Abstract
Streptococcus pneumoniae is one of the common pathogens causing severe invasive infections in children. This study aimed to investigate the serotype distribution and variations of penicillin-binding proteins (PBPs) 2b, 2x and 1a in S. pneumoniae isolates causing invasive diseases in Northeast China. A total of 256 strains were isolated from children with invasive pneumococcal disease (IPD) from January 2000 to October 2014. All strains were serotyped and determined for antibiotic resistance. The amplicons of penicillin-binding domains in pbp1a, pbp2b and pbp2x genes were sequenced for variation identification. The most prevalent serotypes of isolates in IPD children were 19A, 14, 19F, 23F and 6B. 19A and 19F were the most frequent serotypes of penicillin-resistant S. pneumoniae (PRSP), which present with high resistance to amoxicillin, cefotaxime, ceftriaxone and meropenem. The numbers of amino acid substitutions of penicillin-non-susceptible S. pneumoniae (PNSP) isolates were higher than those of penicillin-sensitive S. pneumoniae isolates in all the PBP genes (p < 0.01). The patterns of amino acid mutation in PBP2b, PBP2x and PBP1a were unique and different from those of other countries. All of the serotype 19A and 19F PRSP isolates carried 25 amino acid mutations, including Ala618 → Gly between positions 560 and 675 in PBP2b and Thr338 → Ala substitutions in PBP2x. The amino acid alterations in PBP2b, PBP2x and PBP1a from S. pneumoniae were closely associated with resistance to β-lactam antibiotics. This study provides new data for further monitoring of genetic changes related to the emergence and spread of resistance to β-lactam antibiotics in China.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptococcus pneumoniae is one of the most common pathogens causing severe invasive infections (including sepsis, empyema and meningitis) in infants. Both morbidity and mortality of invasive pneumococcal disease (IPD) are high worldwide, especially in developing countries. The World Health Organization (WHO) estimated that there were ∼8.8 million deaths among children aged <5 years worldwide in 2008, and 476,000 (333,000–529,000) were caused by pneumococcal infections [1]. More recently, a review showed that there were 12,815 cases/100,000/year of all-cause pneumonia and 14 meningitis cases/100,000/year among children aged 1–59 months, with 526 deaths/100,000/year in China between 1980 and 2008 [2].
The β-lactam antibiotics (BLAs) have played a leading role in the treatment of S. pneumoniae infection for a long time. However, in recent years, penicillin-resistant S. pneumoniae (PRSP) and penicillin-intermediate S. pneumoniae (PISP), which are collectively referred to as penicillin-non-susceptible S. pneumoniae (PNSP), were reported to be detected around the world continually [3, 4]. The detection rates of PNSP isolated from patients in mainland China with invasive and non-invasive disease were 27 % and 17 %, respectively, which were significantly higher than those of other Asian areas [5].
BLAs are involved in the synthesis of peptidoglycans by binding to the active site of penicillin-binding protein (PBPs), thus disrupting the formation of normal cell walls and inducing cell death by bacteriolysis [6, 7]. Streptococcus pneumoniae expresses six kinds of PBPs (PBP1a, PBP1b, PBP2a, PBP2b, PBP2x and PBP3). The first three proteins belong to Class A, which have activities of both transglycosylation and transpeptidylation; Class B proteins (PBP2b and PBP2x) only have transpeptidylation function; and PBP3 has D, D-carboxypeptidase activity [8, 9]. The active catalysis structure of each PBP is formed by three conserved amino acid motifs, SXXK, SXN and KT/SG [10]. Resistance to penicillin and other BLAs in S. pneumoniae is mainly due to alterations within or flanking these motifs, which results in alteration of PBPs and lowers the affinity of PBPs to penicillin [10–12].
Although the variants of pbp1a, pbp2b and pbp2x genes of S. pneumoniae have been studied extensively worldwide, few data are available on S. pneumoniae strains from IPD in mainland China. It may be that the improper use of antibiotics results in the low detection rates of S. pneumoniae. Vaccination is important in preventing S. pneumoniae infection due to the rapid spread of bacteria that are resistant to commonly used antibiotics. For the effective control of pneumococcal disease, the composition of the vaccine must closely match the prevalent serotypes in the native area. In view of the regional disparity of serotypes of S. pneumoniae, the coverage rate of pneumococcal conjugated vaccines (PCVs) and the distribution of PNSP, it is necessary to investigate the serotype distribution and resistance pattern of S. pneumoniae isolates from IPD patients in Northeast China. Thus, this study aimed to determine the amino acid variations encoded by pbp1a, pbp2b and pbp2x genes from clinical isolates of IPD patients in Northeast China. It also aimed to analyse the effect of amino acid modification on resistance to penicillin and other BLAs, and explore the associations between amino acid modification and serotypes.
Materials and methods
Strains
A total of 256 strains were collected from patients with IPD admitted to Shengjing Hospital of China Medical University from January 2000 to August 2014. All the strains were isolated from blood (135, 52.7 %) and other sterile body fluid samples, including pleural fluid (57, 22.3 %), cerebral spinal fluid (CSF) (55, 21.5 %), joint cavity fluid (5, 2.2 %) and abdominal fluid (4, 1.8 %). Most of the strains were isolated from infants aged ≤2 years (165, 64.5 %), whilst others were isolated from children aged 2–5 years (52, 20.3 %) and children aged 5–14 years (39, 15.2 %). All isolates were placed in a skimmed milk preservation medium and stored at −80 °C until further analysis. The study was approved by the Shengjing Hospital Ethics Committee.
Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MICs) of penicillin (PEN) were measured using Etest strips (AB Biodisk, Solna, Sweden). The MICs of amoxicillin (AMC), cefotaxime (CTX), ceftriaxone (CRO) and meropenem (MEM) were determined by the VITEK 2 compact system (bioMérieux, Marcy l’Etoile, France) using AST-GP68 cards. Breakpoints were based on the 2013 revised criteria of the Clinical and Laboratory Standards Institute (CLSI) [13]. Streptococcus pneumoniae ATCC 49619 was used as a quality control strain.
Serotyping
All isolates were serotyped by a Quellung test using type-specific antisera (Statens Serum Insitut, Copenhagen, Denmark). Serotyping was performed by phase-contrast microscopy, as previously described [14]. Only one isolate was accepted when two isolates isolated from one patient had the same serotype.
Analysis of pbp1a, pbp2b and pbp2x genes by PCR and sequencing
Bacterial genomic DNA was extracted using the TIANamp Bacteria DNA Kit (TIANGEN Biotech, Beijing, China). The extracted DNA was stored at −20 °C until use. To amplify pbp1a (1299 bp), pbp2b (1504 bp) and pbp2x (1148 bp) genes by polymerase chain reaction (PCR), we used three sets of primers, as described previously [15–17]. PCR was run in a 50-μl reaction mixture containing ∼40 ng DNA template, 0.2 μM each primer, 2.5 mM each dNTP, 1.5 U Taq DNA polymerase and 1 × PCR buffer (2.0 mM Mg2+ plus). An initial denaturation step of 94 °C for 5 min was followed by 35 amplification cycles: 94 °C for 40 s, 60 °C for 40 s and 72 °C for 50 s. A final extension was performed at 72 °C for 7 min. The purified PCR products were sequenced from both directions using an Applied Biosystems 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA). DNA sequences were assembled by DNA Baser software, then the nucleotide and amino acid sequences were compared with those of the reference strain (GenBank accession no. X90527 for pbp1a, X16022 for pbp2b and X16367 for pbp2x) by BioEdit software and the Basic Local Alignment Search Tool (BLAST) online.
Statistical analysis
The antibiotic susceptibility analyses were made using WHONET 5.6 software (WHO, Geneva, Switzerland). The number of amino acid alterations are presented as the mean ± standard deviation (SD). Statistical analyses were performed with SPSS version 16.0. The resistance rates were compared using Fisher’s exact test or Pearson’s χ2 test. One-way analysis of variance (ANOVA) was used to analyse the differences in the mean number of mutations in isolates with different degrees of sensitivity to PEN. A p-value < 0.05 was considered statistically significant.
Results
Serotype distribution and vaccine coverage
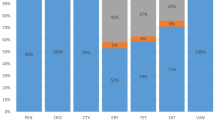
Among the total 256 IPD isolates, serotypes 19A (92, 35.9 %), 14 (44, 17.2 %), 19F (41, 16.0 %), 6B (23, 9.0 %) and 23F (21, 8.2 %) were the most common, which accounted for 86.3 % of all the isolates. The other 13 serotypes only accounted for 13.7 % (35/256) of the isolates. The most frequent serotypes were 14 (20 %), 19F (18.2 %), 19A (16.4 %), 6B (14.8 %) and 23F (10.9 %) in strains isolated from CSF (Fig. 1). All serotypes covered by PCV7, PCV10 and PCV13 were 53.9 %, 56.3 % and 93.8 %, respectively.
Antimicrobial susceptibility testing
According to the recommendations of the CLSI in 2013, the breakpoints of parenteral PEN were an MIC ≤ 0.06 μg/ml for susceptibility and MIC ≥ 0.12 μg/ml for resistance for meningitis isolates [13]. For non-meningitis isolates, the breakpoints of parenteral PEN were ≤2, 4 and ≥8 μg/ml [13]. Of the 256 S. pneumoniae isolates, 55 were meningitis isolates, including penicillin-susceptible S. pneumoniae (PSSP) isolates (11, 20.0 %) and PRSP isolates (44, 80.0 %). More than 60 % of PRSP isolates were susceptible to AMC, while the strains susceptible to CTX, CRO and MEM were relatively few. Among 201 non-meningitis isolates, 67 (33.3 %), 69 (34.3 %) and 65 (32.3 %) were susceptible, intermediate susceptible and resistant to PEN, respectively. For 69 PISP isolates, only the rate of susceptibility to AMC was >50 %. The rate of susceptibility to CTX was <10 %, and that to AMC, CRO and MEM was <5 % for PRSP isolates. PNSP isolates accounted for 69.5 % (178/256) of all the S. pneumoniae isolates, which comprised 44 isolates from CSF and 134 from other samples. All 78 PSSP isolates (including meningitis and non-meningitis isolates) were susceptible to AMC, CTX, CRO and MEM (Table 1).
Association between serotypes and susceptibility to PEN
Among the 256 isolates, the five common serotypes exhibited a significantly higher level of resistance to penicillin than did other serotypes (p < 0.05), especially serotypes 19A, 19F and 14 (Table 2). Serotype 19A had the most PNSP (91.3 %) overall, followed by 19F (85.4 %) and 14 (76.2 %). PNSP of serotype 14 from non-meningitis isolates were all PISP without PRSP. Serotypes 19A (84, 47.2 %), 19F (35, 19.7 %), 14 (32, 17.9 %), 23F (14, 7.9 %) and 6B (8, 4.5 %) were the most common serotypes recovered among 178 PNSP isolates. These five serotypes accounted for 97.2 % (173/178) of all 178 PNSP isolates. The other five isolates contained two of serotype 15 and one each of serotypes 6A, 9V and 20. Among these PNSP isolates, the proportion of serotypes covered by PCV7, PCV10 and PCV13 was 50.6 %, 50.6 % and 98.3 %, respectively. 19A (51, 70.8 %), 19F (19, 26.7 %) and 23F (2, 2.8 %) were the common serotypes among 72 PRSPs, in which the serotype coverage for PCV7, PCV10 and PCV13 was 29.2 %, 29.2 % and 100 %, respectively. Although 19A was the dominant serotype accounting for 51.3 % (39/76) of IPD isolates during 2000–2008, the proportion of serotype 19A isolates decreased over the following years.
