revealed a large plasmid of about 180 kb belonging to the FIIA incompatibility (Inc) type [22]. In another study, genes qnrB4 and qnrS were found in the same strain of K. pneumoniae but on different plasmids. The strain was isolated in 2005 in China [23]. In southern Taiwan, 20 isolates of flomoxef-resistant ESBL-K. pneumoniae, all carrying the blaDHA-1 gene, were identified between March 2004 and November 2005. BlaCTX-M genes were found in 18 of the strains: qnr, aac(6′)-Ib-cr, and armA genes were respectively detected in 100, 61, and 78% of the isolates. Conjugation was successful for only four strains. Genes qnrB4, aac(6′)-Ib-cr, armA, and blaDHA-1 were on the same self-transferable CTX-M-carrying plasmids. They belonged to the FIIA Inc group [24]. Plasmid pKP048, belonging to IncF, was detected in carbapenem-resistant K. pneumoniae in China in 2006. It carried blaKPC-2, blaDHA-1, qnrB4, and armA, and was self-transferable [25]. Plasmid p1220-CTXM, a pKP048-related IncFII(k) plasmid, was recently sequenced in China; it carried blaCTX-M-14 and qnrB4 [26]. A hospital in the northwest of Belgium recorded 22 clinical isolates of K. pneumoniae expressing plasmid-mediated DHA-1 AmpC. and chromosomal SHV-11 beta-lactamases between August and December 2006 [27]. Thirty Enterobacteriaceae (15 E. coli, 10 K. pneumoniae, 4 K. oxytoca, and 1 Proteus mirabilis strains) encoding blaDHA-1 or blaDHA-1 plus blaCTX-M-14, -15 were collected between 2005 and 2007 in Barcelona, Spain. All isolates co-harbored blaDHA-1 and qnrB genes on the same plasmid. Conjugation was possible for 26/30 isolates, of which all but one belonged to the L/M group [28]. Twenty-six DHA-1-producing K. pneumoniae were isolated between June 2007 and January 2008 in Barcelona, Spain. Three of them also had blaCTX-M-15 and aac(6′)-Ib-cr and 25 contained qnrB. Transconjugants were obtained with one isolate of each pulsed-field gel electrophoresis (PFGE) profile. The blaDHA-1 gene was present in a plasmid of 180 kb belonging to the L/M Inc group. Exceptionally, one gene was located in a plasmid of 120 kb. The FII replicon was also detected. The plasmids were transferable and the strains belonged to sequence type (ST) 17, ST13, ST427, ST416, ST37, ST326, and ST428 [29]. In September 2007, a strain of S. enterica Thompson co-harboring blaDHA-1 and blaTEM-1b was isolated from the stool sample of a pediatric inpatient (aged 3 years) with diarrhea in China [30]. Three strains of C. freundii were isolated from wastewater treatment plants in Vancouver, British Columbia (Canada), in June 2007. They carried large plasmids (> 60 kb) with resistance genes such as qnrB4, aac(6′)-Ib-cr, blaDHA-1, and blaOXA-1 or blaOXA-10 [31]. Two K. pneumoniae isolates carrying blaDHA-1 and qnrB4 genes were identified in 2008 in Tortosa, Catalonia (Spain). They belonged to the same MLST, ST483, and harbored the intl1, blaDHA-1, blaOXA-1, blaSHV-1, aac(6′)-Ib-cr, qnrB4, and qnrS2 genes [32]. A SFO-1-producing strain of K. pneumoniae isolated in 2008 in Shanghai (China) harbored a large DHA-1-bearing IncFII plasmid in a ST11 clone (SFO-1 is an ESBL enzyme). The plasmid also carried a qnrB4 gene [33]. K. pneumoniae KPS30 strain (ST11) was isolated from urine in 2008 in Paris (France). KPS30 carries the ß-lactamase genes blaSHV-11, blaOXA-30, and blaDHA-1. pKPS30 is a 61,228-bp plasmid of the IncR group. It carries blaOXA-30 and blaDHA-1 genes and tetA, aac6′-Ib-cr, catB3, arr-3, sul1 (two copies), and mph(A) genes. It was not transferable [34]. At the same time, a similar plasmid of 57,382, KpQ3 was isolated in Madrid (Spain) in a ST37 K. pneumoniae strain [35]. Seventeen K. pneumoniae strains producing DHA-1 were collected between 2006 