Abstract
Treatment choice for patients with malaria in Israeli hospitals is based on microscopy and rapid diagnostic tests (RDTs). Here, we demonstrate the cumulative value of real-time polymerase chain reaction (PCR) in optimizing the treatment of malaria. Between January 2009 and December 2015, 451 samples from 357 patients were tested in our laboratory using a real-time PCR assay. Hospital laboratory results (without real-time PCR) were compared to those obtained in our laboratory. A total of 307 patients had a malaria-positive laboratory finding in the hospital. Out of those, 288 were confirmed positive and 19 negative using real-time PCR. Two negative hospital results were found to be positive by real-time PCR. More specifically, of 153 cases positive for Plasmodium falciparum by real-time PCR, only 138 (90%) had been correctly identified at the hospitals. Similarly, 66 (67%) of 99 cases positive for P. vivax, 2 (11%) of 18 cases positive for P. ovale, and 3 (30%) of 10 cases positive for P. malariae had been correctly identified. Of 10 cases of mixed infection, only one had been identified as such at the hospital. Thus, real-time PCR was required for correct identification in 81 (28%) out of 290 positive cases. In 52 (18%) of those, there was an erroneous categorization of relapsing versus non-relapsing parasites. In a nationwide study, we found that the use of real-time PCR is definitely beneficial and may change the decision regarding the choice of treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malaria, formerly endemic in Israel, was eradicated 50 years ago, but new cases have been increasingly reported of malaria imported by returning travelers, immigrants, and foreign workers arriving from endemic areas [1–3].
Rapid diagnosis and immediate treatment of malaria is desired, especially for patients with Plasmodium falciparum infection, which is associated with a higher incidence of severe malaria and life-threatening complications, particularly in travelers with no immunity. Thus, the identification and discrimination of falciparum versus non-falciparum species is essential. Nevertheless, discrimination among non-falciparum species is also important, despite the fact that these infections are generally less severe, since patients infected with P. vivax or P. ovale require anti-relapsing treatment to eliminate the hypnozoites and prevent relapse (while P. malariae and P. knowlesi do not need it).
In Israel, most of the hospital laboratories combine microscopy and rapid diagnostic tests (RDTs). Microscopy may be challenging to use in a non-endemic setting. The lack of skilled technologists may lead to poor interpretation of data, particularly at low-density parasitemia (<50 parasites/μl blood) [4–6]. RDTs help to identify malaria infection, mainly P. falciparum, but are much less sensitive for non-falciparum species, especially for P. ovale and P. malariae [7].
Molecular diagnosis of malaria has developed in recent years. Benefits include earlier diagnosis, precise species identification, and improved identification in cases of low parasitemia as well as in mixed infections. Molecular tests, particularly real-time polymerase chain reaction (PCR) are increasingly used as confirmatory tests in reference labs [8–11].
In this report, we demonstrate the cumulative clinical value of real-time PCR in a non-endemic national setting.
Materials and methods
Malaria tests are performed in the hospitals, and almost all malaria-positive cases are hospitalized. It is mandated that blood samples found to be positive by microscopy or by rapid tests (or both) are sent for further confirmation of status to the reference parasitology laboratory of the Ministry of Health in Jerusalem. In rare cases, when malaria is highly suspected but laboratory findings at hospitals are repeatedly negative, negative samples are also sent to the reference laboratory.
Between January 2009 and December 2015, 451 samples from 357 patients were tested at the reference parasitology laboratory using an implemented real-time PCR [12]. This test employs a general assay (Plasmodium spp.) able to detect all five species that infect humans, as well as specific assays targeting each species (P. falciparum, P. vivax, P. malariae, P. ovale, or P. knowlesi) [12, 13]. Briefly, DNA is extracted from 200 μl of blood with EDTA, either manually, using the QIAGEN Blood & Tissue Kit (cat. # 69506), or via the NucliSENS easyMAG platform (bioMérieux), according to the manufacturers’ protocols. The elution volume is 200 μl with QIAGEN or 110 μl and adjusted to 200 μl when using easyMAG. Real-time PCR is performed essentially as described by Shokoples et al. [12], except for the use of a monoplex format in our lab. The assay is run on an Applied Biosystems 7500 Real-Time PCR machine, using 5 μl of eluted DNA in a total reaction volume of 20 μl. In cases of inconsistency with hospital results, additional tests are performed, which include one or more of the following: repeating the test on the same sample, repeating the test on a different sample, performing nested PCR, or sequencing the real-time PCR products. In all cases tested, the additional work corroborated the real-time PCR result (data not shown). In this work, real-time PCR was considered the definitive test.
