Abstract
The distribution and public health significance of Cryptosporidium species and genotypes in humans and bovine differ across geographical areas. Cryptosporidium species causes a disease known as cryptosporidiosis in humans and animals. To characterize the prevalence of cryptosporidiosis in humans in southern Assam, India, stool samples (n = 1119) of diarrhea patients were collected from different hospitals and from the community during the period January 2014 to July 2016. Fecal smears were examined microscopically for Cryptosporidium species using modified acid fast staining and were screened to ascertain the presence of Cryptosporidium antigen by enzyme-linked immunosorbent assay (ELISA). The genomic DNA of positive fecal samples were analyzed by nested polymerase chain reaction (PCR), which were subsequently genotyped by PCR-restriction fragment length polymorphism (RFLP), based on small subunit (SSU) 18S rRNA. It was found that the prevalence of Cryptosporidium spp. was high during the monsoon season. The average infection rate of Cryptosporidium spp. was found to be 2.4% (27/1119) microscopically. When subjected to nested PCR using amplification of the 18S rRNA gene, Cryptosporidium was found to be 8.57% (98/1119). Based on the 18S rRNA gene, two Cryptosporidium spp., namely Cryptosporidium andersoni (6.97%: 78/1119) and Cryptosporidium parvum (1.7%: 20/1119), were identified. Cryptosporidium andersoni infections were found to be of either zoonotic or anthroponotic origin. The prevalence was statistically significant (p = 0.03, R2 = 0.042) considering age, gender, and cast.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cryptosporidium species is the unicellular, microscopic, spore-forming, obligate intracellular organism of the apicomplexa family and is living like a parasite in the microvillus border of the gastrointestinal tract of a wide range of vertebrates, including man [1]. It is a common zoonotic enteric pathogen responsible for diarrheal diseases in humans and a variety of animals around the world [2]. Cryptosporidium has emerged as a significant human pathogen, particularly in children, elderly people, and immune-compromised patients, where an untreated infection might be life-threatening [3]. The parasite spreads through the fecal-oral route, through contaminated water or food, and often through contact with animals [3, 4]. However, previous studies suggested that cryptosporidiosis is a zoonotic as well as anthroponotic infection caused by different species. including C. meleagridis, C. felis, C. canis, C. cuniculus, C. ubiquitum, C. viatorum, C. suis, C. muris, and C. andersoni [5–9].
The detection of oocysts of Cryptosporidium in stool using the Ziehl–Neelsen modified acid fast stain is generally known as Kinyoun, and it is the most commonly used conventional technique in detecting the Cryptosporidium species [10, 11]. So far, 30 Cryptosporidium species and over 70 subtypes of Cryptosporidium have been recognized in a variety of vertebrate hosts [7, 12]. There is a need to develop fast and high-throughput molecular techniques for the detection and identification of Cryptosporidium species and their subtypes infecting humans. Molecular techniques like restriction fragment length polymorphism (RFLP) analysis based upon the gene of the small subunit rRNA (SSU rRNA) has been applied for the detection of Cryptosporidium species [13–15].
In India, inconceivable discrepancy persists in the characteristic ethnicity, culture, food habit, educational background, and standard of living within the nation [16]. The 2001 survey explains that 72.2% of the inhabitants lived in villages, and over 70% of the countryside inhabitants own domestic animals. Thus, domestic animals play an essential role in the socio-economic life of India. Since Cryptosporidium is recognized as one of the organisms which transmit by both anthroponotically and zoonotically, therefore, parasitic zoonoses affect human and animal health directly, and, consequently, affect livestock production [17]. The earliest traces of bovine cryptosporidiosis in India dates back to the late 1980s [18] and, subsequently, the incidence of C. andersoni has been reported from cattle [19].