and 2010 in Paris (France). ST48 and ST1263 were predominant and the plasmids belonged to IncL/M or IncHI2 groups; 8/14 were transferable [36]. Seven K. pneumoniae strains were isolated from dogs and cats in Spain between 2008 and 2010. All isolates were typed as ST11 and had SHV-11 ß-lactamase. The IncR plasmids of 50 kb carried blaDHA-1, armA, and qnrB4 genes and were not transferable [37]. A carbapenemase NDM-1 (for New Delhi metallo-carbapenemase) E. coli strain was isolated in October 2009 in Hong Kong. The plasmid named pNDM-HK, 88,803 bp in size, was self-transferable and belonged to the IncL/M group. It also harbored genes associated with resistance to ß-lactams (blaTEM-1, blaDHA-1), aminoglycosides (aac2, armA), sulfonamides (sul1), and macrolides (mel, mph2) [38]. A ST11 K. pneumoniae strain co-expressing the OXA-162 carbapenemase and DHA-1 was isolated in Serres (Greece) in 2010 [39]. The blaOXA-162 gene was harbored on a plasmid of 62 kb belonging to IncL/M while the blaDHA-1 gene was carried on an IncFII plasmid of 140 kb. The plasmids were not self-transferable [39]. A carbapenem-resistant Enterobacter aerogenes was isolated from the sputum of a 69-year-old patient in China in July 2010. The strain harbored a plasmid of 56 kb carrying carbapenemase KPC-2, DHA-1, and TEM-1; the plasmid was self-transferable by conjugation [40]. Fourteen ST274 K. pneumoniae were recovered from dogs, cats, sheep, and a hedgehog between July 2011 and June 2012 in Paris (France). They harbored a plasmid of 253,984 bp named pENVA carrying blaCTX-M-15, blaTEM-1, blaDHA-1, aacA2, aadA1, qnrB4, dfrA15, and sul1 (two copies) genes. Two replicons, those of IncFIB and IncHIB-like plasmids, were found [41]. Between December 2009 and December 2013, 312 DHA-1-producing K. pneumoniae strains of ST11 were isolated in Hungary. The blaCTX-M-15 was present in 90% of isolates and three isolates were also resistant to colistin by inactivation of mgrB [42]. In July 2016, a colistin- and tigecycline-resistant K. pneumoniae strain was isolated from a dog in Japan. Insertional inactivation of mrgB and ramR by IS10R could have been involved in these resistances. The strain belonged to ST37, carried blaSHV-12 and blaDHA-1, and was positive for qnrB and armA [43].
Complete plasmids (Table 2)
Twenty-one complete DHA-1 plasmids are available on GenBank (www.ncbi.nlm.nih.gov/genbank/). They mainly belong to the Enterobacteriaceae family: 13 were found in K. pneumoniae, 2 in E. coli, 3 in C. freundii, 1 in Raoultella ornithinolytica, and 1 in Cronobacter sakazakii strains. One plasmid was found in a strain of Vibrio cholerae. We looked at their Inc groups with PlasmidFinder [44] and at their profiles of resistance with Card [45]. They belong to different Inc groups: R (2), A/C2 (4), R and A/C2 (1), L/M (1), FII(k) (2), FII(k) and R (2), R and FII(k) (2), HIB and FIB(Mar) (1), HI2 (1), H12A and H112 (1). Their sizes varied from ca. 22 to 312 kb. They carried many antibiotic resistance genes: to beta-lactams with bla OXA -type, bla SHV -type, bla CTX-M -type, bla TEM -type, bla IMP -type, bla NDM -type, bla KPC -type, blaDHA-1, to aminoglycosides with aph(3′)-type, aac(6′)-Ib-cr, aadA1, aacA4, strA, strB, armA, to fluoroquinolones with aac(6′)-Ib-cr and qnrB4, to phenicols with catB3, floR, cmlA, to rifampicine with arr, to sulfonamides with sul1 and sul2, to trimethoprim with dfrA, to tetracyclines with tetA, and to macrolides with mph. In some of them, blaDHA-1 and ampR genes were part of a ISCR1-linked resistance gene region associated with a class 1 integron. Fifteen of the 21 plasmids had an ISCR1 element. The genetic organization of the multidrug resistance regions is described in Table 2.
