Abstract
Streptococcus pyogenes (GAS) is one of the major human pathogenic bacteria that cause a wide range of diseases. Currently, increased incidence of streptococcal invasive infections is observed worldwide. In this study, we focused on the prevalence of genes encoding superantigens and type M proteins in the population of GAS strains from invasive versus non-invasive infections. We tested 253 GAS strains: 48 strains from patients with invasive infections (18 from wound/deep skin localization, 30 from women in labour) and 205 strains from non-invasive forms (147 from common infections of the upper respiratory, 49 from the vagina of females with genital tract infections and 9 from non-invasive wound and superficial skin infections). Significant differences were found in the occurrence of genes: speG, speI, speJ and smeZ, which were more common in GAS isolated from invasive than from non-invasive strains; speJ and smeZ occurred more frequently in strains from invasive perinatal infections versus strains from women without symptoms of invasive infection; speH and speI in strains from invasive skin/wound infection versus strains isolated from non-invasive wound and superficial skin infections. Emm types 1 and 12 predominated in the group of strains isolated from superficial infections and type 28 in those from puerperal fever. Occurrence of genes encoding virulence factors is common in genomic DNA of most of S. pyogenes, regardless whether these streptococcal infections are invasive or non-invasive. On the other hand, it appears that strains with speG, speI, speJ and smeZ genes may have a particular potential for virulence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Group A Streptococcus (GAS) (Streptococcus pyogenes) infections vary from superficial infection of the pharynx (strep throat, pharyngitis) to serious skin and soft tissue infections that can lead to lethal invasive disease, despite antibiotic treatment [1]. Currently, increased incidence of invasive infections such as puerperal fever, streptococcal toxic shock syndrome and sepsis caused by S. pyogenes is observed worldwide [2–4]. Protein M is considered the main virulence factor, which limits phagocytosis, disturbs the function of complement and is responsible for adhesion [5]. Furthermore, the emm gene, which encodes this protein, forms the basis for epidemiological typing of GAS, to correlate serotype with pathogenicity [6]. Although GAS possess many virulence factors that engage a wide variety of host defences, the streptococcal superantigens play a pivotal role in triggering potent inflammatory responses. GAS strains that cause invasive infections usually produce one or more superantigens: SpeA, Spe C, Spe G, Spe H, Spe I, Spe J, Spe K, Spe L, SpeM, SmeZ, and Ssa [6–8]. Epidemiological studies, which provide the distributions of the types of streptococci prevalent in communities, are of basic importance for the identification and control of streptococcal infections. The US Centres for Disease Control and Prevention (CDC) reported an average of 3.5 cases of invasive GAS infections per 100,000 population in the United States in 2000–2004 [9]. During nearly the same period of time, a European epidemiological survey of GAS infection was reported. The ten most predominant M/emm types were M/emm type 1 (M/emm1), M/emm28, M/emm3, M/emm89, M/emm87, M/emm12, M/emm4, M/emm83, M/emm81, and M/emm5, in descending order, but the M/emm type distribution varied broadly between participating countries. Unfortunately, no data on GAS strains isolated in Poland were included in this survey [10]. It should be stressed that in the last 10 years there has been an increase in invasive GAS infections with associated mortality, and these data are especially alarming from an epidemiological point of view; however, the factors underlying the worldwide resurgence of this pathogen remain unknown [11, 12].
Aim
The primary objective of this study was to compare virulence factors of S. pyogenes strains isolated both from invasive and non-invasive infections, in Polish and German centres, in the years 2009–2011. Additionally, we focused on the prevalence genes encoding superantigens and M protein type in the population of invasive GAS strains.