Results
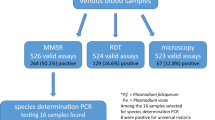
Over the 7 years studied, a total of 451 samples from 357 patients suspected of malaria have been sent for confirmation (Fig. 1). Among them, 307 patients had a positive laboratory finding at the hospital. Of those, 288 were confirmed positive (94% true-positive at the hospital level) using real-time PCR and 19 were negative (amounting to 6% false-positive results at the hospital level). Two of 50 additional patients, negative by hospital testing but who had been in endemic areas and had fever, were found to be positive by our lab (4% false-negative at the hospital level) (Fig. 1).
In total, 290 (81%) patients were found positive for malaria. Demographic information was available for 287 of those cases. All had acquired the parasite outside Israel, 169 (59%) were Israeli citizens, travelers, and immigrants, and the other 118 patients were foreigners, including 57 foreign workers and tourists (20%) and 61 African refugees (21%). Plasmodium speciation within each population group is depicted in Fig. 2.
Species-specific diagnosis
Detailed results for positive patients are summarized in Table 1. Considering real-time PCR as the definitive test, 153 cases were positive for P. falciparum, of which 138 (90%) had been correctly identified at the hospital (identification as “P. falciparum or mixed” was considered correct). Twelve had been identified as Plasmodium spp. without speciation and three wrongly as P. vivax or non-falciparum.
Sixty-six (67%) of 99 cases positive for P. vivax by real-time PCR had been correctly identified, as well as 2 (11%) of 18 positive for P. ovale, and 3 (30%) of 10 P. malariae.
Of ten cases of mixed infection identified by real-time PCR, only one had been identified as such at the hospital level.
Nineteen patients who had a positive laboratory finding in the hospital’s laboratory were not confirmed positive by real-time PCR. Two had other infections misidentified as P. falciparum, a severe Babesia infection and an African trypanosomiasis. Three patients had a negative RDT and an inconclusive blood smear, while three additional patients were not clinically compatible with malaria. Six patients in this group had a positive RDT for P. falciparum and negative microscopy. Two of these patients were known to have begun treatment a week before entering the hospital and probably didn’t have any intact parasites or DNA in blood. The information regarding the other four could not be retrieved. None of those found negative by real-time PCR subsequently developed malaria.
Discussion
Treatment is rapidly initiated on the basis of malaria diagnosis in the hospital. Blood samples are then transported to the reference parasitology laboratory and real-time PCR is performed. Results from the reference laboratory are usually obtained within 2–3 days.
Plasmodium falciparum is the most life-risking species of malaria, requiring rapid initiation of treatment in order to prevent complications. Assuming real-time PCR as the gold standard, 138 out of 153 cases (90%) were correctly diagnosed for this parasite. In 12 cases, real-time PCR was needed for speciation. Three cases were identified mistakenly to be non-falciparum, which could have led to unnecessary treatment of liver stages. Two additional cases were missed in mixed infections (identified as P. vivax only or not identified to the species level).
Plasmodium malariae is also a non-relapsing species that does not require treatment of the liver stage. Although usually causing a less severe disease than P. falciparum or P. vivax, P. malariae is considered unique in its ability to become quiescent, and patients may harbor low levels of parasites in blood that can lead to recrudescence after many years [14]. Of ten cases of P. malariae, one was found negative in the hospital’s laboratory. Two were identified as non-falciparum, potentially leading to unnecessary liver stage treatment. One was determined to be positive for plasmodium but real-time PCR was needed for speciation. Taken together, of 163 patients with P. falciparum or P. malariae, 21 (13%) were mistakenly identified as harboring relapsing parasites. In a mixed infection of them both, P. malariae was missed.
Plasmodium ovale and P. vivax are relapsing species that require additional treatment with primaquine to eliminate the liver stages. Failure to distinguish between them would not significantly affect the treatment given to the patient. Also, cases categorized as “non-falciparum” would direct physicians to give treatment against the liver stages, as P. vivax is more prevalent (Fig. 2) than P. malariae, which does not require such treatment. Thus, excluding the above mistakes (identifying P. vivax infection as “non-falciparum”), in 28 of 125 patients (22%), an infection with relapsing species failed to be detected; among these were six patients with mixed infections. Eleven patients (9%) were falsely identified as P. falciparum or P. malariae, thus potentially at risk of missing the liver stage treatment. In 16 patients (13%), Plasmodium was indicated but real-time PCR was needed for speciation. One P. vivax patient was found negative for Plasmodium in the hospital.
Genetic polymorphisms that exist in P. ovale have been shown to influence PCR detection accuracy [15]. While implementing real-time PCR in our lab, both subspecies of P. ovale, namely, P.o. curtisi and P.o. wallikeri, were included and demonstrated acceptable validation results. Indeed, sequencing of P. ovale-positive PCR products from blood samples of patients indicated that both subtypes were identified (data not shown). This is in accordance to the evaluation made in the paper by Calderaro et al. [15], predicting successful hybridization of the specific ovale probe to both species. However, the sensitivity of the P. ovale-specific assay for the variant species P.o. wallikeri was not systematically investigated within the scope of this work.