The aim of the present study is to examine Cryptosporidium infection in patients with diarrhea, using conventional techniques such as microscopy, serological techniques like enzyme-linked immunosorbent assay (ELISA), and molecular tools such as polymerase chain reaction (PCR) and PCR-RFLP. In our study, samples were collected from various hospitals as well as from local communities of southern Assam, India. We then further examined if any feasible relationship exists between epidemiological data and clinical manifestations of the predominant Cryptosporidium species collected from the different study areas.
Materials and methods
Study design
A cross-sectional study was conducted to determine the prevalence of Cryptosporidium species among the patients from January 2014 to July 2016. The study was carried out in southern Assam, India (Cachar, Karimganj, and Hailakandi districts of the Barak Valley) at the level of community and district hospitals (Fig. 1). We used a systematic approach for the determination of the prevalence rate of Cryptosporidium species in southern Assam, India (Fig. 2).
Consent and ethical consideration
All stool samples were collected with the consent of each patient/subject, in the form of questionnaires. All study participants had given written consent before enrollment into the study. Parents/guardians provided consent on behalf of all infant participants. The study protocol was reviewed and approved by the Institutional Ethical Committee (ref. no. GCC/9440) of Gurucharan College, Silchar, Assam, before the commencement of the study and Silchar Medical College and Hospital, Silchar, Assam.
Collection of stool samples
A total of 1119 stool samples were obtained randomly from various hospitals and the locality of Cachar, Karimganj, and Hailakandi of southern Assam, India. The stool samples were collected in fresh, broad-mouthed, twist-cap disposable plastic containers. Within 3 h of collection, each stool sample was divided into three parts, each having equal quantity; one part was directly used for microscopic scrutiny; the second part was stored at 4°C in formal ether, which was later used for the serological analysis (ELISA); and the third part was stored at −20°C for PCR analysis.
Microscopy
This analysis was carried out according to standard techniques described earlier [20]. Slides were observed at 40× and then confirmed in oil immersion at 100× magnification under a phase contrast microscope (CX31, Japan).
ELISA
The Cryptosporidium antigen in the sample was distinguished by microwell ELISA. The ELISA was carried out with an existing kit according to the manufacturer’s instructions (Cryptosporidium 2nd Generation, Fecal, Diagnostic Automation, Inc., cat #8301–3).
Extraction of DNA
The DNA was isolated from the frozen stool samples using a Nucleopore™ stool DNA kit (Genetix Biotech Asia Pvt. Ltd., New Delhi, India), according to the procedure given by the provider, with minor modifications. In addition to seven freeze-thawing cycles, the samples were vortexed for 30 min in lysis buffer FL containing thrashing beads (Genetix Biotech Asia Pvt. Ltd., New Delhi, India). The samples were then processed according to the manufacturer’s instructions. The eluted DNA was quantified spectrophotometrically (BioPhotometer plus) and stored at −20°C for further use.
Nested PCR
The PCR amplification of the 18S rRNA gene was accomplished from individual samples using the oligonucleotide primers for species- and strain-specific identification of Cryptosporidium species. The sequences unique to all Cryptosporidium species were detected from the multiple alignments (CLUSTALW) and used as primers in a nested PCR procedure. For the major PCR step, a PCR product that was about 1325 bp long was amplified by using primers 5′-TTCTAGAGCTAATACATGCG-3′ and 5′-CCCTAATCCTTCGAAACAGGA-3′ [21] (Table 1). PCR amplifications were carried out in a volume of 20 μL with 100 ng of sample DNA, 1 μM of each primer, 1× PCR buffer, 2 mM MgCl2, 1× BSA, 0.2 Mm dNTPs, and 1U of Taq DNA Polymerase (Thermo Scientific, USA) in the MJ Mini™ (Bio-Rad Laboratories, Hercules, CA, USA) thermal cycler. A total of 35 cycles, each consisting of 94°C for 45 s, 55°C for 45 s, and 72°C for 90 s, were performed; an initial hot start at 94°C for 3 min and a final extension step at 72°C for 7 min were also incorporated. For the secondary PCR step, a PCR product that was 819 to 825 bp long (depending upon the species) was amplified by using 2 mL of the primary PCR product and primers F-5′-GGAAGGGTTGTATTTATTAGATAAAG-3′ and R-5′-AAGGAGTAAGGAACAACCTCCA-3′ [21]. The PCR mixture and the standard PCR conditions were kept the same as the conditions used for the primary PCR step, except that 3 mM MgCl2 was used for the PCR mixture and the annealing temperature was 57.8°C. The amplification of specific PCR product was checked by gel electrophoresis using 1% agarose.