Variations in or close to three conserved motifs (SXXK, SXN and KT/SG) in PBP2b, PBP2x and PBP1a
Three conserved amino acid motifs in PBPs were 385SVVK, 442SSN and 614KTG in PBP2b; 337STMK, 395SSN and 547KSG in PBP2x; and 370STMK, 428SRN and 557KTG in PBP1a. No amino acid substitutions were found in these conserved motifs from 45 isolates with MICs to PEN (0.016–0.25 mg/L). Fewer amino acid alterations (<10/gene) were found in these isolates. The number of amino acid substitutions and variation of conserved motifs forming or surrounding the active PBP binding sites in PBP2b, PBP2x and PBP1a in the other 211 S. pneumoniae isolates are shown in Table 3.
Variations of the pbp 2b gene
Compared with PSSP isolate R6 (GenBank accession no. X16022), 42 isolates with MICs to PEN (0.016–0.25 mg/L) had highly similar sequence to that of the R6 isolate. In these isolates, no variations were found in the three conserved motifs (385SVVK, 442SSN and 614KTG), and only two amino acid substitutions were identified in the non-conserved domains.
The variations were mainly concentrated in 454 amino acids between codons 224 and 678 in the other 214 isolates, and 14.31 % (65/454) of amino acid substitutions were identified in PBP2b. Compared with pbp2b sequences of the control R6 isolate, an average of 8.22 ± 7.73 mutations in PSSP, 30.88 ± 14.37 in PISP and 42.94 ± 4.11 in PRSP isolates were found. The number of variations in PRSP and PISP isolates was higher than in PSSP isolates (p < 0.01). Although there were no mutations in the three conserved motifs, an Ala618 → Gly substitution and a high frequency of substitutions containing 25 amino acids between positions 560 and 675 were identified in 115 PNSP isolates. In addition, the Thr445 → Ala next to SSN (442–444) motif and Glu475 → Gly mutation were the most common mutations found in 214 isolates.
A total of 214 isolates were grouped into eight different variants according to Baek et al. [16] and Granger et al. [18] based on the amino acid sequences of the transpeptidase binding domain in PBP2b (Table 4). Groups I (58 isolates), II (115 isolates) and III (23 isolates) were the main groups in all the isolates. Eighteen isolates (11 PSSP and 7 PISP) were classified in Baek et al.’s group I, which showed the same 15 amino acid substitutions. Additional amino acid changes of F381L and S411P were found in our group I isolates besides 13 amino acid substitutions described by Granger et al. [18]. Another 40 isolates (14 PSSP and 26 PISP) showed additional amino acid alterations Lys365 → Asn, but without Ile360 → Leu mutation; they were named as subgroup Ia. Isolates (115) belonging to group II were all PNSP isolates, which contained 97.2 % (70 /72) of PRSP (MICs to PEN ≥8 mg/L). These isolates showed that not only did the mutations belong to group I, but there were also another 25 amino acid substitutions between positions 560 and 675. Among these isolates, 87 isolates containing the short mosaic sequence VDT between residues 248 and 250 were assembled in subgroup IIa. Their serotypes were all 19A, except for two which were 14. The other 28 isolates were named as subgroup IIb, which did not have the short mosaic sequence VDT but showed the amino acid alteration Val224 → Ile and Glu368 → Asp. Twenty-three isolates containing the group I mutation block (except Ile360 → Leu), Ala515 → Ser and Ala314 → Ser, belonged to group III, which included 12 PSSP and 11 PISP isolates. Four isolates shared 20 amino acid alterations containing another distinct block (314–365) in addition to the mutation block (421–488) were assembled into group IV. The isolates belonging to groups V, VII and VIII were all PSSP isolates. No isolate with a similar sequence to Baek et al.’s group VI was found in our study.
Variations of the pbp2x gene
Compared with the pbp2x gene sequence of the R6 strain, 45 isolates (MIC to PEN ≤ 0.25 mg/L) were highly prototype-like, and the remaining 211 isolates (MIC to PEN ≥ 0.5 mg/L) had 79 (17.14 %) amino acid mutations concentrated within 461 amino acids between codons 254 and 715. We found an average of 29.26 ± 27.88 mutations in PSSP, 63.37 ± 2.51 in PISP and 65.55 ± 2.93 in PRSP isolates. The numbers of mutations in PISP and PRSP were significantly increased compared with those of PSSP (p < 0.01). A Thr338 → Ala mutation in the STMK motif (337–340) and Leu546 → Val mutation just before the KSG motif (547–549) were found in 208 isolates (MIC to PEN ≥ 0.5 mg/L). Only one isolate of serotype 14 from CSF showed a His394 → Leu alteration next to the SSN motif (395–397), without the two mutations mentioned above.