Patients with P. knowlesi were not encountered. A P. knowlesi infection would be identified by the general assay that is based on sequences common to all species [12, 13]. As this species is rare and found only in South East Asia, in countries seldom visited by Israeli travelers, we did not include the P. knowlesi-specific assay in our routine testing. Thus, the risk that P. knowlesi may have existed as a mixed infection with another species and overlooked is small.
As detailed in Table 1, two cases without positive laboratory findings at the hospital (using microscopy as well as RDT), despite testing of several blood samples taken several hours apart, were positive for malaria by real-time PCR. Both of these patients (positive for P. vivax and P. malariae) had taken prophylactic treatment while traveling and had a low parasite load. In addition, at least six of the patients which were eventually identified as positive for malaria in hospitals (five P. falciparum and one P. vivax) had been tested and found negative earlier in the course of disease at the hospital but were positive when testing those earlier samples using real time-PCR. These examples indicate that real-time PCR may facilitate earlier identification in some cases, consistently with the higher sensitivity of molecular methods [6, 11, 16–19]. This work does not fully assess the scope of this phenomenon, as submission of negative samples to the reference lab has not been recommended.
Our experience is in line with the account of a four-year real-time PCR testing study by Shokoples et al. [10], who reported accurate detection of 90% of the P. falciparum cases by microscopy, with the most significant contribution of real-time PCR being species identification of the non-falciparum group. In their experience, 18% of the cases could not be identified by microscopy alone and another 4% were misidentified using microscopy.
In our setting, real-time PCR was required for identification or correct speciation in 81 (28%) out of 290 positive cases. Excluding cases in which P. vivax and P. ovale were cross-identified leaves 72 (25%) such patients. If we further consider the identification of P. vivax and P. ovale as “non-falciparum” to be sufficient for patient treatment, we are left with 52 (18%) patients. In these 52 patients, there was an erroneous categorization of the relapsing versus non-relapsing parasites and the use of real-time PCR had an impact on the choice of optimal treatment (Table 1).
Another interesting observation arising from our analysis is of the poor success rate of RDTs in recognizing patients with P. malariae and P. ovale. Sixteen patients positive for P. ovale had an RDT test performed. Of those, six were reported as non-falciparum (correct), while ten were reported negative. Regarding patients with P. malariae, four were reported as non-falciparum (correct), while six reported negative on RDT. These results are compatible with other reports of the poor sensitivity of RDTs for P. ovale (5.5–86.7%) and P. malariae (21.4–45.2%) [7].
Perhaps the contribution of real-time PCR to correct diagnosis reported here should be regarded as an overestimation, to some extent. This is because the compliance with the requirement to send samples for confirmation is generally only 85% (data not shown), while at least some of the hospital laboratories tend to more often send to the reference lab those cases in which they have encountered difficulties in species diagnosis. Those cases are naturally more likely to reveal discrepancies.
Microscopy, a method more than 100 years old now, is still the gold standard of malaria diagnosis in hospitals. It has the advantages of concomitantly screening other infections that might appear in the blood (e.g., Babesia, trypanosomiasis, etc.), as well as providing estimated parasite load and information concerning parasite morphology and gametogenesis. Accordingly, microscopy would ideally be part of a routine workup. But already in 1909, Patrick Manson commented in his paper, “Diagnosis of fever in patients from the tropics”, that “the possession of microscope…does not always imply ability to recognize the malaria parasite” [20]. These days, especially in a non-endemic setting where experience in malaria microscopy is declining, applying this “gold standard” method becomes more challenging. Therefore, molecular techniques that are extensively used in many other areas of laboratory diagnosis should be applied to malaria diagnosis as well.
In conclusion, we have demonstrated that using real-time PCR in malaria diagnosis is beneficial to the patient. In 18% of the patients with proven malaria, real-time PCR changed the decision regarding treatment, while correction for epidemiological purposes occurred in 28% of cases. Had it been used for primary identification, there may have been an even greater advantage for earlier identification of malaria and proper treatment.