For RFLP analysis, the secondary PCR product was digested in a 25-μL reaction mixture containing 5 U of SspI (Thermo Scientific, USA) (for species diagnosis) or 5 U of VspI (Thermo Scientific, USA) (for genotyping of C. parvum) (Table 2), and 2.5 μL of 10× restriction buffer at 37°C for 2.5 h, under conditions recommended by the supplier. The digested products were fractionated on a 2.0% agarose gel and visualized by ethidium bromide in a transilluminator [21, 22].
DNA sequence analysis
Positive secondary PCR products were subjected to sequencing from both directions using the secondary primers by the ABI 3500 genetic analyzer (Applied Biosystems Inc., Foster City, CA, USA) from the Genome Service of the Department of Biotechnology, Assam University, Silchar. Sequences obtained were analyzed and assembled using Sequencher software (Gene Codes Corp.). The obtained nucleotide sequences were used to search the GenBank nucleotide sequence database for sequence similarities using BLAST software (NCBI, Bethesda, MD, USA). Multiple alignments of these sequences were made by using the BioEdit program. The accuracy of the sequencing data was confirmed manually by looking at the overlapping regions. The sequences were deposited in GenBank (accession numbers KJ188438 and KJ719487).
Statistical analysis
All of the statistical analyses were performed using SPSS software v.21 (IBM Software Company, Armonk, NY, USA). The two-factor factorial analysis of variance technique was used to adjudge the effects of gender, caste, and age, along with their possible interactions. The parameters were further compared with Duncan’s test at the 1% level of significance.
Results
We tested a total of 1119 fecal samples: 694 were from males and 425 were from females. The mean age of males and females was 21.34 ± 20.11 and 24.77 ± 21.36 years, respectively. Smears were prepared from fecal material and stained with a modified acid fast technique. The oocysts appeared as pink and round and the size varied from 3 to 5 μm in a blue background of methylene blue stain. The ELISA method (Luminoskan Ascent™ Microplate Luminometer) detected 8.7% (98/1119) of positive samples. The absorbance range of 0.15–2.44 OD indicated that the presence of Cryptosporidium antigen was extremely stable. The nested PCR showed specific amplification of 825 bp of the 18S rRNA gene in 98 samples.
Based on the present study, the overall prevalence of C. andersoni was found to be 6.97%, followed by C. parvum (1.7%). A two-way analysis of variance (ANOVA) implicated that the correlation between the dependent variables like gender, age, and the prevalence of the parasite had no significant effect (p = 0.33, R2 = 0.015) (Table 1). An inverse relationship between the age of the host and infection with Cryptosporidium species was recorded, with maximum prevalence (13.4%) in infants aged <5 years, followed by the age groups 40–49 years (8.3%), 5–12 years (7.6%), >50 years (6.03%), 13–19 years (5.3%), and 20–29 years (4.2%), with the lowest prevalence (3.3%) in adults aged 30–39 years (Table 1). A two-way ANOVA was also conducted to explore the impact of age on the prevalence of C. parvum and C. andersoni. The results indicated that there was a significant mean effect for age on the prevalence of C. parvum [F(1, 1115) = 4.572, p = 0.03] and there was no statistically significant mean effect for age on the prevalence of C. andersoni [F(1, 1115) = 0.044, p = 0.8] (Table 1).