Based on deduced PBP2x amino acid sequences, 211 isolates were grouped referring to the method of Granger et al. [18] (Table 5).Thirty-nine isolates belonged to Granger et al.’s group V, which were all serotype 14 isolates. Another 169 isolates containing one unique amino acid mutation of Asn417 → Lys could be classified in Granger et al.’s group VI, which contained 36 PSSP isolates and the majority (133/178, 74.72 %) of PNSP isolates.
To define further the character of amino acid mutations, we classified Granger et al.’s group VI isolates into four subgroups: VIa, VIb, VIc and VId (Table 5). We named six serotype 6B isolates (MIC to PEN 0.5–1 mg/L) that shared the same 42 amino acid mutations as subgroup VIa. An additional substitution block that contained 20 amino acid alterations was found in 45 isolates (MIC to PEN 1–4 mg/L), which were named as subgroup VIb. Besides these 65 mutations (except M343T), Met339 → Phe, Met400 → Thr, Tyr595 → Phe and Gln629 → Lys were also found in 54 PNSP isolates, which were named as subgroup VIc. Among these isolates, 46 were PRSP strains, which accounted for 63.89 % of PRSP isolates. Sixty-four isolates showing an amino acid mutation ratio of 13.45 % (62/461) were named as subgroup VId, which were all serotype 19A isolates. Compared with other group VI isolates, they had Asn501 → Lys, Lys505 → Glu, Ala507 → Thr, Glu651 → Lys, Glu706 → Ala, Thr709 → Ala and Ala711 → Ser mutations, but without Asp506 → Glu, Asp648 → Glu and Thr678 → Lys mutations. Except for four PSSP isolates, most of these isolates were PNSP isolates that contained 36.11 % (26/72) of PRSP. The remaining three isolates (MIC to PEN 0.5 mg/L) could not be classified into any group.
Variations of the pbp1a gene
Compared with reference PBP1a, a total of 75 amino acid mutations were found in 256 isolates, which were mainly concentrated between codons 316 and 695. Forty-seven (58 %) PSSP (MIC to PEN ≤ 0.5 mg/L) isolates almost had no amino acid mutations (<3/isolate) compared with the PBP1a of the R6 strain. We found an average of 29.26 ± 27.88, 63.37 ± 2.51 and 65.55 ± 2.93 amino acid alterations in PBP1a from PSSP, PISP and PRSP isolates, respectively. The numbers of mutations in PISP and PRSP were significantly increased compared with those of PSSP (p < 0.01). All of the PNSP isolates had a Thr371 → Ala/Ser mutation in the STMK motif (370–373) and a Pro432 → Thr mutation next to the SRN motif (428–430). A short mosaic sequence NTGY instead of TSQF between positions 574 and 577 was found in all the PNSP isolates. However, these mutations were also observed in some PSSP isolates. No mutations were found within or close to the KTG motif (557–559).
According to PBP1a, 48 (60.1 %) PSSP isolates characterised by 1–3 amino acid alterations were assembled in Granger et al.’s group I. Six amino acid substitutions of Thr371 → Ala, Ile393 → Met, His395 → Asn, Glu397 → Ile, Asn405 → Ser and Pro432 → Thr were shared by 27 isolates, which were classified within group II (16 PSSP and 11 PISP isolates). Thirteen isolates of serotype 6B showed the same 53 amino acid substitutions as group IIb, and the other isolates were named as group IIa. Most isolates (179), including 29 PSSP and 150 PNSP isolates, had the characteristic mutations of Thr371 → Ser, Thr392 → Ala, Glu397 → Val, Asn405 → Asp and Val408 → Leu, which belonged to Granger et al.’s group V. According to the characteristic amino acid mutations in our study, we further classified the isolates into three subgroups, Va, Vb and Vc (Table 6). It is worth noting that all the PRSP isolates belonged to subgroup Vc. Glu388 → Asp was shown in all the isolates in this study.
Discussion
The morbidity of invasive disease induced by S. pneumoniae has shown an increasing trend in the past 15 years, especially for children aged <5 years, who comprised 84.8 % (217/256) of the patients with IPD in our region. The increasing morbidity of IPD in children may be partly because of their immature organs and immune system, or due to the variation in virulence of S. pneumoniae isolates caused by misuse of antibiotics. Vaccination is the most effective and economic way to prevent IPD and reduce its incidence and mortality in children, as well as being one of the important ways to prevent the spread of antibiotic resistance [19].
The serotypes associated with IPD vary with time, geographic regions, race, age and vaccination. In the present study, the most prevalent serotypes were 19A, 19F, 14, 23F and 6B. The results were similar to a multicentre surveillance study from China. That study showed that 19A (22.1 %), 19F (21.7 %), 14 (7.5 %), 3 (7.1 %) and 23F (5.4 %) were the most common pneumococcal serotypes among 218 IPD strains collected from 2005 to 2011 [20]. Nevertheless, the serotype distribution was completely different from that of the USA, in which 33F, 22F, 15B/15C and 35B instead of 19A and 7F became the most common serotypes after PCV13 implementation during 2011–2013 [21]. The spread of serotype 19A S. pneumoniae in the USA was after the large-scale use of PCV7 (including pneumococcal serotypes 4, 6B, 9V, 14, 18C, 19F and 23F). During 1998–1999, the serotype 19A isolates only accounted for 2.5 % of all isolates causing IPD, but this ratio increased to 36 % in 2005 [22]. Now, serotype 19A has become common in isolates causing IPD and is associated with multidrug resistance [23]. However, some researchers have suggested that the prevalence of serotype 19A was not the result of the introduction of PCV7, but associated with antibiotic selection pressure [24]. A multicentre study [25] showed that, before PCV7 was introduced into China, serotype 19A was the most common serotype in paediatric patients in the cities in Northern China, while it rarely appeared in Southern and Eastern China. Although PCV7 was introduced to China in 2008, it was still a for-fee vaccine. Because of the relatively high price, the coverage of PCV7 remained low [25, 26]. Thus, the potential cause of prevalence of serotype 19A in our region was that 19A, as the multidrug-resistant serotype, was easier to spread under the pressure of antibiotics. Among the 72 PRSP isolates, the serotype coverage rates of PCV7, PCV10 and PCV13 were 29.2 %, 29.2 % and 100 %, respectively. It was obvious that the protection of PCV7 and PCV10 against S. pneumoniae IPD was limited. Therefore, PCV13 was urgently needed in our region for protecting more children from S. pneumoniae IPD in Northeast China.