Patric database (Table 3)
We looked in the Patric database [46] for genomes of Klebsiella strains that were resistant to cefoxitin by synthesis of a class C beta-lactamase from the ‘DHA/MOR’ family. We found 26 genome assemblies and 1 complete plasmid (data supplement). The strains were collected from urine, blood, sputum, rectum, canal, pus, and river water between 2008 and 2017 in the USA, Europe, Asia, and Africa. The genomes were mainly sequenced by Illumina technology. We looked for the presence of plasmids with PlasmidFinder [44], for antibiotic resistance genes with Card [45], and for ST, wzi, and virulence genes on the Pasteur MLST site (http://bigsdb.pasteur.fr/). Contigs were compared with known DHA-1 plasmids by Mauve [47].
The strains harbored several plasmids with two to six different Inc groups. Plasmids related to pKPS30 were the most frequently recovered (ten times). pKPS30 plasmid of the Inc-R group was not self-transferable by conjugation [36]. It was associated with K. pneumoniae strains of ST11 [36] and ST37 [35]. It was also found associated with other STs such as ST895, ST29, and ST1307. Sequence types ST11, ST37, ST29, and ST895 were the most frequently found. No homology was found for eight plasmids. Five strains produced carbapenemase (NDM-type or OXA-48).
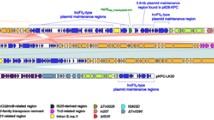
Most of the strains were of classical pathogenesis with type 3 fimbriae (mrk operon). Usually, hypervirulent isolates belong to serotypes K1 or K2. In this study, just one strain was of serotype K2 but possessed only the iron transport kfu operon as extra virulent genes and therefore could not be considered as hypervirulent. Four strains had a yersiniabactin, which is a siderophore that increases the virulence of the strain [48]. One strain of serotype K54 could be considered as hypervirulent with several siderophores (yersiniabactin and aerobactin [49]) and the presence of RmpA, a regulator of the mucoid phenotype [50]. The K. pneumoniae AR_048 strain (Accession: CP021950.1) of ST11 was sequenced by PacBio technology with a genome coverage of 120. The isolate belongs to a collection of carbapenemase-producing Enterobacteriaceae. It has four plasmids, one of which is a DHA-1 plasmid of 176,349 bp: tig00000169_pilon (Accession: CP021952.1). PacBio technology allows the complete sequencing of plasmids. DNAplotter (Sanger Institute) [51] was used to construct a schematic plasmid map (Fig. 1). Plasmid tig00000169_pilon belongs to the IncA/C2 group. The backbone part included genes for replication, transfer, stabilization, and transfer: the plasmid, therefore, is self-transferable by conjugation. Several antibiotic genes were found in the variable region: resistant to ß-lactams with blaSHV-11, blaDHA-1, blaCMY-6, blaNDM-1; resistant to aminoglycosides with aac(6′)-Ib-cr, aac(3)-IId, rmtC; resistant to fluoroquinolones with aac(6′)-Ib-cr and qnrB9; and to sulfonamides with sul1. The co-occurrence of two genes of class C ß-lactamase has already been reported [52]. However, the presence of blaSHV-11, blaDHA-1, blaCMY-6, and blaNDM-1 on the same plasmid is very surprising. Genes blaDHA-1 and blaNDM-1 belong to a class 1 integron of sul1-type with ISCR1 (see below) (Fig. 2).