The prevalence rate of Cryptosporidium species was compared between the different districts of Assam; the rate was highest in the district of Cachar (10.80%), followed by Hailakandi (9.7%) and Karimganj (8.20%) districts. The two-way ANOVA indicated that there was a significant mean effect for different districts on the prevalence of C. andersoni [F(1, 1116) = 4.128, p = 0.04] and there was no significant mean effect for different districts on the prevalence of C. parvum [F(1, 1115) = 3.601, p = 0.05] (Table 2). The prevalence of the disease was highest during the monsoon season (14.48%), followed by pre-monsoon (7.36%) and post-monsoon months (4.1%). The two-way ANOVA indicated that there was no statistically significant difference for season in the prevalence of C. parvum [F(1, 1115) = 0.333, p = 0.56] and C. andersoni [F(1, 1115) = 1.403, p = 0.14] (Table 3).
The samples collected from the community included members of three different communities, i.e., Hindus, Muslims and Christians, with a mean participant age of 24.77 ± 21.36. The prevalence of the Cryptosporidium species was maximum in the Muslim population (12.79%), followed by the Christians (12.24%) and the Hindus (7.2%). The prevalence of C. andersoni found in the Hindu population (7.14%) was highest, followed by the Christian (10.20%) and Muslim (5.8%) individuals. Similarly, the prevalence of C. parvum was highest in the Muslim population (6.9%) followed by the Christian (2.04%) and the Hindu (0.12%) populations. The two-way ANOVA results indicated that there was a significant mean effect for different communities in the prevalence of C. parvum [F(1, 1115) = 5.903, p = 0.01] and there was no statistically significant mean effect for different communities in the prevalence of C. andersoni [F(1, 1115) = 0.000, p = 0.9] (Table 4).
In this study, the patients were divided into five groups in different districts (Group 1: 0–12 years; Group 2: 12–24 years; Group 3: 24–36 years; Group 4: 36–48 years; Group 5: 48–60 years). The prevalence of C. andersoni in Cachar district was highest in Group 2 (1.5%), followed by Group 1 (1.34%), Group 3 (0.80%), Group 4 (0.44%), and Group 5 (0.35%). The prevalence of C. parvum was only found in Group 4 (0.08%). The two-way ANOVA implicated that there was a significant mean effect for Cachar district in the prevalence of C. andersoni [F(1, 1109) = 12.801, p = 0.000] and there was no statistically significant mean effect for Cachar district in the prevalence of C. parvum [F(1, 1109) = 0.480, p = 0.48] (Table 5). The prevalence of C. andersoni in Karimganj district was highest in Group 1 (0.26%), followed by Group 2 (0.17%), Group 3 (0.08%), Group 5 (0.08%), and Group 4 (0.0%). Similarly, the prevalence of C. parvum was found to be highest in Group 1 (0.71%), followed by Group 2 (0.44), Group 3 (0.17), Group 4 (0.0), and Group 5 (0.0). The two-way ANOVA results indicated that there was a significant mean effect for Karimganj district in the prevalence of C. parvum [F(1, 1116) = 18.175, p = 0.000] and there was no statistically significant mean effect for Karimganj district in the prevalence of C. andersoni [F(1, 1116) = 2.727, p = 0.09] (Table 5). The prevalence of C. andersoni in Hailakandi district was highest in Group 1 (1.16%), followed by Group 2 (0.35%), Group 3 (0.26%), Group 4 (0.08%), and Group 5 (0.0%). Similarly, the prevalence of C. parvum was found to be highest in Group 1 (0.26%), followed by Group 4 (0.08), Group 2 (0.0), Group 3 (0.0), and Group 5 (0.0). The two-way ANOVA results indicated that there was a significant mean effect for Hailakandi district in the prevalence of C. andersoni [F(1, 1116) = 23.813, p = 0.000] and there was no statistically significant mean effect for Karimganj district in the prevalence of C. parvum [F(1, 1116) = 3.324, p = 0.06] (Table 5).