Correctly identifying pathogens and effective antibiotic treatment at an early stage, which can reduce the hospital stay and mortality of IPD, are important in the prognosis of IPD. All the PRSP isolates belonged to serotypes 19A, 19F and 23F, especially serotype 19A, which accounted for 70.8 % of PRSP isolates. In addition, for all the PRSP from non-meningitis isolates, the susceptibility rates to AMC, CTX, CRO and MEM were all low, which was possibly related to the high frequency of application of these BLAs in this area. It is suggested that CTX, CRO and MEM should be chosen carefully according to the results of susceptibility testing when treating IPD. Our data showed that antibiotic resistance had been a serious problem in Northeast China, but, fortunately, overall, the serotypes of PRSP strains in this study could be covered by PCV13. As confirmed in other countries, PCV13 can prevent IPD in children and control the spread of antibiotic-resistant strains [27–29].
In the present study, the genetic diversity of pbp2b, pbp2x and pbp1a genes were analysed in 256 IPD strains by comparing with the reference sequence of the R6 strain. The mean numbers of amino acid substitutions in PNSP isolates were significantly higher than those of PSSP isolates in all the PBP genes (p < 0.01).This was similar to the results reported previously [10, 16, 18] and confirmed that the increase in resistance to BLAs was associated with the increase in amino acid alterations of PBP2b, PBP2x and PBP1a.
The amino acid mutations of PBP2b were classified into eight subgroups. Some subgroups were associated with a specific serotype, such as subgroup Ia and serotype 14, and subgroup IIb and serotype 19F. In this study, the Thr445 → Ala alteration adjacent to the SSV motif and Glu475 → Gly were the most common mutations found in 214 isolates, including some PSSP isolates. An in vitro study showed that the Thr445 → Ala mutation displayed a 60 % reduction affinity to penicillin G compared with wild-type protein [30], and many researchers have found this mutation in PNSP isolates [31–33]. In contrast, other researchers have also found this mutation in PSSP isolates, as in our study [10, 18]. Whether this mutation plays a key role in PEN resistance is worthy of exploration.
Group II was the most prevalent mutation type in our study, which carried the largest number of amino acid mutations. All the isolates (115) belonging to this group were PNSP isolates. The Ala618 → Gly mutation next to the KTG (614–616) motif was found in these isolates. This mutation has also been reported in PRSP strains previously [16, 18, 30]. This suggests that the mutation is closely related to the mechanism of resistance of S. pneumoniae to BLAs. All the subgroup IIa isolates were serotype 19A, except for two that were serotype 14, and all the subgroup IIb isolates were serotype 19F. The amino acid sequence of these isolates was the same as Korean J77, Korean J83 or Canadian 14907 [16, 18]. The spread of these high-level-resistant strains may be due to the limited spread of serotypes 19A and 19F clones. Some antibiotic-resistant isolates could be imported to China with the increasingly frequent international exchanges, which could be one of the major causes of increasing high-level-resistant strains. To the best of our knowledge, Phe 381 → Leu was a novel mutation that was identified in 198 isolates. There are no data demonstrating that the mutation was directly related to PEN resistance, and the biochemical function of this mutation needs further research.
Variation of pbp2x is also one of the important causes of BLA resistance in S. pneumoniae. Amino acid substitutions of Thr338 → Ala and Met339 → Phe in the STMK motif simultaneously occurred in subgroup VIc isolates, which were not only resistant to PEN but also to AMC, CTX, CRO and MEM. The Thr338 → Ala mutation drastically reduced acylation efficiency for BLAs due to the absence of a hydroxyl group at this position [34]. The T338A variation was frequently observed in clinical BLA-resistant isolates (208) in our study. The crystal structure of the double mutant T338A/M339F showed a distortion of the active site and an alternative conformation of the active site Ser337, which reduced 20-fold the efficiency of acylation of PEN and CTX [35, 36]. These two mutations were also found in BLA-resistant strains in other countries [37–39]. These isolates also carried amino acid substitutions of Met400 → Thr, Tyr595 → Phe and Gln629 → Lys compared with other isolates, suggesting that these mutations are responsible for the high level of resistance. Met339 → Phe, Met400 → Thr and Tyr595 → Phe substitutions have not been reported in previous studies from our region [33].
The His394 → Leu mutation just before the conserved SSN motif was found in one serotype 14 PRSP isolate from CSF (MIC to PEN 0.5 mg/L), which could not be classified in any group. This isolate did not carry Thr338 → Ala and Leu546 → Val mutations, which is consistent with previous studies [25, 35, 36]. The Leu546 → Val mutation adjacent to the KSG motif was found in 208 isolates, which contained 168 isolates with MICs to CTX and CRO ≥ 1 mg/L [39, 40], showing that the Leu546 → Val substitution is associated with high-level BLA resistance, especially with resistance to cephalosporins. However, it was also found in some cephalosporin-sensitive isolates, which suggests that the single mutation is not enough to cause high-level resistance. Granger et al.’s group VId was another prevalent group with a high level of BLA resistance, which contained 60 PNSP isolates with serotype 19A. This PBP2x pattern may present as a new prognostic indicator of resistance to BLAs in serotype 19A isolates, which could be applied to clinical practice using PCR.