Materials and methods
A total of 253 GAS strains were tested in that 48 originated from patients with clinical signs of invasive infection and 205 from non-invasive cases. In the group with invasive infections, 18 S. pyogenes strains were isolated from wound and deep skin infections, half of which required surgical intervention due to development of necrotising fasciitis. Additionally, 30 invasive S. pyogenes strains originated from women in labour with clinical symptoms of puerperal fever, sepsis and four cases of streptococcal toxic shock syndrome. The control group consisted of 147 S. pyogenes strains isolated from common infections of the upper respiratory tract in children and in adults associated with S.pyogenes, 49 S. pyogenes strains isolated from the vagina of females manifesting clinical symptoms of genital tract infection, and nine GAS originated from non-invasive wound and superficial skin infections (Table 1). All S. pyogenes strains were collected in the years 2009–2011 in outpatient and inpatient clinical centres in southern and northern Poland and in different healthcare institutions in Germany. The strains were initially characterized in the local microbiological laboratories and then sent to the Department of Bacteriology, Microbial Ecology and Parasitology, Chair of Microbiology, Jagiellonian University Medical College, Kracow, Poland for further testing. The strains originating from Germany were characterized and collected in the German National Reference Center for Streptococci in Aachen. Speciation of the strains was performed using phenotypic methods (API, bioMérieux, France) and latex agglutination test for serological grouping of β-haemolytic streptococci (Oxoid, UK). In case of inconclusive results, polymerase chain reaction (PCR) was performed with species-specific primers, i.e. spy1258F and spy1258R, constructed for transcriptional regulator gene spy1258. Amplification was performed according to the methodology described by Liu et al. [13].
Polymerase chain reaction-based gene detection of streptococcal exotoxins
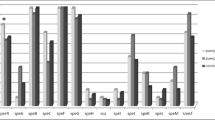
Presence or absence of the genes coding for different streptococcal pyrogenic exotoxins (SpeA, SpeC, and SpeG through SpeM), streptococcal superantigen (SSA), and streptococcal mitogenic exotoxin Z (SmeZ) were evaluated using PCR (for speI, speJ, speK, speL, speM, smeZ,) and multiplex PCR (for speA, speB, speC, speF, speG, speH, and ssa). Primers used in the reactions are shown in Table 2. Amplification was performed according to methodologies previously described [13, 15–17], and the obtained results are presented in Fig. 1 and Table 3.
Detection of emm gene serotype by PCR
The presence of the emm gene encoding protein M was verified in all strains from the study group. The PCR method, as described by Podbielski et al. [14] designed for the N-terminal region of the emm gene, was used. The PCR reaction products were sequenced using an ABI Prism® 310 Genetic Analyser (Applied Biosystems, Weiterstadt, Germany). The sequences obtained were compared with all available reference sequences on the United States (US) Center for Disease Control and Prevention (CDC) website (http://www2a.cdc.gov/ncidod/biotech/strepblast.asp).
The emm gene was considered if the degree of homology between the sequences reached 95%. Studies related to detection and sequencing of the emm gene were performed at the Department of Medical Microbiology, National Reference Center for Streptococci and Institute of Medical Statistics, RWTH Aachen University, Aachen, Germany.
Statistical analysis
Statistical analyses were performed to demonstrate significant differences in the distribution of the genes encoding superantigens and M protein types in the population of invasive vs. non-invasive GAS strains. The Kołmogorow-Smirnov test was used. Calculations were made in the R environment (version 3.1.0).
Results
The prevalence of the genes encoding superantigens (pyrogenic exotoxins, superantigen, mitogenic exotoxin) in the S. pyogenes population was high and they were commonly found in the GAS genomes, regardless of the type of infection. Based on statistical analysis, we found significant differences in the occurrence of only four genes: speG, speI, speJ and smeZ in GAS isolated from total invasive versus total non-invasive infections (Fig. 1).
A more detailed analysis of these results (Table 3) showed that in case of 18 S. pyogenes strains isolated from patients with invasive wound, skin and soft tissue infections, percentage distribution of genes was as follows: speA–none, speB–77.8%, speC–44.4%, speF–100%, speG–83.3%, speH–50%, speI–61.1%, speJ–5.6%, speK–16.7%, speL–5.6%, speM–5.6%, smeZ–77.8% and ssa–16.7%, with two genes speH and speI significantly more often present in the genome of GAS strains isolated from invasive skin/wound infection compared with strains isolated from non-invasive wound and superficial skin infections.In case of 30 women who developed invasive perinatal streptococcal infections, isolated S. pyogenes had the following profile of genes encoding superantigens: speA–26.7%, speC–56.7%, speG–93.3%, speH–10%, speI–10%, speJ–60%, speK–36.7%, speL–6.7%, speM–33.3%, smeZ–83.3% and ssa–6.7%. Two genes, speJ and smeZ, were significantly more often present in the genome of S. pyogenes strains isolated in this patient group compared with S. pyogenes strains isolated from the genital tract of women without symptoms of invasive infection. Two genes coding for enzymes related to virulence were also detected in a vast majority of the strains: speB–96.7% and speF–100%.