All of the 98 ELISA-positive samples showed amplification when nested PCR was carried out. The species identification was performed by nested PCR using the 18S rRNA gene, which yielded 1325-bp fragment in primary PCR and a 825-bp fragment in secondary PCR (Table 6) [21]. To identify the species and genotype of Cryptosporidium isolates present in the sample, the nested PCR products were digested with SspI and VspI endonuclease enzymes. Cryptosporidium species and genotypes were diagnosed by comparing band patterns with those published previously [21, 22]. The secondary PCR-RFLP analysis using SspI digestion of nested PCR products yielded three bands of about 448 bp, 247 bp, and 106 bp, for which the banding pattern of 247 bp suggested that C. parvum (human genotype) was present, and the two bands of about 448 bp and 106 bp in different areas suggested that C. parvum in the bovine genotype originated from humans and cattle, respectively. Cryptosporidium andersoni generated two visible bands of 448 bp and 397 bp in three areas, which indicates the bovine and human genotypes, respectively (Table 7). In the PCR-RFLP analysis of the nested PCR products of Cryptosporidium-positive samples, the VspI digestion showed bands of 556 bp and 104 bp, which indicates the presence of C. parvum human genotype, and 628 bp and 104 bp for most samples, which also indicates the presence of C. parvum bovine genotype. In another case, the samples of VspI digestion showed bands of 730 bp and 115-bp, confirming the identification of C. andersoni. Some variation has been shown in the bands, as one is very dark and another is light. The dark band indicates the C. andersoni (bovine genotype), while the light band indicates the C. andersoni (human genotype) (Table 7).
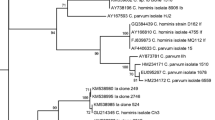
The RFLP analysis yielded typical restriction patterns for C. andersoni in 78 cases and C. parvum in 20 cases. The prevalence of C. andersoni and C. parvum infection stratified by three districts, viz., Cachar, Karimganj, and Hailakandi, is shown in Fig. 3. Restriction analysis of secondary PCR products with SspI and VspI revealed the presence of C. andersoni and C. parvum, and further confirmed the presence of human and bovine genotypes (eight samples were of the C. parvum human genotype and 12 samples were of the bovine genotype) in the districts of Cachar, Karimganj, and Hailakandi. The presence of C. andersoni was found predominantly in the samples of these regions under study and the C. parvum human genotype was found only in a single isolate of Cachar district. In addition, of the total C. andersoni-positive samples, 31 samples were of the bovine genotype and 19 were of the human genotype. Cryptosporidium andersoni was the major species found in Cachar in comparison to the other districts.
Intra-species variations were also found in C. parvum and C. andersoni. The most significant observation was found with C. parvum, where two genotypes of C. parvum differed from each other in the three different areas. The human genotypes were isolated only from humans, whereas the bovine genotypes were isolated from both cattle and humans. Cryptosporidium andersoni isolates from the human and bovine genotypes were found to be different from that of the C. parvum genotypes isolated from the three districts of Barak Valley, southern Assam. It was also evident from the sequencing results that the differences between the two genotypes (bovine and human) of C. parvum as well as C. andersoni were confirmed by sequence analysis of the 18S rRNA gene locus.
The sequences of C. andersoni and C. parvum have been deposited in GenBank under accession numbers KJ188438 and KJ719487, respectively. These samples showed 100% similarity with the C. andersoni sequence (GenBank accession number KF826294) and 99% similarity with the C. parvum sequence (GenBank accession number KF146224) (Table 8).