The variation of the PBP1a pattern of 209 isolates was classified into six groups: Granger et al.’s group I and subgroups IIa, IIb, Va, Vb and Vc. Subgroups IIb, Vb and Vc were associated with specific serotypes 6B, 14, and 19A/19F, respectively. In all PNSP isolates (MICs for PEN ≥ 4 mg/L and CTX ≥2 mg/L), a Thr371 → Ala/Ser mutation close to the STMK motif, a Pro432 → Thr substitution next to the SRN motif and TSQF(574–577)NTGY were found, which were identified with high-level resistance to PEN and cephalosporins [18, 39, 41]. A site-directed mutagenesis study demonstrated that the Thr371 → Ala mutation reduced the efficiency of acylation of R6-PBP1a by cefotaxime (2.4-fold) and penicillin G (26-fold), and the decrease of acylation efficiency induced by the TSQF(574–577)NTGY mutation was even greater [42]. An in vivo study confirmed that only the double mutant conferred increased resistance [42]. This suggested that the impact of mutation on antibiotic susceptibility might be more complicated than we expected.
Our data showed that the different serotype isolates with similar levels of BLA resistance often share the same pattern of amino acid substitution in PBP2b, PBP2x and PBP1a, such as serotypes 19A, 19F, 14, 23F and 6. Before 2008, the breakpoints of parenteral PEN for non-meningitis strains were ≤0.125, 0.125–1 and ≥2 mg/L for susceptible, intermediate and resistant strains, respectively [43]. In the present study, CLSI standards (2013) were used as the criteria. Some PSSP isolates (MIC for PEN 0.5–2 mg/L) that belonged to PNSP according to old standards showed the same amino acid substitution pattern as the PNSP isolates.
We investigated variation in pbp2b, pbp2x and pbp1a genes and serotype distribution from IPD S. pneumoniae isolates with a large sample size (256) from 2000 to 2014 in Northeast China. We analysed the association among the amino acid substitutions, resistance to BLAs and serotype distribution. This was a serious problem for the widespread prevalence of BLA-resistant S. pneumoniae isolates in Northeast China. Inappropriate use of antibiotics may provide the necessary selective advantage for the expansion of drug-resistant serotypes. Based on our data, PCVs, especially PCV13, were necessary in our region to reduce the occurrence of IPD and prevent the spread of antibiotic-resistant strains. The limitation of this study was that we did not analyse the multilocus sequence typing of isolates because of the high costs. So, the prevalent antibiotic-resistant clones could not be definitely identified and the results could not be directly compared with the data of some international studies.
References
World Health Organization (2012) Pneumococcal vaccines WHO position paper—2012. Wkly Epidemiol Rec 87(14):129–144
Chen Y, Deng W, Wang SM, Mo QM, Jia H, Wang Q, Li SG, Li X, Yao BD, Liu CJ, Zhan YQ, Ji C, Lopez AL, Wang XY (2011) Burden of pneumonia and meningitis caused by Streptococcus pneumoniae in China among children under 5 years of age: a systematic literature review. PLoS One 6(11), e27333. doi:10.1371/journal.pone.0027333
Jones RN, Sader HS, Moet GJ, Farrell DJ (2010) Declining antimicrobial susceptibility of Streptococcus pneumoniae in the United States: report from the SENTRY antimicrobial surveillance program (1998–2009). Diagn Microbiol Infect Dis 68(3):334–336. doi:10.1016/j.diagmicrobio.2010.08.024
Kim SH, Song JH, Chung DR, Thamlikitkul V, Yang Y, Wang H, Lu M, So TM, Hsueh PR, Yasin RM, Carlos CC, Pham HV, Lalitha MK, Shimono N, Perera J, Shibl AM, Baek JY, Kang CI, Ko KS, Peck KR; ANSORP Study Group (2012) Changing trends in antimicrobial resistance and serotypes of Streptococcus pneumoniae isolates in Asian countries: an Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Antimicrob Agents Chemother 56(3):1418–1426. doi:10.1128/AAC.05658-11
Dai LH, Dong L, Li HY, Su MS (2015) Control study on antimicrobial resistance of invasive and non-invasive Streptococcus pneumoniae in children. Zhongguo Dang Dai Er Ke Za Zhi 17(4):303–307. doi:10.7499/j.issn.1008-8830.2015.04.002
Jacobs MR (2008) Antimicrobial-resistant Streptococcus pneumoniae: trends and management. Expert Rev Anti Infect Ther 6(5):619–635. doi:10.1586/14787210.6.5.619
Zapun A, Contreras-Martel C, Vernet T (2008) Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol Rev 32(2):361–385. doi:10.1111/j.1574-6976.2007.00095.x
Navarre WW, Schneewind O (1999) Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev 63(1):174–229
Grebe T, Hakenbeck R (1996) Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of beta-lactam antibiotics. Antimicrob Agents Chemother 40(4):829–834