In the 147 S. pyogenes strains from the control group, derived from patients with non-invasive infections of the upper respiratory tract, the profile of genes encoding superantigens was as follows: speA–17.7%, speC–57.1%, speG–69.4%, speH–21.1%, speI–23.1%, speJ–29.9%, speK–17.7%, speL–10.9%, speM–16.3%, smeZ–49.7% and ssa–9.5%. Two genes coding for enzymes related to virulence were also detected: speB–91.2% and speF–100%.The speF (100%) and speB (91.2%) genes were commonly present in the genome of S. pyogenes colonizing the throat, in contrast to ssa (9.5%), which occurred least frequently in the genomic DNA.
Distribution of the emm types among all tested GAS strains belonged to 23 different types (Table 4). Only in case of emm type 28, statistically significant differences were observed, namely, the type 28 M protein was much more frequently isolated in the population of GAS causing invasive streptococcal infections of women in childbirth, as compared to non-invasive GAS originating from genital tract infections in women. Among 147 S. pyogenes strains isolated from non-invasive upper respiratory tract infections (control group), emm type 1 (31 /147), emm type 12 (20/147), emm type 2 (18/147) and emm type 28 (16/147) predominated over the other types. S. pyogenes isolated from invasive wound infections belonged to ten different emm types; the most frequent were types 44 (3/18) and 89 (3/18), while those from puerperal sepsis were mostly of types 28 and 1, but five other types (12, 77, 89, 2, 75) were also noted. The strains from invasive infections belonged more frequently to types 28 and 1, while those isolated from non-invasive GAS infections to types 2, 1 and 28.
Discussion
A special role in pathomechanisms of the invasive infections caused by S. pyogenes is played by the superantigens: a group of proteins that cause excessive activation of T lymphocytes. Rapid stimulation of the immune system leads to both the secretion of proinflammatory cytokines such as IL-1, TNF alpha, IL-2, as well as sudden symptoms of acute inflammation manifested by high fever, altered respiration, heart failure, multiple organ dysfunction ending in shock and death of the patient [1, 5]. At present, in the tested populations of S. pyogenes strains, there are several genes that encode superantigens including, among others, speA, speC, speG, speH, speI, speJ, speK, speL, speM, ssa, smez, and enzymes speB and speF. At the turn of the 1980s and the 1990s, it was proven that in case of speB and speF genes, there are more genes encoding proteins of cysteine protease and streptococcal DNase, that were not included in the group of superantigens [18, 19]. The above conclusion was confirmed in our results, as indeed speB and speF were the most common genes in genomic DNA of group A streptococci and both types of infections—invasive and non-invasive. We can, therefore, say that the distribution of genes encoding the superantigens is a feature common in most of the genomic DNA of S. pyogenes, regardless of whether these streptococcal infections caused invasive or non-invasive inflammation of the skin or respiratory system. Interestingly, non-invasive S. pyogenes strains isolated from women of childbearing potential during routine diagnosis of inflammatory conditions of the genital tract, also have most of the genes encoding superantigens. According to our results of such genes, including speG, speI, speJ and smeZ, they occurred significantly more often in the genomic DNA of streptococci isolated from invasive GAS infections. Based on the publication of Unnikrisnan et al. [20] we suppose that the gene smeZ encoding streptococcal mitogenic exotoxin Z (SmeZ) plays a special role in stimulating the secretion of pro-inflammatory cytokines. These authors indicated that human mouse HLA-DQ transgenic cells stimulated with supernatant from the S. pyogenes without smeZ gene (−) led to a complete inability to elicit cytokine production (TNF-alpha, lymphotoxin-alpha, IFN-gamma, IL-1 and -8) from cells. According to other authors, S. pyogenes strains capable of producing mitogenic exotoxin Z, have particular potential for virulence but also the ability to colonize and proliferate in the human body [20–22]. The gene smeZ demonstrates extensive allelic variation (smeZ1, smeZ2), thereby producing polymorphic protein that displays antigenic variation, especially detected during invasive infection [23].
It seems that the SmeZ protein is often released into the extracellular space by streptococci responsible for the invasive forms of infection. This information may be important from an epidemiological point of view.