An overview of the tig00000169_pilon plasmid (Accession: NZ_CP021952.1). The open reading frames were annotated in the second circle with arrows representing the direction of transcription. Mobile elements are indicated in green. Antibiotic resistance genes are indicated in red. The third circle indicates the functional sequence blocks. The G + C plot is indicated in the inner circle. The figure was made with DNAplotter [51]
Genetic organization (Fig. 3)
Isolates carrying ISCR1
The blaDHA-1 gene has been observed in a particular class 1 integron of sul1-type which contains two partial copies of the 3′-conserved segment qacEΔ1sul1 surrounding a common region ISCR1 and antibiotic resistance genes [53, 54]. Different antibiotic resistance genes (VR1) have been detected between the int gene, encoding the integrase, (the 5′-conserved segment) and the first qacEΔ1sul1 segment (3′-conserved segment). For example, aadB and aadA2 were detected in the integron from pKp760 plasmid [15]; dhfr, arr2, cmlA7, and a fused gene cassette bla OXA10 -aadA1 were detected in the integron from pRDDHA plasmid (the clmA7 and bla OXA10 genes were inactivated by insertion sequences IS); aac(6′)-Ib-cr, blaOXA-30, catB3, and arr-3 were detected in the integron from pRBDHA and pKPS30 plasmids [36]; and aac(6′)-II, ereA2, aac, and arr were detected in the integron from pMPDHA plasmid (ereA2 was interrupted by IS1247) [15]. The blaDHA-1 and ampR region was mobilized from the Morganella morganii chromosome and inserted downstream of ISCR1 [15]. This region was followed by the second qacEΔ1sul1 segment (3′-conserved segment). In pMPDHA, IS26 followed the second qacEΔ1sul1 segment, whereas a partial tniB was found in pRDDHA and pKp760. In pRBDHA, the end of the second qacEΔ1sul1 segment was deleted by IS4321. In pMPDHA, pRBDHA, pHS7, and pKPS30, genes of the sap operon (ABC transporter family) and genes of the psp operon (phage shock proteins) were found downstream of ISCR1 and upstream of ampC [15, 23, 36]. In pRBDHA and pHS7, a qnrB4 gene was inserted between genes of the sap and psp operons; for pHS7, partial IS26 was located between aphA1 and partial sapB.
In pKP048, IS26 was located at the 5′ and 3′ ends [25]. In Gram negative, most class I transposon are bounded by two copies of IS26 [55]. ISEc28, armA, ISEc29, and the mel and mph genes were found upstream of ISCR1. A qacEΔ1sul1 segment was detected downstream of ISCR1 followed by the M. morganii region with ampR and blaDHA-1, the psp operon, and qnrB4. Plasmid pB1025-1 shares the same genetic structure but without the qnrB4 gene.
Isolates not carrying ISCR1 but IS26
In pTN60013, IS26 was detected at the 5′ and 3′ ends [15]. A partial qacEΔ1sul1 region was found downstream of ampR and the psp operon upstream of the M. morganii region. In C1911, the integron was very close to the integron of pRBDHA but the qacEΔ1sul1 region, the ISCR1 element, and the sap operon were missing, to the benefit of IS26 and qnrB4. IS26 moves via a replicative mechanism that deletes sequences immediately adjacent [55]. In pENVA, ISCR1 was also missing and IS26 was detected between the first qacEΔ1sul1 region and qnrB4 [41].
Conclusion
The first plasmid-mediated DHA-1 β-lactamase was isolated from a strain of Salmonella spp. in Saudi Arabia (Western Asia) in 1992 [4]. The following plasmids were reported in China and Korea (East Asia), then in the USA and finally in London, France, Belgium, and Spain (Europe). In the 1990s, the cases were sporadic, localized, and could be counted on the fingers of one hand. In the 2000s, the cases were worldwide and counted in tens. The strains were mainly isolated from human beings but also from pets (dogs, cats), sheep, and the environment (wastewater, river water). The first plasmid was isolated from a strain of Salmonella sp. but Klebsiella sp. strains, in particular K. pneumoniae, are its favorite host. The DHA-1 plasmids carried several antibiotic resistance genes and the strains could be considered as multi-resistant (MDR). The DHA-1 plasmid is rarely the only one in the strain and can be associated with one to six other Inc groups. The number of antibiotic resistance genes that could be present under an integron element in a strain is impressive and concerns all the antibiotic families. The genes can be located on the same region of the plasmid as that of the blaDHA-1 gene or on different regions or other plasmids. The increasing presence of carbapenemase enzymes in these strains is worrying and gives the feeling that these plasmids could harbor any kind of antibiotic resistance genes. Twenty-one complete DHA-1 plasmids have been sequenced: 13 from K. pneumoniae, 2 from E. coli, 3 from C. freundii, 1 from Raoultella ornithinolytica, 1 from Cronobacter sakazakii, and 1 from Vibrio cholerae, the only non-Enterobacteriaceae. Their size is variable and some of them are not self-transferable by conjugation. Seven Inc groups were found: R, A/C2, L/M, FII(k), HIB, FIB, and H12 sometimes with the presence of two hybrids. However, many plasmid-mediated DHA-1 β-lactamases remain unknown. Twenty-four STs have been listed, with ST11 and ST37 being predominant. ST11 is the most frequent clone of K. pneumoniae and is related to MDR strains [42]. The virulence of these strains has been little studied. They are mainly of classical pathogenesis and generally not hypervirulent [56]. DHA-1-producing K. pneumoniae strains are mainly responsible for nosocomial infections that affect people with weak health. The higher mortality observed with these infections was due to delay in initiating adequate antibiotic treatment [57, 58]. MDR and hypervirulent strains were usually non-overlapping [59] but these very diffusible plasmids can be found in a hypervirulent strain under antibiotic pressure, as seen for one of the strains from the Patric database. Some cases have already been reported [60] but it is to be hoped that the fitness cost of such an event is too high for the bacteria to be able to diffuse efficiently.