Nagai K, Davies TA, Jacobs MR, Appelbaum PC (2002) Effects of amino acid alterations in penicillin-binding proteins (PBPs) 1a, 2b, and 2x on PBP affinities of penicillin, ampicillin, amoxicillin, cefditoren, cefuroxime, cefprozil, and cefaclor in 18 clinical isolates of penicillin-susceptible, -intermediate, and -resistant pneumococci. Antimicrob Agents Chemother 46(5):1273–1280
Smith AM, Klugman KP (1998) Alterations in PBP 1A essential-for high-level penicillin resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother 42(6):1329–1333
Asahi Y, Takeuchi Y, Ubukata K (1999) Diversity of substitutions within or adjacent to conserved amino acid motifs of penicillin-binding protein 2X in cephalosporin-resistant Streptococcus pneumoniae isolates. Antimicrob Agents Chemother 43(5):1252–1255
Clinical and Laboratory Standards Institute (CLSI) (2013) Performance standards for antimicrobial susceptibility testing; Twenty-second informational supplement. CLSI document M100-S22. CLSI, Wayne, PA
Sørensen UB (1993) Typing of pneumococci by using 12 pooled antisera. J Clin Microbiol 31(8):2097–2100
Zhanel GG, Wang X, Nichol K, Nikulin A, Wierzbowski AK, Mulvey M, Hoban DJ (2006) Molecular characterisation of Canadian paediatric multidrug-resistant Streptococcus pneumoniae from 1998–2004. Int J Antimicrob Agents 28(5):465–471. doi:10.1016/j.ijantimicag.2006.08.005
Baek JY, Ko KS, Oh WS, Jung SI, Kim YS, Chang HH, Lee H, Kim SW, Peck KR, Lee NY, Song JH (2004) Unique variations of pbp2b sequences in penicillin-nonsusceptible Streptococcus pneumoniae isolates from Korea. J Clin Microbiol 42(4):1746–1750
Kosowska K, Jacobs MR, Bajaksouzian S, Koeth L, Appelbaum PC (2004) Alterations of penicillin-binding proteins 1A, 2X, and 2B in Streptococcus pneumoniae isolates for which amoxicillin MICs are higher than penicillin MICs. Antimicrob Agents Chemother 48(10):4020–4022. doi:10.1128/AAC.48.10.4020-4022.2004
Granger D, Boily-Larouche G, Turgeon P, Weiss K, Roger M (2006) Molecular characteristics of pbp1a and pbp2b in clinical Streptococcus pneumoniae isolates in Quebec, Canada. J Antimicrob Chemother 57(1):61–70. doi:10.1093/jac/dki401
Pineda V, Fontanals D, Larramona H, Domingo M, Anton J, Segura F (2002) Epidemiology of invasive Streptococcus pneumoniae infections in children in an area of Barcelona, Spain. Acta Paediatr 91(11):1251–1256
Zhao C, Zhang F, Chu Y, Liu Y, Cao B, Chen M, Yu Y, Liao K, Zhang L, Sun Z, Hu B, Lei J, Hu Z, Zhang X, Wang H (2013) Phenotypic and genotypic characteristic of invasive pneumococcal isolates from both children and adult patients from a multicenter surveillance in China 2005–2011. PLoS One 8(12), e82361. doi:10.1371/journal.pone.0082361
Metcalf BJ, Gertz RE Jr, Gladstone RA, Walker H, Sherwood LK, Jackson D, Li Z, Law C, Hawkins PA, Chochua S, Sheth M, Rayamajhi N, Bentley SD, Kim L, Whitney CG, McGee L, Beall B; Active Bacterial Core surveillance team (2015) Strain features and distributions in pneumococci from children with invasive disease before and after 13-valent conjugate vaccine implementation in the USA. Clin Microbiol Infect. doi:10.1016/j.cmi.2015.08.027
Beall B (2007) Vaccination with the pneumococcal 7-valent conjugate: a successful experiment but the species is adapting. Expert Rev Vaccines 6(3):297–300. doi:10.1586/14760584.6.3.297
Reinert R, Jacobs MR, Kaplan SL (2010) Pneumococcal disease caused by serotype 19A: review of the literature and implications for future vaccine development. Vaccine 28(26):4249–4259. doi:10.1016/j.vaccine.2010.04.020
Choi EH, Kim SH, Eun BW, Kim SJ, Kim NH, Lee J, Lee HJ (2008) Streptococcus pneumoniae serotype 19A in children, South Korea. Emerg Infect Dis 14(2):275–281. doi:10.3201/eid1402.070807
Xue L, Yao K, Xie G, Zheng Y, Wang C, Shang Y, Wang H, Wan L, Liu L, Li C, Ji W, Xu X, Wang Y, Xu P, Liu Z, Yu S, Yang Y (2010) Serotype distribution and antimicrobial resistance of Streptococcus pneumoniae isolates that cause invasive disease among Chinese children. Clin Infect Dis 50(5):741–744. doi:10.1086/650534
Wagner AL, Sun X, Montgomery JP, Huang Z, Boulton ML (2014) The impact of residency and urbanicity on Haemophilus influenzae type b and pneumococcal immunization in Shanghai children: a retrospective cohort study. PLoS One 9(5), e97800. doi:10.1371/journal.pone.0097800
Imöhl M, Möller J, Reinert RR, Perniciaro S, van der Linden M, Aktas O (2015) Pneumococcal meningitis and vaccine effects in the era of conjugate vaccination: results of 20 years of nationwide surveillance in Germany. BMC Infect Dis 15:61. doi:10.1186/s12879-015-0787-1
Greenberg D, Givon-Lavi N, Ben-Shimol S, Ziv JB, Dagan R (2015) Impact of PCV7/PCV13 introduction on community-acquired alveolar pneumonia in children <5 years. Vaccine 33(36):4623–4629. doi:10.1016/j.vaccine.2015.06.062