Of course, the presence of a pool of genes in genomic DNA of S. pyogenes does not necessarily lead to the production and release of proteins/superantigens. There are probably many mechanisms that regulate the process of protein synthesis, and these are both strain-dependent factors, as well as factors related to the individual characteristics of the patient, which may include diabetes, immunosuppression, alcoholism, or extensive surgery with the discontinuity of the skin, including caesarean section. All these risk factors were accumulated in a group of homeless people and injectable drug abusers who were involved in an outbreak of invasive GAS infection in England and Wales [24]. However, there has been limited investigation into how combinations of transcriptional regulators control gene expression during infection despite the clear importance of regulatory networks to microbial pathogenesis. The data generated by Shelbourne et al. [25] demonstrate that the global metabolic gene regulator CcpA and the virulence factor regulator CovR act together to control expression of diverse GAS genes. The streptococcal pyrogenic exotoxins (SpeI, SpeJ and SpeH) bind to the beta-chain of CD4 T cells and MHC class II molecules on B cells, monocytes and dendritic cells, resulting in the overstimulation of the inflammatory response and subsequent systemic toxicity, tissue necrosis, organ failure and shock. Despite the increasing knowledge on GAS virulence factors and their role in disease pathogenesis [8, 23, 26], there is no clear view on their involvement in the pathomechanisms of different forms of invasive infections. We have recently reported [8] on the main virulence factors of GAS strains of emm28 serotype (less commonly emm1, emm12, emm75, and emm89) responsible for invasive perinatal infections which possessed speF, speG, and speB genes as well as genes responsible for mechanisms allowing for the binding and metabolism of iron ions. In the European study [10], a high rate of occurrence of speA was found among isolates of emm1 and emm3, types that were often involved in severe infections, and also for the less frequent type emm43. In the same study, GAS strains from puerperal sepsis harboured mostly speG, speF and speC genes, while our strain of this origin had predominantly smeZ and speJ genes. Similarly, cellulitis derived strains in the European study had mostly speG and speF genes, while our strains from wound infections clinically presenting as cellulitis possessed more frequently speI and speH genes. These discrepancies confirm differences among GAS strains isolated in different regions and countries related to local epidemiology, which, unfortunately, pose many problems in the ongoing attempts at creating a universal vaccine against GAS disease [27]. Distribution of the M types among GAS strains tested in this study was compared with epidemiological data summarized by Walker et al. [5]. Types 1 and 12 predominated in the group of strains isolated from superficial infections and type 28 in those from puerperal fever. On the other hand, types 2, 77 and 89, which were present more often than others in our GAS, were not mentioned in this review. It is known that epidemiological studies have shown a remarkable difference in the distribution of emm types in geographically and socioeconomically distinct regions of the world [3, 4]. In general, frequency of the types found by us seems to be similar to that reported for other European countries [10], with some exceptions, such as types 2 or 77 more frequently present in our GAS from non-invasive infections. We cannot offer an explanation for this discrepancy. Our strains were isolated from infections in three centres distributed in different regions of Poland and many centres in Germany. However, it should be emphasized here that emm types found in different outbreaks vary considerably and, for example, GAS strains isolated from very recent outbreaks described in Canada and England, were different and uncommon, namely, emm59 and emm66, respectively [24, 28]. Szczypa et al. [29] found an association between severe invasive GAS diseases and M type emm1 GAS isolates bearing the speA2 gene in their study on virulence factors of 41 GAS strains isolated in Poland between 1997 and 2005 from various invasive infections. Our data obtained several years later and reported in this study showed no difference between GAS isolated from invasive infections versus those from non-invasive infections in occurrence of speA gene and emm1 type. In fact, the speA was quite rarely present. Also, invasive GAS strains from streptococcal toxic shock syndrome post-caesarean section outbreak characterized by us [30], belonged to emm type 28 and had speC gene.