Discussion
The coexistence of Cryptosporidium with severe and constant diarrhea in children is prominent in the developing countries. A number of cross-sectional studies in children with diarrhea revealed that cryptosporidiosis was common in developing countries with a prevalence of up to 26% in Mexican and 16.5% in Brazilian children with diarrhea [23, 24]. The prevalence of Cryptosporidium in Asian countries, African countries, and Central and South American countries is greater than in Europe and North America [25]. In our study, the prevalence of Cryptosporidium in infants (<5 years old) was 13.4% and it was 7.6% in children (aged 5–12 years). In Pakistan and Indonesia, the parasite has been reported in 10.3 and 8.2% of children with diarrhea, respectively [26, 27]. The parasite was detected in pediatric diarrhea in 1.4% of cases in North India, 5.5% from East India, 5.6% from West India, and 13.1% from South Indian children [28–30]. In a study from Hyderabad, the prevalence was reported to be as low as 2.99% in children and 0.12% in adults [31]. Previous studies also stated that the prevalence of Cryptosporidium was 7.9% in Indian adults [32], which is supported by our current study (6.06%). In our study area, the highest prevalence was found to be (14.5%) in the monsoon season, followed by pre-monsoon (7.36%) and post-monsoon (4.1%) months. Water may be the major route of transmission of C. andersoni, C. hominis, and C. parvum genotypes, which are evident to the existence of high prevalences of C. parvum and C. andersoni during the rainy season [33]. From the present study, the prevalence of C. andersoni (6.97%) is higher than that of C. parvum (1.7%). No differences were observed morphologically between the dimensions of the C. andersoni and C. parvum oocysts isolated from the humans in our current study and those reported previously [34]. However, our study showed that the rate of Cryptosporidium prevalence was 6.06% in elderly patients (>50 years age), but earlier studies reported that the prevalence was 7.9% in Indian adults [32]. We also found an infection rate of 13.4% in infants in our study, which was higher than those from studies (4.3–13.3%) [29, 30, 35]. Cryptosporidium andersoni is also a major etiological agent of human cryptosporidiosis in India, especially in infants and the elderly (>50 years age) [32]. However, the results from our study clearly showed that C. andersoni has become a novel predominant species (78 positive for C. andersoni out of 98 cases) among the hospital-based and community-level patients with diarrhea in southern Assam, India. Only a few cases of C. andersoni infection in humans have been reported in France [36], Malawi [37], and England [38]. As per our knowledge, this might be the first report on the prevalence of C. andersoni in southern Assam, India, using molecular techniques. We have attempted to study the prevalence and distribution of two species in the study population and found significant impacts of age and season on the prevalence of these species. Therefore, screening for this pathogen should obviously be done among animals for national or international trade. The findings also clearly suggest the existence of two genotypes, including two of potential animal origin. Further genotyping, subtyping, and phylogenetic analysis with large sample sizes and extensive collection of epidemiological data are needed for a better understanding of cryptosporidiosis transmission in India.
References
Spano F, Putignani L, Guida S, Crisanti A (1998) Cryptosporidium parvum: PCR-RFLP analysis of the TRAP-C1 (thrombospondin-related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Exp Parasitol 90(2):195–198
Liu A, Zhang J, Zhao J, Zhao W, Wang R, Zhang L (2015) The first report of Cryptosporidium andersoni in horses with diarrhea and multilocus subtype analysis. Parasit Vectors 8(1):1–4