References
Jacoby GA (2009) AmpC beta-lactamases. Clin Microbiol Rev 22:161–182, Table of Contents. https://doi.org/10.1128/CMR.00036-08
Papanicolaou GA, Medeiros AA, Jacoby GA (1990) Novel plasmid-mediated beta-lactamase (MIR-1) conferring resistance to oxyimino- and alpha-methoxy beta-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother 34:2200–2209
Philippon A, Arlet G, Jacoby GA (2002) Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemother 46:1–11
Gaillot O, Clément C, Simonet M, Philippon A (1997) Novel transferable beta-lactam resistance with cephalosporinase characteristics in Salmonella enteritidis. J Antimicrob Chemother 39:85–87
Ambler RP (1980) The structure of beta-lactamases. Philos Trans R Soc Lond Ser B Biol Sci 289:321–331
Bush K, Jacoby GA, Medeiros AA (1995) A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother 39:1211–1233
Nourrisson C, Tan RN, Hennequin C, Gibold L, Bonnet R, Robin F (2015) The MAST® D68C test: an interesting tool for detecting extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol 34:975–983. https://doi.org/10.1007/s10096-014-2305-6
Maraskolhe DL, Deotale VS, Mendiratta DK, Narang P (2014) Comparision of three laboratory tests for detection of AmpC β lactamases in Klebsiella species and E. coli. J Clin Diagn Res JCDR 8:DC05–DC08. https://doi.org/10.7860/JCDR/2014/8256.4432
Reuland EA, Hays JP, de Jongh DMC, Abdelrehim E, Willemsen I, Kluytmans JAJW et al (2014) Detection and occurrence of plasmid-mediated AmpC in highly resistant gram-negative rods. PLoS One 9:e91396. https://doi.org/10.1371/journal.pone.0091396
Edquist P, Ringman M, Liljequist BO, Wisell KT, Giske CG (2013) Phenotypic detection of plasmid-acquired AmpC in Escherichia coli—evaluation of screening criteria and performance of two commercial methods for the phenotypic confirmation of AmpC production. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol 32:1205–1210. https://doi.org/10.1007/s10096-013-1869-x
Pérez-Pérez FJ, Hanson ND (2002) Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 40:2153–2162
Alvarez M, Tran JH, Chow N, Jacoby GA (2004) Epidemiology of conjugative plasmid-mediated AmpC beta-lactamases in the United States. Antimicrob Agents Chemother 48:533–537
Moland ES, Black JA, Ourada J, Reisbig MD, Hanson ND, Thomson KS (2002) Occurrence of newer beta-lactamases in Klebsiella pneumoniae isolates from 24 U.S. hospitals. Antimicrob Agents Chemother 46:3837–3842
Liebana E, Batchelor M, Clifton-Hadley FA, Davies RH, Hopkins KL, Threlfall EJ (2004) First report of Salmonella isolates with the DHA-1 Amp. beta-lactamase in the United Kingdom. Antimicrob Agents Chemother 48:4492. https://doi.org/10.1128/AAC.48.11.4492.2004
Verdet C, Benzerara Y, Gautier V, Adam O, Ould-Hocine Z, Arlet G (2006) Emergence of DHA-1-producing Klebsiella spp. in the Parisian region: genetic organization of the ampC and ampR genes originating from Morganella morganii. Antimicrob Agents Chemother 50:607–617. https://doi.org/10.1128/AAC.50.2.607-617.2006
Pai H, Kang C-I, Byeon J-H, Lee K-D, Park WB, Kim H-B et al (2004) Epidemiology and clinical features of bloodstream infections caused by AmpC-type-beta-lactamase-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 48:3720–3728. https://doi.org/10.1128/AAC.48.10.3720-3728.2004
Pai H, Seo M-R, Choi TY (2007) Association of QnrB determinants and production of extended-spectrum beta-lactamases or plasmid-mediated AmpC beta-lactamases in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother 51:366–368. https://doi.org/10.1128/AAC.00841-06