References
Cunningham MW (2000) Pathogenesis of group a streptococcal infections. Clin Microb Rev 13(3):470–511
Efstratiou A, Lamagni T (2016) Epidemiology of streptococcus pyogenes. In: Ferretti JJ, Stevens DL, Fischetti VA (eds) Source streptococcus pyogenes: basic biology to clinical manifestations [internet]. University of Oklahoma Health Sciences Center, Oklahoma City (OK)
Naseer U, Steinbakk M, Blystad H, Caugant DA (2016) Epidemiology of invasive group a streptococcal infections in Norway 2010-2014: a retrospective cohort study. Eur J Clin Microbiol Infect Dis 35:1639–1648
Rudolph K, Bruce MG, Bruden D, Zulz T, Reasonover A, Hurlburt D, Hennessy T (2016) Epidemiology of invasive group a streptococcal disease in Alaska, 2001 to 2013. J Clin Microbiol 54(1):134–141. doi:10.1128/JCM.02122-15
Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V (2014) Disease manifestations and pathogenic mechanisms of group a streptococcus. Clin Microbiol Rev 27(2):264–301
Tan LK, Eccersley LR, Sriskandan S (2014) Current views of haemolytic streptococcal pathogenesis. Curr Opin Infect Dis 27(2):155–164
Oehmcke S, Shannon O, Mörgelin M, Herwald H (2010) Streptococcal M proteins and their role as virulence determinants. Clin Chim Acta 7:1172–1180
Golińska E, van der Linden M, Więcek G, Mikołajczyk D, Machul A, Samet A, Piórkowska A, Dorycka M, Heczko PB, Strus M (2016) Virulence factors of streptococcus pyogenes strains from women in peri-labor with invasive infections. Eur J Clin Microbiol Infect Dis 35(5):747–754. doi:10.1007/s10096-016-2593-0
O’Loughlin RE, Roberson A, Cieslak PR, Lynfield R, Gershman K, Craig A, Albanese BA, Farley MM, Barrett NL, Spina NL, Beall B, Harrison LH, Reingold A, Van Beneden C, Active Bacterial Core Surveillance Team (2007) The epidemiology of invasive group a streptococcal infection and potential vaccine implications: United States, 2000–2004. Clin Infect Dis 45:853–862
Luca-Harari B, Darenberg J, Neal S, Siljander T, Strakova L, Tanna A, Creti R, Ekelund K, Koliou M, Tassios PT, van der Linden M, Straut M, Vuopio-Varkila J, Bouvet A, Efstratiou A, Schalén C, Henriques-Normark B, Strep-EURO Study Group, Jasir A (2009) Clinical and microbiological characteristics of severe streptococcus pyogenes disease in Europe. J Clin Microbiol 47(4):1155–1165. doi:10.1128/JCM.02155-08
Watanabe S, Takemoto N, Ogura K, Miyoshi-Akiyama (2016) Severe invasive streptococcal infection by streptococcus pyogenes and streptococcus dysgalactiae subsp. equisimilis. Microbiol Immunol 60(1):1–9
Parks T, Barrett L, Jones N (2015) Invasive streptococcal disease: a review for clinicians. Br Med Bull 115(1):77–89
Liu D, Hollingshead S, Swiatlo E (2005) Rapid identification of streptococcus pyogenes with PCR primers from a putative transcriptional regulator gene. Res Microbiol 156(4):564–567
Podbielski A, Melzer B, Lutticken R (1991) Application of the polymerase chain reaction to study the M protein (−like) gene family in beta-hemolytic streptococci. Med Microbiol Immunol 180:213–227
Luca-Harari B, Ekelund K, van der Linden M, Staum-Kaltoft M, Hammerum AM, Jasir A (2008) Clinical and epidemiological aspects of invasive streptococcus pyogenes infections in Denmark during 2003 and 2004. J Clin Microbiol 46:79–86. doi:10.1128/JCM.01626-07
Rivera A, Rebollo M, Miró E, Mateo M, Navarro F, Gurguí M, Mirelis B, Coll P (2006) Superantigen gene profile, emm type and antibiotic resistance genes among group a streptococcal isolates from Barcelona, Spain. J Med Microbiol 55(8):1115–1123. doi:10.1099/jmm.0.46481-0
Banks DJ, Lei B, Musser JM (2003) Prophage induction and expression of prophage-encoded virulence factors in group a streptococcus serotype M3 strain MGAS315. Infect Immun 71:7079–7086. doi:10.1128/IAI.71.12.7079-7086.2003
Bohach GA, Hauser AR, Schlievert PM (1998) Cloning of the gene, speB, for streptococcal pyrogenic exotoxin type B in Escherichia coli. Infect Immun 56(6):1665–1667