Fayer R (2004) Cryptosporidium: a water-borne zoonotic parasite. Vet Parasitol 126(1–2):37–56
Xiao L, Ryan UM (2004) Cryptosporidiosis: an update in molecular epidemiology. Curr Opin Infect Dis 17(5):483–490
Peng MM, Xiao L, Freeman AR, Arrowood MJ, Escalante AA, Weltman AC, Ong CS, Mac Kenzie WR, Lal AA, Beard CB (1997) Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg Infect Dis 3(4):567–573
Sulaiman IM, Xiao L, Yang C, Escalante L, Moore A, Beard CB, Arrowood MJ, Lal AA (1998) Differentiating human from animal isolates of Cryptosporidium parvum. Emerg Infect Dis 4(4):681–685
Xiao L (2010) Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol 124(1):80–89
Berrilli F, D’Alfonso R, Giangaspero A, Marangi M, Brandonisio O, Kaboré Y, Glé C, Cianfanelli C, Lauro R, Di Cave D (2012) Giardia duodenalis genotypes and Cryptosporidium species in humans and domestic animals in Côte d’Ivoire: occurrence and evidence for environmental contamination. Trans R Soc Trop Med Hyg 106(3):191–195
Gatei W, Wamae CN, Mbae C, Waruru A, Mulinge E, Waithera T, Gatika SM, Kamwati SK, Revathi G, Hart CA (2006) Cryptosporidiosis: prevalence, genotype analysis, and symptoms associated with infections in children in Kenya. Am J Trop Med Hyg 75(1):78–82
Ma P, Soave R (1983) Three-step stool examination for cryptosporidiosis in 10 homosexual men with protracted watery diarrhea. J Infect Dis 147(5):824–828
Clarke SC, McIntyre M (2001) Acid-fast bodies in faecal smears stained by the modified Ziehl–Neelsen technique. Br J Biomed Sci 58(1):7–10
Šlapeta J (2013) Cryptosporidiosis and Cryptosporidium species in animals and humans: a thirty colour rainbow? Int J Parasitol 43(12–13):957–970
Trotz-Williams LA, Martin DS, Gatei W, Cama V, Peregrine AS, Martin SW, Nydam DV, Jamieson F, Xiao L (2006) Genotype and subtype analyses of Cryptosporidium isolates from dairy calves and humans in Ontario. Parasitol Res 99(4):346–352
Pereira MGC, Li X, McCowan B, Phillips RL, Atwill ER (2010) Multiple unique Cryptosporidium isolates from three species of ground squirrels (Spermophilus beecheyi, S. beldingi, and S. lateralis) in California. Appl Environ Microbiol 76(24):8269–8276
Pedraza-Díaz S, Amar C, Iversen AM, Stanley PJ, McLauchlin J (2001) Unusual Cryptosporidium species recovered from human faeces: first description of Cryptosporidium felis and Cryptosporidium ‘dog type’ from patients in England. J Med Microbiol 50(3):293–296
Singh G, Prabhakar S, Ito A, Cho SY, Qiu D-C (2002) Taenia solium taeniasis and cysticercosis in Asia. In:Singh G, Prabhakar S (eds) Taenia solium cysticercosis: from basic to clinical science. CAB International, Oxon, pp 111–127
Parija SC (2004) Review of emerging parasitic zoonoses in India. Summer school “Current trends and strategies in diagnosis and control of parasitic diseases of domestic animals and poultry”, vol 25, pp 140–159
Nooruddin M, Sarma DK (1987) Role of Cryptosporidium in calf diarrhoea. Livest Advis 12:49
Kumar D, Sreekrishana R, Das SS (2004) Cryptosporidiosis in man and animals in Pondicherry. Indian Journal of Animal Sciences (India)
Ash LR, Orihel TC (1987) Parasites: a guide to laboratory procedures and identification. ASCP Press, Chicago
Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, Fayer R, Lal AA (1999) Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol 65(4):1578–1583
Feng Y, Ortega Y, He G, Das P, Xu M, Zhang X, Fayer R, Gatei W, Cama V, Xiao L (2007) Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet Parasitol 144(1–2):1–9
Newman RD, Sears CL, Moore SR, Nataro JP, Wuhib T, Agnew DA, Guerrant RL, Lima AA (1999) Longitudinal study of Cryptosporidium infection in children in northeastern Brazil. J Infect Dis 180(1):167–175
Jiang Y, Ren J, Yuan Z, Liu A, Zhao H, Liu H, Chu L, Pan W, Cao J, Lin Y, Shen Y (2014) Cryptosporidium andersoni as a novel predominant Cryptosporidium species in outpatients with diarrhea in Jiangsu Province, China. BMC Infect Dis 14(1):1