Yan J-J, Ko W-C, Jung Y-C, Chuang C-L, Wu J-J (2002) Emergence of Klebsiella pneumoniae isolates producing inducible DHA-1 beta-lactamase in a university hospital in Taiwan. J Clin Microbiol 40:3121–3126
Kim J-Y, Park Y-J, Lee S-O, Song W, Jeong SH, Yoo YA et al (2004) Case report: bacteremia due to Salmonella enterica serotype Montevideo producing plasmid-mediated AmpC beta-lactamase (DHA-1). Ann Clin Lab Sci 34:214–217
Yan J-J, Ko W-C, Wu H-M, Tsai S-H, Chuang C-L, Wu J-J (2004) Complexity of Klebsiella pneumoniae isolates resistant to both cephamycins and extended-spectrum cephalosporins at a teaching hospital in Taiwan. J Clin Microbiol 42:5337–5340. https://doi.org/10.1128/JCM.42.11.5337-5340.2004
Roh KH, Uh Y, Kim J-S, Kim H-S, Shin DH, Song W (2008) First outbreak of multidrug-resistant Klebsiella pneumoniae producing both SHV-12-type extended-spectrum beta-lactamase and DHA-1-type AmpC beta-lactamase at a Korean hospital. Yonsei Med J 49:53–57. https://doi.org/10.3349/ymj.2008.49.1.53
Tamang MD, Seol SY, Oh J-Y, Kang HY, Lee JC, Lee YC et al (2008) Plasmid-mediated quinolone resistance determinants qnrA, qnrB, and qnrS among clinical isolates of Enterobacteriaceae in a Korean hospital. Antimicrob Agents Chemother 52:4159–4162. https://doi.org/10.1128/AAC.01633-07
Hu F, Xu X, Zhu D, Wang M (2008) Coexistence of qnrB4 and qnrS1 in a clinical strain of Klebsiella pneumoniae. Acta Pharmacol Sin 29:320–324. https://doi.org/10.1111/j.1745-7254.2008.00757.x
Lee C-H, Liu J-W, Li C-C, Chien C-C, Tang Y-F, Su L-H (2011) Spread of ISCR1 elements containing bla DHA- 1 and multiple antimicrobial resistance genes leading to increase of flomoxef resistance in extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 55:4058–4063. https://doi.org/10.1128/AAC.00259-11
Jiang Y, Yu D, Wei Z, Shen P, Zhou Z, Yu Y (2010) Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying bla KPC-2 , bla DHA-1 , qnrB4, and armA. Antimicrob Agents Chemother 54:3967–3969. https://doi.org/10.1128/AAC.00137-10
Zhang D, Yin Z, Zhao Y, Feng J, Jiang X, Zhan Z et al (2017) p1220-CTXM, a pKP048-related IncFIIK plasmid carrying bla CTX-M-14 and qnrB4. Future Microbiol 12:1035–1043. https://doi.org/10.2217/fmb-2017-0026
Vanwynsberghe T, Verhamme K, Raymaekers M, Cartuyvels R, Boel A, De Beenhouwer H (2007) Outbreak of Klebsiella pneumoniae strain harbouring an AmpC (DHA-1) and a bla SHV-11 in a Belgian hospital, August–December 2006. Eur Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull 12:E070201.3
Mata C, Miró E, Toleman M, Rivera MA, Walsh TR, Navarro F (2011) Association of bla(DHA-1) and qnrB genes carried by broad-host-range plasmids among isolates of Enterobacteriaceae at a Spanish hospital. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis 17:1514–1517. https://doi.org/10.1111/j.1469-0691.2011.03539.x
Diestra K, Miró E, Martí C, Navarro D, Cuquet J, Coll P et al (2011) Multiclonal epidemic of Klebsiella pneumoniae isolates producing DHA-1 in a Spanish hospital. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis 17:1032–1036. https://doi.org/10.1111/j.1469-0691.2010.03319.x
Yu F, Chen C, Chen Q, Yu X, Ding B, Yang L et al (2012) Identification of transferable DHA-1 type AmpC β-lactamases and two mutations in quinolone resistance-determining regions of Salmonella enterica serovar Thompson. J Med Microbiol 61:460–462. https://doi.org/10.1099/jmm.0.040691-0