Sriskandan S, Unnikrishnan M, Krausz T, Cohen J (2000) Mitogenic factor (MF) is the major DNase of serotype M89 streptococcus pyogenes. Microbiology 146(Pt 11):2785–2792
Unnikrishnan M, Altmann DM, Proft T, Wahid F, Cohen J, Fraser JD, Sriskandan S (2002) The bacterial superantigen streptococcal mitogenic exotoxin Z is the major immunoactive agent of streptococcus pyogenes. J Immunol 169(5):2561–2569
Kamezawa Y, Nakahara T, Nakano S, Abe Y, Nozaki-Renar J, Isono T (1997) Streptococcal mitogenic exotoxin Z, a novel acidic superantigenic toxin produced by a T1 strain of streptococcus pyogenes. Infect Immun 65:3828
Proft T, Moffat SL, Weller KD, Paterson A, Martin D, Fraser JD (2000) The streptococcal superantigen SMEZ exhibits wide allelic variation, mosaic structure, and significant antigenic variation. J Exp Med 191:1765
Proft T, Arcus VL, Handley V, Baker EN, Fraser JD (2001) Immunological and Biochemicl characterization of streptococcal pyrogenic Exootoxins I and J (SPE-I and SPE-J) from streptococcus pyogenes. J Immunol 166(11):6711–6719
Bundle N, Bubba L, Coelho J, Kwiatkowska R, Cloke R, King S, Rajan-Iyer J, Courtney-Pillinger M, Beck CR, Hope V, Lamagni T, Brown CS, Jermacane D, Glass R, Desai M, Gobin M, Balasegaram S, Anderson C (2017) Ongoing outbreak of invasive and non-invasive disease due to group A Streptococcus (GAS) type emm66 among homeless and people who inject drugs in England and Wales, January to December 2016. Eurosurveillance 22(3) pii: 30446
Shelburne SA, Olsen RJ, Suber B, Sahasrabhojane P, Sumby P, Brennan RG, Musser JM (2010) A combination of independent transcriptional regulators shapes bacterial virulence gene expression during infection. PLoS Pathog 19 6(3):e1000817. doi:10.1371/journal.ppat.1000817
Olsen RJ, Shelburne SA, Musser JM (2009) Molecular mechanism underlying group a streptococcal pathogenesis. Cell Microbiol 11(1):1–12
Dale JB, Niedermeyer SE, Agbaosi T, Hysmith ND, Penfound TA, Hohn CM, Pullen M, Bright MI, Murrell DS, Shenep LE, Courtney HS (2015) Protective immunogenicity of group a streptococcal M-related proteins. Clin Vaccine Immunol 22(3):344–350. doi:10.1128/CVI.00795-14
Athey TB, Teatero S, Sieswerda LE, Gubbay JB, Marchand-Austin A, Li A, Wasserscheid J, Dewar K, McGeer A, Williams D, Fittipaldi N (2016) High incidence of invasive group a streptococcus disease caused by strains of uncommon emm types in Thunder Bay, Ontario, Canada. J Clin Microbiol 54(1):83–92. doi:10.1128/JCM.02201-15
Szczypa K, Sadowy E, Izdebski R, Strakova L, Hryniewicz W (2006) Group a streptococci from invasive-disease episodes in Poland are remarkably divergent at the molecular level. J Clin Microbiol 44(11):3975–3979 . doi:10.1128/JCM.01163-06 Published online 2006 Sep 6
Strus M, Drzewiecki A, Chmielarczyk A et al (2010) Microbiological investigation of a hospital outbreak of invasive group a streptococcal disease in Krakow, Poland. Clin Microbiol Infect 16:1442–1447
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
National Science Centre Poland, no. N N401618040, and Jagiellonian University Medical College grant, no. K/ZDS/005465.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Since this study was based on bacterial strains isolated from human cases for routine diagnostic purposes and collected over several years, for this type of study formal consent is not required.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Supported by the National Science Centre Poland, no. N N401618040 and Jagiellonian University Medical College grant, no. K/ZDS/005465
Rights and permissions
About this article
Cite this article
Strus, M., Heczko, P.B., Golińska, E. et al. The virulence factors of group A streptococcus strains isolated from invasive and non-invasive infections in Polish and German centres, 2009–2011. Eur J Clin Microbiol Infect Dis 36, 1643–1649 (2017). https://doi.org/10.1007/s10096-017-2978-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-017-2978-8