Garcia LS, Bruckner DA, Brewer TC, Shimizu RY (1983) Techniques for the recovery and identification of Cryptosporidium oocysts from stool specimens. J Clin Microbiol 18(1):185–190
Iqbal J, Munir MA, Khan MA (1999) Cryptosporidium infection in young children with diarrhea in Rawalpindi, Pakistan. Am J Trop Med Hyg 60(5):868–870
Khubnani H, Sivarajan K, Khubnani AH (1997) Study of cryptosporidiosis in a rural area of Maharashtra. Indian J Pathol Microbiol 40:33–36
Sehgal R, Rai SK, Sharma P, Malla N, Ganguly NK, Walia BN, Mahajan RC (1989) Cryptosoridium as a cause of diarrahoea in children. Indian J Med Microbiol 7(03):130
Saraswathi K, Pandit DV, Deodhar LP, Bichile LS (1988) Prevalence of Cryptosporidia in patients with diarrhoea in Bombay. Indian J Med Res 87:221–224
Mathan MM, Venkatesan S, George R, Mathew M, Mathan VI (1985) Cryptosporidium and diarrhoea in southern Indian children. Lancet 326(8465):1172–1175
Nagamani K, Rajkumari GA (2001) Cryptosporidiosis in a tertiary care hospital in Andhra Pradesh. Indian J Med Microbiol 19(4):215–216
Nath G, Choudhury A, Shukla BN, Singh TB, Reddy DC (1999) Significance of Cryptosporidium in acute diarrhoea in North-Eastern India. J Med Microbiol 48(6):523–526
Xiao L, Feng Y (2008) Zoonotic cryptosporidiosis. FEMS Immunol Med Microbiol 52(3):309–323
Wang L, Zhang H, Zhao X, Zhang L, Zhang G, Guo M, Liu L, Feng Y, Xiao L (2013) Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J Clin Microbiol 51(2):557–563
Das P, Roy SS, MitraDhar K, Dutta P, Bhattacharya MK, Sen A, Ganguly S, Bhattacharya SK, Lal AA, Xiao L (2006) Molecular characterization of Cryptosporidium spp. from children in Kolkata, India. J Clin Microbiol 44(11):4246–4249
Guyot K, Follet-Dumoulin A, Lelièvre E, Sarfati C, Rabodonirina M, Nevez G, Cailliez JC, Camus D, Dei-Cas E (2001) Molecular characterization of Cryptosporidium isolates obtained from humans in France. J Clin Microbiol 39(10):3472–3480
Morse TD, Nichols RA, Grimason AM, Campbell BM, Tembo KC, Smith HV (2007) Incidence of cryptosporidiosis species in paediatric patients in Malawi. Epidemiol Infect 135(8):1307–1315
Leoni F, Amar C, Nichols G, Pedraza-Díaz S, McLauchlin J (2006) Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J Med Microbiol 55(Pt 6):703–707
Acknowledgments
The help of Prof. S.K. Ghosh, Dean, School of Life Sciences, Assam University in the sequencing is greatly acknowledged. The authors acknowledge the help from the Department of Medicine, Silchar Medical College and Hospital in the collection of the stool samples. JP acknowledges the funds received through UGC Resource Networking and DST PURSE grant given to the faculty of the School of Life Sciences, Jawaharlal Nehru University, New Delhi.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
The research leading to these results has received funding from UGC Resource Networking and DST PURSE grant given to the faculty of the School of Life Sciences, Jawaharlal Nehru University, New Delhi.
Conflict of interest
No conflict of interest.
Ethics approval
The study protocol was reviewed and approved by the Institutional Ethical Committee (ref. no. GCC/9440) of Gurucharan College, Silchar, Assam, before the commencement of the study and Silchar Medical College and Hospital, Silchar, Assam.
Informed consent
All stool samples were collected with the consent of each patient/subject, in the form of questionnaires. All study participants had given written consent before enrollment into the study. Parents/guardians provided consent on behalf of all infant participants.
Rights and permissions
About this article
Cite this article
Hussain, G., Roychoudhury, S., Singha, B. et al. Incidence of Cryptosporidium andersoni in diarrheal patients from southern Assam, India: a molecular approach. Eur J Clin Microbiol Infect Dis 36, 1023–1032 (2017). https://doi.org/10.1007/s10096-016-2887-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2887-2