Yim G, Kwong W, Davies J, Miao V (2013) Complex integrons containing qnrB4-ampC (bla(DHA-1)) in plasmids of multidrug-resistant Citrobacter freundii from wastewater. Can J Microbiol 59:110–116. https://doi.org/10.1139/cjm-2012-0576
Pérez-Moreno MO, Estepa V, Sáenz Y, Cortell-Ortolá M, Fort-Gallifa I, Ruiz J et al (2012) Intrahospitalary dissemination of Klebsiella pneumoniae carrying bla(DHA-1) and qnrB4 genes within a novel complex class 1 integron. Diagn Microbiol Infect Dis 73:210–211. https://doi.org/10.1016/j.diagmicrobio.2012.02.011
Guo Q, Wang P, Ma Y, Yang Y, Ye X, Wang M (2012) Co-production of SFO-1 and DHA-1 β-lactamases and 16S rRNA methylase ArmA in clinical isolates of Klebsiella pneumoniae. J Antimicrob Chemother 67:2361–2366. https://doi.org/10.1093/jac/dks244
Compain F, Frangeul L, Drieux L, Verdet C, Brisse S, Arlet G et al (2014) Complete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniae. Antimicrob Agents Chemother 58:4207–4210. https://doi.org/10.1128/AAC.02773-13
Tobes R, Codoñer FM, López-Camacho E, Salanueva IJ, Manrique M, Brozynska M et al (2013) Genome sequence of Klebsiella pneumoniae KpQ3, a DHA-1 β-lactamase-producing nosocomial isolate. Genome Announc 1. https://doi.org/10.1128/genomeA.00167-12
Compain F, Decré D, Fulgencio J-P, Berraho S, Arlet G, Verdet C (2014) Molecular characterization of DHA-1-producing Klebsiella pneumoniae isolates collected during a 4-year period in an intensive care unit. Diagn Microbiol Infect Dis 80:159–161. https://doi.org/10.1016/j.diagmicrobio.2014.06.009
Hidalgo L, Gutierrez B, Ovejero CM, Carrilero L, Matrat S, Saba CKS et al (2013) Klebsiella pneumoniae sequence type 11 from companion animals bearing ArmA methyltransferase, DHA-1 β-lactamase, and QnrB4. Antimicrob Agents Chemother 57:4532–4534. https://doi.org/10.1128/AAC.00491-13
Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I et al (2011) Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989. https://doi.org/10.1371/journal.pone.0017989
Voulgari E, Poulou A, Dimitroulia E, Politi L, Ranellou K, Gennimata V et al (2015) Emergence of OXA-162 carbapenemase- and DHA-1 Amp. cephalosporinase-producing sequence type 11 Klebsiella pneumoniae causing community-onset infection in Greece. Antimicrob Agents Chemother 60:1862–1864. https://doi.org/10.1128/AAC.01514-15
Kuai S, Shao H, Huang L, Pei H, Lu Z, Wang W et al (2014) KPC-2 carbapenemase and DHA-1 Amp. determinants carried on the same plasmid in Enterobacter aerogenes. J Med Microbiol 63:367–370. https://doi.org/10.1099/jmm.0.054627-0
Schlüter A, Nordmann P, Bonnin RA, Millemann Y, Eikmeyer FG, Wibberg D et al (2014) IncH-type plasmid harboring bla CTX-M-15 , bla DHA-1 , and qnrB4 genes recovered from animal isolates. Antimicrob Agents Chemother 58:3768–3773. https://doi.org/10.1128/AAC.02695-14
Kis Z, Tóth Á, Jánvári L, Damjanova I (2016) Countrywide dissemination of a DHA-1-type plasmid-mediated AmpC β-lactamase-producing Klebsiella pneumoniae ST11 international high-risk clone in Hungary, 2009-2013. J Med Microbiol 65:1020–1027. https://doi.org/10.1099/jmm.0.000302
Taniguchi Y, Maeyama Y, Ohsaki Y, Hayashi W, Osaka S, Koide S et al (2017) Co-resistance to colistin and tigecycline by disrupting mgrB and ramR with IS insertions in a canine Klebsiella pneumoniae ST37 isolate producing SHV-12, DHA-1 and FosA3. Int J Antimicrob Agents 50:697–698. https://doi.org/10.1016/j.ijantimicag.2017.09.011
Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L et al (2014) In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. https://doi.org/10.1128/AAC.02412-14
McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ et al (2013) The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. https://doi.org/10.1128/AAC.00419-13
Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C et al (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45:D535–D542. https://doi.org/10.1093/nar/gkw1017
Darling AE, Mau B, Perna NT (2010) progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. https://doi.org/10.1371/journal.pone.0011147
Lawlor MS, O’connor C, Miller VL (2007) Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect Immun 75:1463–1472. https://doi.org/10.1128/IAI.00372-06
Nassif X, Sansonetti PJ (1986) Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect Immun 54:603–608
Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL (2010) RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol 192:3144–3158. https://doi.org/10.1128/JB.00031-10
Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J (2009) DNAPlotter: circular and linear interactive genome visualization. Bioinforma Oxf Engl 25(1):119–120. https://doi.org/10.1093/bioinformatics/btn578
Chérif T, Saidani M, Decré D, Boutiba-Ben Boubaker I, Arlet G (2015) Cooccurrence of multiple AmpC β-lactamases in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis in Tunisia. Antimicrob Agents Chemother 60:44–51. https://doi.org/10.1128/AAC.00828-15
Toleman MA, Bennett PM, Walsh TR (2006) ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev MMBR 70:296–316. https://doi.org/10.1128/MMBR.00048-05
Cheng C, Sun J, Zheng F, Lu W, Yang Q, Rui Y (2016) New structures simultaneously harboring class 1 integron and ISCR1-linked resistance genes in multidrug-resistant Gram-negative bacteria. BMC Microbiol 16:71. https://doi.org/10.1186/s12866-016-0683-x
Harmer CJ, Hall RM (2016) IS26-mediated formation of transposons carrying antibiotic resistance genes. mSphere 1. https://doi.org/10.1128/mSphere.00038-16
Hennequin C, Robin F, Cabrolier N, Bonnet R, Forestier C (2012) Characterization of a DHA-1-producing Klebsiella pneumoniae strain involved in an outbreak and role of the AmpR regulator in virulence. Antimicrob Agents Chemother 56:288–294. https://doi.org/10.1128/AAC.00164-11
Luk S, Wong W-K, Ho AY-M, Yu KC-H, To W-K, Ng T-K (2016) Clinical features and molecular epidemiology of plasmid-mediated DHA-type AmpC β-lactamase-producing Klebsiella pneumoniae blood culture isolates, Hong Kong. J Glob Antimicrob Resist 7:37–42. https://doi.org/10.1016/j.jgar.2016.06.006
Ku Y-H, Chuang Y-C, Chen C-C, Lee M-F, Yang Y-C, Tang H-J et al (2017) Klebsiella pneumoniae isolates from meningitis: epidemiology, virulence and antibiotic resistance. Sci Rep 7:6634. https://doi.org/10.1038/s41598-017-06878-6
Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard A-S et al (2014) Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 20:1812–1820. https://doi.org/10.3201/eid2011.140206
Cheong HS, Chung DR, Lee C, Kim SH, Kang C-I, Peck KR et al (2016) Emergence of serotype K1 Klebsiella pneumoniae ST23 strains co-producing the plasmid-mediated AmpC beta-lactamase DHA-1 and an extended-spectrum beta-lactamase in Korea. Antimicrob Resist Infect Control 5:50. https://doi.org/10.1186/s13756-016-0151-2
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(ODS 7 kb)
Rights and permissions
About this article
Cite this article
Hennequin, C., Ravet, V. & Robin, F. Plasmids carrying DHA-1 β-lactamases. Eur J Clin Microbiol Infect Dis 37, 1197–1209 (2018). https://doi.org/10.1007/s10096-018-3231-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-018-3231-9