Ceyhan M, Ozsurekci Y, Gürler N, Öksüz L, Aydemir S, Ozkan S, Yuksekkaya S, Emiroglu MK, Gültekin M, Yaman A, Kiremitci A, Yanık K, Karli A, Ozcinar H, Aydin F, Bayramoglu G, Zer Y, Gulay Z, Gayyurhan ED, Gül M, Özakın C, Güdücüoğlu H, Perçin D, Akpolat N, Ozturk C, Camcıoğlu Y, Öncel EK, Çelik M, Şanal L, Uslu H (2015) Serotype distribution of Streptococcus pneumoniae in children with invasive diseases in Turkey: 2008–2014. Hum Vaccin Immunother. doi:10.1080/21645515.2015.1078952
Pagliero E, Chesnel L, Hopkins J, Croizé J, Dideberg O, Vernet T, Di Guilmi AM (2004) Biochemical characterization of Streptococcus pneumoniae penicillin-binding protein 2b and its implication in beta-lactam resistance. Antimicrob Agents Chemother 48(5):1848–1855
Granger D, Boily-Larouche G, Turgeon P, Weiss K, Roger M (2005) Genetic analysis of pbp2x in clinical Streptococcus pneumoniae isolates in Quebec, Canada. J Antimicrob Chemother 55(6):832–839. doi:10.1093/jac/dki118
Izdebski R, Rutschmann J, Fiett J, Sadowy E, Gniadkowski M, Hryniewicz W, Hakenbeck R (2008) Highly variable penicillin resistance determinants PBP 2x, PBP 2b, and PBP 1a in isolates of two Streptococcus pneumoniae clonal groups, Poland 23F-16 and Poland 6B-20. Antimicrob Agents Chemother 52(3):1021–1027. doi:10.1128/AAC.01082-07
Tian SF, Chu YZ, Chen BY (2008) Molecular characteristics of penicillin-binding protein 2b, 2x, and 1a sequences in penicillin-nonsusceptible Streptococcus pneumoniae isolates in Shenyang, China. Can J Microbiol 54(6):489–494. doi:10.1139/w08-030
Mouz N, Gordon E, Di Guilmi AM, Petit I, Pétillot Y, Dupont Y, Hakenbeck R, Vernet T, Dideberg O (1998) Identification of a structural determinant for resistance to beta-lactam antibiotics in Gram-positive bacteria. Proc Natl Acad Sci U S A 95(23):13403–13406
Chesnel L, Pernot L, Lemaire D, Champelovier D, Croizé J, Dideberg O, Vernet T, Zapun A (2003) The structural modifications induced by the M339F substitution in PBP2x from Streptococcus pneumoniae further decreases the susceptibility to beta-lactams of resistant strains. J Biol Chem 278(45):44448–44456. doi:10.1074/jbc.M305948200
Lu WP, Kincaid E, Sun Y, Bauer MD (2001) Kinetics of beta-lactam interactions with penicillin-susceptible and -resistant penicillin-binding protein 2x proteins from Streptococcus pneumoniae. Involvement of acylation and deacylation in beta-lactam resistance. J Biol Chem 276(34):31494–31501. doi:10.1074/jbc.M102499200
Sanbongi Y, Ida T, Ishikawa M, Osaki Y, Kataoka H, Suzuki T, Kondo K, Ohsawa F, Yonezawa M (2004) Complete sequences of six penicillin-binding protein genes from 40 Streptococcus pneumoniae clinical isolates collected in Japan. Antimicrob Agents Chemother 48(6):2244–2250. doi:10.1128/AAC.48.6.2244-2250.2004
du Plessis M, Bingen E, Klugman KP (2002) Analysis of penicillin-binding protein genes of clinical isolates of Streptococcus pneumoniae with reduced susceptibility to amoxicillin. Antimicrob Agents Chemother 46(8):2349–2357
Davies TA, Flamm RK, Lynch AS (2012) Activity of ceftobiprole against Streptococcus pneumoniae isolates exhibiting high-level resistance to ceftriaxone. Int J Antimicrob Agents 39(6):534–538. doi:10.1016/j.ijantimicag.2012.02.016
Nichol KA, Zhanel GG, Hoban DJ (2002) Penicillin-binding protein 1A, 2B, and 2X alterations in Canadian isolates of penicillin-resistant Streptococcus pneumoniae. Antimicrob Agents Chemother 46(10):3261–3264
Reichmann P, König A, Marton A, Hakenbeck R (1996) Penicillin-binding proteins as resistance determinants in clinical isolates of Streptococcus pneumoniae. Microb Drug Resist 2(2):177–181
Job V, Carapito R, Vernet T, Dessen A, Zapun A (2008) Common alterations in PBP1a from resistant Streptococcus pneumoniae decrease its reactivity toward beta-lactams: structural insights. J Biol Chem 283(8):4886–4894. doi:10.1074/jbc.M706181200
Clinical and Laboratory Standards Institute (CLSI) (2007) Performance standards for antimicrobial susceptibility testing; Seventeenth informational supplement. CLSI document M100-S17. CLSI, Wayne, PA
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No funds were received for the realisation of this work.
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
X. Zhou and J. Liu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhou, X., Liu, J., Zhang, Z. et al. Molecular characteristics of penicillin-binding protein 2b, 2x and 1a sequences in Streptococcus pneumoniae isolates causing invasive diseases among children in Northeast China. Eur J Clin Microbiol Infect Dis 35, 633–645 (2016). https://doi.org/10.1007/s10096-016-2582-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2582-3