References
Dan M, Costin C, Slater PE (1996) Malaria imported by travelers: the Israeli experience. J Travel Med 3(3):182–185
Kopel E, Schwartz E, Amitai Z, Volovik I (2010) Relapsing vivax malaria cluster in Eritrean refugees, Israel, June 2010. Euro Surveill 15(26). pii: 19601
Anis E, Pener H, Goldmann D, Leventhal A (2004) The reemergence of malaria in Israel?. Harefuah 143(11):815–819, 838, 837
Bowers KM, Bell D, Chiodini PL, Barnwell J, Incardona S, Yen S, Luchavez J, Watt H (2009) Inter-rater reliability of malaria parasite counts and comparison of methods. Malar J 8:267
Kilian AH, Metzger WG, Mutschelknauss EJ, Kabagambe G, Langi P, Korte R, von Sonnenburg F (2000) Reliability of malaria microscopy in epidemiological studies: results of quality control. Trop Med Int Health 5(1):3–8
Britton S, Cheng Q, McCarthy JS (2016) Novel molecular diagnostic tools for malaria elimination: a review of options from the point of view of high-throughput and applicability in resource limited settings. Malar J 15:88
Maltha J, Gillet P, Jacobs J (2013) Malaria rapid diagnostic tests in travel medicine. Clin Microbiol Infect 19(5):408–415
Morassin B, Fabre R, Berry A, Magnaval JF (2002) One year’s experience with the polymerase chain reaction as a routine method for the diagnosis of imported malaria. Am J Trop Med Hyg 66(5):503–508
Rougemont M, Van Saanen M, Sahli R, Hinrikson HP, Bille J, Jaton K (2004) Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J Clin Microbiol 42(12):5636–5643
Shokoples S, Mukhi SN, Scott AN, Yanow SK (2013) Impact of routine real-time PCR testing of imported malaria over 4 years of implementation in a clinical laboratory. J Clin Microbiol 51(6):1850–1854
Roth JM, Korevaar DA, Leeflang MM, Mens PF (2016) Molecular malaria diagnostics: a systematic review and meta-analysis. Crit Rev Clin Lab Sci 53(2):87–105
Shokoples SE, Ndao M, Kowalewska-Grochowska K, Yanow SK (2009) Multiplexed real-time PCR assay for discrimination of Plasmodium species with improved sensitivity for mixed infections. J Clin Microbiol 47(4):975–980
Divis PC, Shokoples SE, Singh B, Yanow SK (2010) A TaqMan real-time PCR assay for the detection and quantitation of Plasmodium knowlesi. Malar J 9:344
Collins WE, Jeffery GM (2007) Plasmodium malariae: parasite and disease. Clin Microbiol Rev 20(4):579–592
Calderaro A, Piccolo G, Gorrini C, Rossi S, Montecchini S, Dell’Anna ML, De Conto F, Medici MC, Chezzi C, Arcangeletti MC (2013) Accurate identification of the six human Plasmodium spp. causing imported malaria, including Plasmodium ovale wallikeri and Plasmodium knowlesi. Malar J 12:321
Lek D, Popovici J, Ariey F, Vinjamuri SB, Meek S, Bruce J, Taylor WR, Socheat D, Menard D, Rogers WO (2016) National malaria prevalence in Cambodia: microscopy versus polymerase chain reaction estimates. Am J Trop Med Hyg 95(3):588–594
Mahende C, Ngasala B, Lusingu J, Yong TS, Lushino P, Lemnge M, Mmbando B, Premji Z (2016) Performance of rapid diagnostic test, blood-film microscopy and PCR for the diagnosis of malaria infection among febrile children from Korogwe District, Tanzania. Malar J 15(1):391
Walk J, Schats R, Langenberg MC, Reuling IJ, Teelen K, Roestenberg M, Hermsen CC, Visser LG, Sauerwein RW (2016) Diagnosis and treatment based on quantitative PCR after controlled human malaria infection. Malar J 15(1):398
Iglesias N, Subirats M, Trevisi P, Ramírez-Olivencia G, Castán P, Puente S, Toro C (2014) Performance of a new gelled nested PCR test for the diagnosis of imported malaria: comparison with microscopy, rapid diagnostic test, and real-time PCR. Parasitol Res 113(7):2587–2591
Manson P (1909) An address on the diagnosis of fever in patients from the tropics: delivered at a meeting of the Westminster Division of the Metropolitan Counties Branch. Br Med J 1(2516):704–706
Acknowledgments
We thank Dr. Stephanie Yanow from Alberta, Canada and Dr. Katia Jaton from Lausanne, Switzerland for their help and support in implementing real-time PCR for Plasmodium into our lab. We thank Dr. Heather Schnur for her constructive comments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
Tests were performed by the Israeli Ministry of Health as part of the public health service.
Ethical approval
All procedures performed were part of routine healthcare and reference lab duty regulated by law (Public Health Ordinance, 1940), with no need for informed consent.
Rights and permissions
About this article
Cite this article
Grossman, T., Schwartz, E., Vainer, J. et al. Contribution of real-time PCR to Plasmodium species identification and to clinical decisions: a nationwide study in a non-endemic setting. Eur J Clin Microbiol Infect Dis 36, 671–675 (2017). https://doi.org/10.1007/s10096-016-2844-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2844-0